Abstract
Tumour-derived mutant p53 proteins promote invasion, in part, by enhancing Rab coupling protein (RCP)-dependent receptor recycling. Here we identified MET as an RCP-binding protein and showed that mutant p53 promoted MET recycling. Mutant p53-expressing cells were more sensitive to hepatocyte growth factor, the ligand for MET, leading to enhanced MET signalling, invasion and cell scattering that was dependent on both MET and RCP. In cells expressing the p53 family member TAp63, inhibition of TAp63 also lead to cell scattering and MET-dependent invasion. However, in cells that express very low levels of TAp63, the ability of mutant p53 to promote MET-dependent cell scattering was independent of TAp63. Taken together, our data show that mutant p53 can enhance MET signalling to promote cell scattering and invasion through both TAp63-dependent and -independent mechanisms. MET has a predominant role in metastatic progression and the identification of mechanisms through which mutations in p53 can drive MET signalling may help to identify and direct therapy.
Keywords: mutant p53, MET, recycling
Introduction
The p53 tumour suppressor protein has a critical role in preventing malignant progression and is mutated in more than half of all human cancers, frequently resulting in the expression of p53 proteins that carry a single amino acid substitution. The majority of these mutations affect the central DNA-binding domain of p53 and lead to the loss of wild-type p53 functions, such as the ability to promote cell cycle arrest and apoptosis in response to oncogenic stress. However, several of these p53 mutants, including the hotspot mutants R175H and R273H, show a gain of oncogenic function in driving invasion and metastasis.1, 2, 3 This novel gain of function is related, at least in part, to the ability of mutant p53 to cause changes in gene expression.4, 5 Mutant p53 controls the expression of various genes both directly6, 7, 8 and indirectly by interfering with the function of other transcription factors, including NF-Y, VDR and the p53 family members p63 and p73.6, 9, 10, 11 Various molecules can enhance or antagonize the function of mutant p53 in regulating these transcription factors, including TopBP1, Pin1 and ANKRD11.12, 13, 14 The ability of mutant p53 to regulate the activity of TAp63 is particularly interesting, as TAp63 can function as a suppressor of tumour development and metastasis15, 16 and can regulate the expression of anti-metastatic genes, such as Dicer,16 Cyclin G2 and Sharp-1.17 However, it is clear that mutant p53 can also function through TAp63-independent mechanisms12, 18 and mutant p53 drives invasive behaviour in cells that do not express detectable TAp63.19, 20
We found previously that one consequence of mutant p53 expression is the increased recycling and signalling of integrins and EGFR (epidermal growth factor receptor)20 mediated by Rab coupling protein (RCP), which itself can act as an oncogene in breast cancers.21 Similar effects were induced by depletion of TAp63, suggesting that one mechanism by which mutant p53 functions is by preventing this function of TAp63.
In this study, we show that RCP-dependent MET (the receptor for hepatocyte growth factor, HGF) signalling is enhanced by mutant p53 expression, resulting not only in increased invasion but also in cell scattering. In cells expressing TAp63, this activity can be mediated through inhibition of TAp63, although mutant p53 also shows p63-independent activities. The importance of MET recycling in tumorigenesis has recently been demonstrated for tumour-derived MET mutants.22 However, our work is the first to demonstrate the importance of RCP in MET signalling and the modulation of this mechanism by mutant p53.
Results
Mutant p53 expression promotes cell scattering
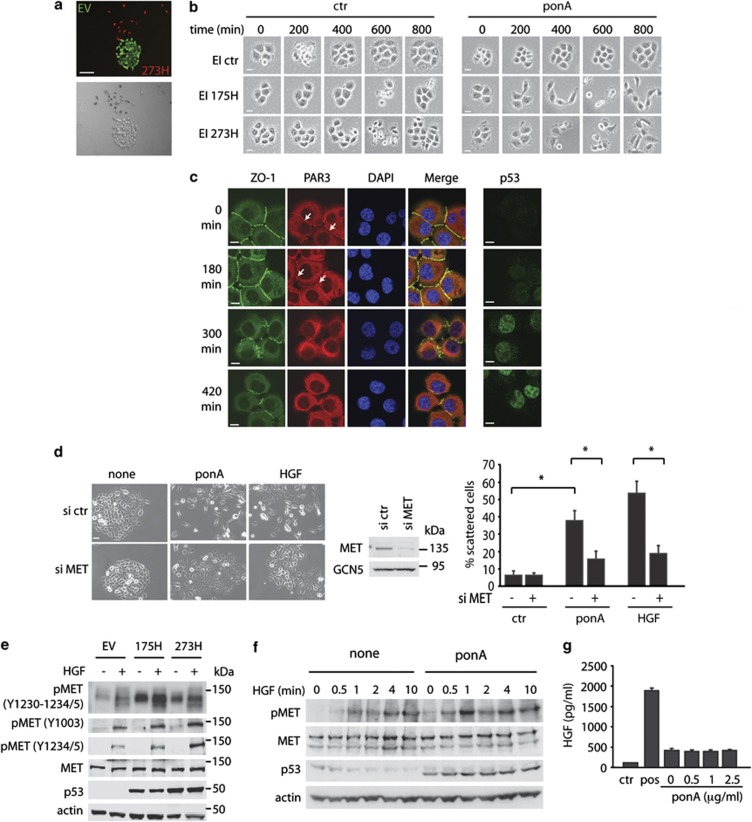
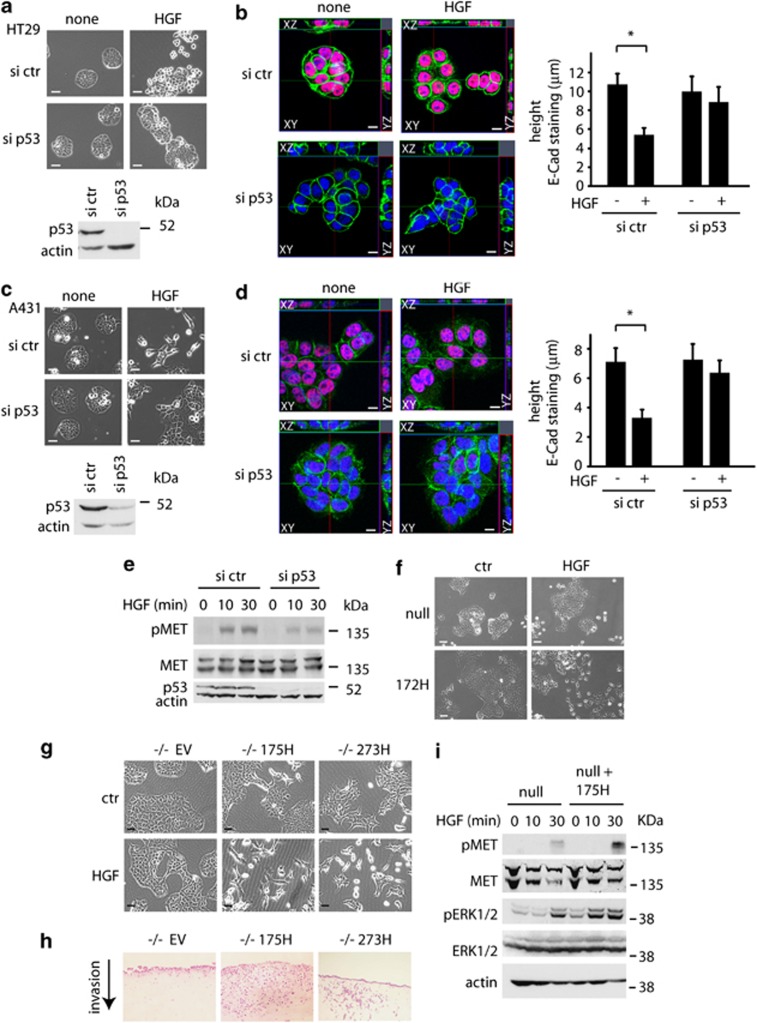
Several studies have shown that the ability of mutant p53 to promote enhanced metastasis in mouse models is paralleled by enhanced invasion of mutant p53-expressing cells in various tissue culture systems. To examine the effects of mutant p53 expression on cell growth characteristics, we generated a p53 null small cell lung cancer cell line (H1299) tagged with green fluorescent protein (GFP) and a mutant p53 (273H)-expressing H1299 cell line tagged with Cherry. Co-culture of p53 273H and control cells showed that the mutant p53 cells did not grow in colonies, but adopted a more scattered appearance compared with empty vector cells (red compared with green cells in Figure 1a). Using Ecdysone inducible (EI) H1299 cells, in which ponasterone A (ponA) can induce expression of either the 175H or 273H mutant p53 to levels observed for MDA MB231 cells that express endogenous mutant p53 (Supplementary Figure S1a), we found that induction of either p53 mutant resulted in dissociation of established cell colonies (Figure 1b), an effect that was not seen in the EI control cells lacking mutant p53. Although we were unable to detect either of the most commonly expressed adherens junction proteins, E-Cadherin or P-Cadherin, in H1299 cells, tight junctions proteins, such as ZO-123 and PAR3,24 were detected at points of cell–cell contacts. PonA induction in EI 175H cells (Figure 1c) and EI 273H cells, but not EI ctr cells (Supplementary Figure S1b), led to a time-dependent loss of both ZO-1 and PAR3 from the cellular junctions that coincided with increased nuclear mutant p53 expression.
Figure 1.
Mutant p53 enhances MET signalling to drive scattering. (a) Merged red and green channel microscope images and bright field images of control (EV) and mutant p53 (273H) H1299 cells tagged with, respectively, GFP or Cherry. Scale bars indicate 50 μm. (b) EI H1299 cells expressing a vector (ctr) or mutant p53 (175H or 273H) were allowed to form discrete colonies. A total of 2.5 μg/ml ponA was then added to the cells and scattering was monitored using time lapse microscopy. Scale bars indicate 20 μm. (c) Immunostaining for ZO-1, PAR3, DAPI and a merge (left panels) or p53 (right panels) in EI 175H cells after ponA induction. Scale bars indicate 10 μm. Arrows indicate PAR3 staining in the cell–cell junctions. (d) EI 175H cells were transfected with MET siRNA and after 16 h induced with ponA or HGF for 48 h and analysed for scattering (left panels, quantification right panels). Knockdown of MET was verified using immunoblot analysis for MET expression (middle panel) with GCN5 as loading control. Scale bars indicate 50 μm. * indicates statistical significant changes (P<0.05) as determined by a T-test. (e) Mutant p53 (175H or 273H) or control (EV) H1299 cells were analysed for MET expression and MET phosphorylation as assessed by western blot using different pMET antibodies with or without 10 min HGF stimulation. Total MET and actin were used as loading control. (f) Immunoblot analysis showing MET phosphorylation in EI 175H H1299 cells uninduced (none) or induced to express mutant p53 (ponA), incubated with HGF for 0.5, 1, 2, 4 or 10 min. Actin and total MET expression were used as loading controls. (g) ELISA measurement of HGF excretion in the medium of ponA-treated EI 175H cells. Ctr indicates medium that has not been exposed to cells. Pos indicates a positive HGF lysate as included in the ELISA kit.
The ability of mutant p53 to promote dissociation or ‘scattering'25, 26of epithelial cell colonies is reminiscent of the effect of activation of the MET receptor by its ligand HGF. The scattering of H1299 cells in response to mutant p53 was similar to that seen following treatment of cells with HGF (Figure 1d), although mutant p53 was somewhat less potent than HGF in inducing this response. Furthermore, HGF and ponA treatment decreased levels of ZO-1 at cellular junctions (Supplementary Figure S1b). Both mutant p53- and HGF-induced scattering (Figure 1d) and loss of ZO-1 from cell junctions (Supplementary Figure S1c) was dependent on MET expression, suggesting that signalling through MET is important for the scattering response to mutant p53. Importantly, the reduction in mutant p53-induced scattering resulting from small interfering RNA (siRNA)-mediated depletion of MET was rescued by MET overexpression, confirming the specificity of MET knockdown (Supplementary Figure S1d). By contrast, EGF treatment of control cells did not induce either scattering (Supplementary Figure S1e) or loss of ZO-1 from cellular junctions (Supplementary Figure S1f). Furthermore, inhibition of the EGFR with PD153035 did not prevent HGF- or mutant p53-induced driven scattering (data not shown), indicating that EGFR activity is not required for scattering.
Mutant p53 enhances MET activation and promotes MET recycling
The scattering behaviour of cells in response to mutant p53 expression prompted us to test whether mutant p53-expressing cells showed any increase in MET activation. Using H1299 cells stably expressing mutant p53, we found that the activating phosphorylations on tyrosine residues within the catalytic domain of MET (1230–1235) were enhanced in mutant p53 (175H and 273H) cells (Figure 1e). Mutant p53 expression also increased the sensitivity of cells to phosphorylation on these sites following treatment with exogenous HGF (Figure 1e). This enhanced phosphorylation was much less apparent when using an antibody that recognizes phosphorylation of MET at residue Y1003, which is a negative regulatory site (Figure 1e). Similarly, induction of mutant p53 by ponA treatment in the inducible H1299 cells resulted in enhanced phosphorylation of MET and increased sensitivity to HGF stimulation (Figure 1f). Notably, although both wild-type p53 and mutant p53 have been shown to regulate MET transcription and expression,27, 28 we did not observe a mutant p53-induced change in total MET protein levels under these conditions. Furthermore, mutant p53 expression did not increase HGF secretion by H1299 cells, although we were able to detect a basal level of HGF production in these cells (Figure 1g). Taken together, the data indicate that expression of mutant p53 enhances the sensitivity of MET to HGF, either produced by the cell or added to the medium.
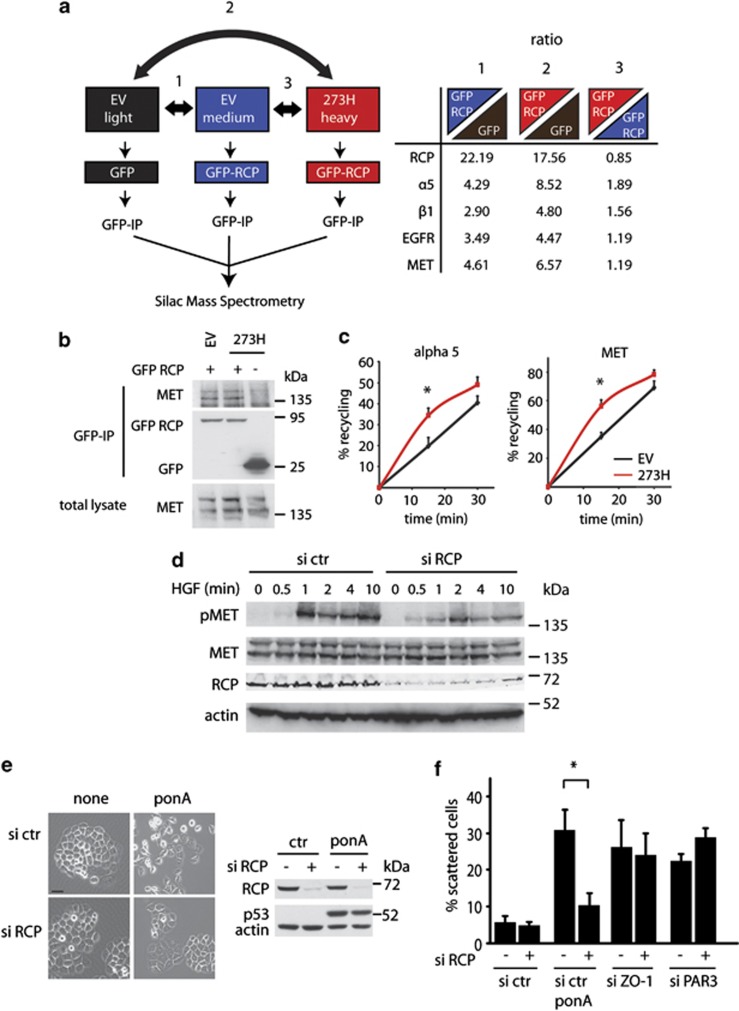
Our previous studies demonstrated that mutant p53 can promote EGFR signalling by enhancing receptor recycling through an RCP-dependent mechanism. Using a SILAC-based approach to identify proteins co-immunoprecipitating with RCP in control and mutant p53-expressing H1299 cells, we confirmed the interaction of RCP with α5β1 integrin and the enhancement of this by mutant p53 (Figure 2a). As seen previously, RCP also binds to EGFR, although this interaction was not enhanced in mutant p53-expressing cells.20 Interestingly, among the proteins that were detected, we identified MET as a novel RCP-binding protein (Figure 2a, right panel). Like EGFR, the interaction of MET with RCP was not enhanced by mutant p53, and we were able to confirm the interaction in independent co-immunoprecipitation experiments (Figure 2b). The interaction of MET with RCP suggested that, like EGFR, MET may be recycled to the cell surface in mutant p53-expressing cells. Indeed, internalized MET and α5 integrin were recycled to the plasma membrane more rapidly in mutant p53 cells than in control cells (Figure 2c). By analogy with previous observations with the EGFR, mutant p53-dependent enhanced recycling of MET would be predicted to result in enhanced MET activation and scattering. Indeed, we were able to show that both the sensitivity of mutant p53-expressing cells to HGF-induced MET phosphorylation (Figure 2d) and the scattering of mutant p53-expressing cells (Figure 2e) were dependent on RCP. Importantly, induction of cell scattering through direct depletion of cell junction proteins, such as ZO-1 and PAR3, was not opposed by RCP depletion (Figure 2f), demonstrating a specificity of the requirement of RCP in mutant p53-induced scattering.
Figure 2.
RCP-dependent recycling of MET contributes to MET signalling in mutant p53 cells. (a) Mutant p53 (273H) or control (EV) H1299 cells were SILAC-labelled with ‘light', ‘medium' or ‘heavy' amino acids and transfected with GFP or GFP-RCP as indicated in the left diagram. Cells were lysed and a co-immunoprecipitation with GFP was used to precipitate RCP-binding proteins. Quantitative mass spectrometry data analysis revealed a ratio of RCP-binding proteins in EV cells and 273H cells compared with GFP-transfected EV cells (comparison 1 and 2), or differential binding to RCP in 273H cells compared with EV cells (comparison 3, right table). (b) MET co-immunoprecipitation with RCP in EV control or 273H H1299 cells that were transfected with GFP or GFP-RCP. Cell lysates were immunoprecipitatedfor GFP and analysed for MET binding as assessed by immunoblot analyses for MET. (c) Recycling assays for alpha 5 integrin and MET in EV and 273H H1299 cells. * indicates statistical significant changes (P<0.05) as determined by a T-test. (d) Immunoblot analysis showing MET phosphorylation in mutant p53 (273H) H1299 that were transfected with siRNA targeting RCP and incubated with HGF for indicated times. Actin and total MET expression were used as loading controls. (e) Scattering of EI 175H cells that were transfected with RCP siRNA and after 16 h induced with ponA (2.5 μg/ml) for 48 h (left panel). Scale bars indicate 50 μm. Knockdown of RCP expression and expression of p53 after ponA (2.5 μg/ml) induction was verified by western blot analysis using actin as a loading control (right panel). (f) Quantification of scattering of EI 175H cells that were transfected with RCP siRNA in combination with or without ZO-1 or PAR3 siRNA and after 16 h induced with ponA for 48 h.
Mutant-p53 expressing cells show enhanced MET-dependent signalling
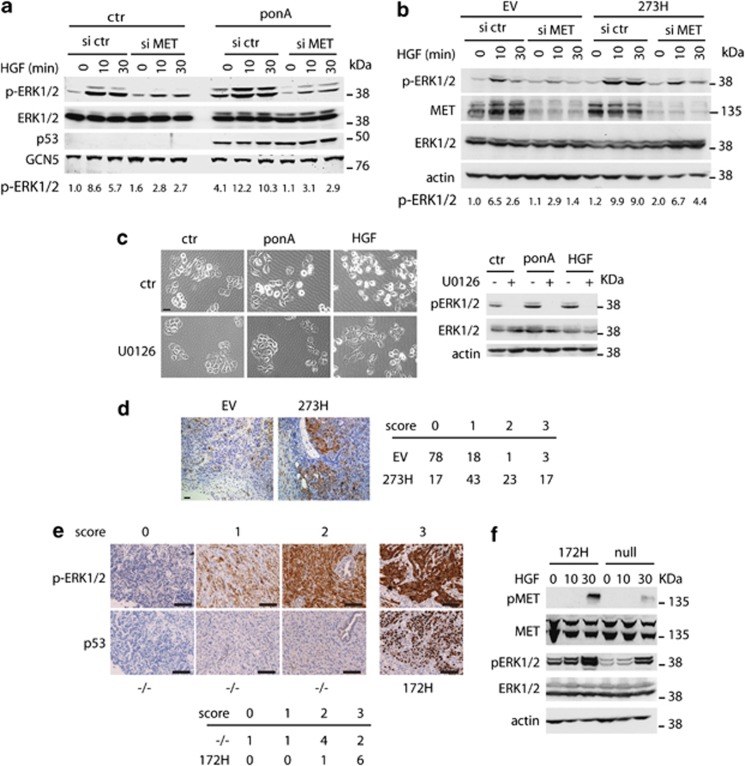
To determine whether signalling downstream of MET was enhanced in mutant p53-expressing cells, we examined MAPK activation.29, 30 Induction of mutant p53 (in EI 175H cells) resulted in an increase in ERK1/2 phosphorylation compared with control cells, and enhanced ERK1/2 phosphorylation in response to HGF treatment (Figure 3a). A titration of ponA revealed a dose-dependent increase in ERK1/2 phosphorylation in response to p53, although again no increase in overall MET or ERK1/2 proteins were detected (Supplementary Figure S2a). Similarly, transient expression of either 175H or 273H mutant p53 constructs enhanced ERK1/2 phosphorylation (Supplementary Figure S2b). Mutant p53 also enhances EGFR signalling, but importantly we were able to show that the induction of ERK1/2 phosphorylation was largely dependent on MET in these cells, as MET depletion substantially reduced phospho-ERK1/2 accumulation (Figure 3a). In H1299 cells stably expressing mutant p53, the basal levels of phospho-ERK1/2 were the same as those seen in p53 null control cells (Figure 3b), suggesting that prolonged growth in culture stimulates the known negative feedback mechanisms function to limit ERK1/2 activation.31 However, mutant p53-expressing cells retained enhanced sensitivity to exogenous HGF, showing enhanced phosphorylation of ERK1/2 that was blunted by MET depletion (Figure 3b) or treatment with a MET inhibitor (Supplementary Figure S2c). Treatment of cells with a MEK inhibitor to inhibit ERK1/2 phosphorylation blocked both HGF and mutant p53-driven cell scattering (Figure 3c), demonstrating the importance of ERK1/2 signalling to this response. To determine whether mutant p53 signals leads to activation of ERK1/2 phosphorylation in vivo, we stained sections of xenografts that had been generated by injecting H1299-EV and H1299 p53 273H cells in the mammary fat pad of mice20 for pERK1/2 expression (Figure 3d). H1299 p53 273H xenografts showed more staining that was localized primarily at the invasive front of the tumour. In a pancreatic ductal carcinoma (PDAC) mouse model, gain-of-function expression of mutant p53 with K-Ras mutation has previously been shown to result in liver metastases,32 which are not seen from tumours arising in the absence of p53. Sections of pancreatic tumours originating from p53172H/172H and p53loxp/loxp mice showed the most intense pERK1/2 expression in almost all tumours derived from mutant p53 mice (Figure 3e). More importantly, cell lines derived from the mutant p53 PDAC had increased basal pERK1/2 levels and increased phosphorylation of both MET and ERK1/2 after HGF exposure (Figure 3f). Together these data demonstrate that mutant p53 can enhance MET-dependent ERK1/2 phosphorylation, resulting in enhanced scattering and invasion.
Figure 3.
ERK1/2 phosphorylation is required for mutant p53-dependent scattering and invasion. (a) Activation of ERK1/2 in EI 175H cells using phospho-specific ERK1/2 antibodies was determined after induction of mutant p53 with ponA for 24 h. Total ERK1/2 levels are shown as control for equal ERK1/2 expression in all samples and GCN5 was used as loading control. The numbers indicate quantification of pERK1/2 levels as compared with ctr cells transfected with ctr siRNA and not treated with HGF and normalized for total ERK1/2 levels. (b) Activation of ERK1/2 after HGF stimulation in p53 273H and control EV H1299 cells, assessed by western blot using a pERK1/2 antibody. The numbers indicate quantification of pERK1/2 levels as compared with EV cells transfected with ctr siRNA and not treated with HGF and normalized for total ERK1/2 levels. (c) Scattering of EI 175H cells 48 h after ponA, HGF and/or U0126 treatment. Scale bars indicate 50 μm. pERK1/2 expression is shown (right panel) with ERK1/2 and actin as loading controls. (d) Staining of pERK1/2 in xenografts of H1299 EV (8 mice) and 273H cells (7 mice) (left panels). pERK1/2 staining was divided into four intensity categories ranging from 0 for no staining and 3 for the highest intensity. The percentage of staining per category at the invasive edge of the tumour in each section was analysed and the overall percentage of staining for each score is depicted in the table. (e) Immunohistochemical images for pERK1/2 and p53 levels in p53 null and mutant p53 172H (mouse equivalent of human 175H) pancreatic tumours. The scoring indicates the strength of the immunohistochemical signal in which 0 is no staining and 3 is the highest signal. Scale bars indicate 100 μm. The table indicates the numbers of tumours of independent mice in which each score was found. (f) MET and ERK1/2 phosphorylation in cell lines established from the p53 null or mutant p53 (172H) pancreatic tumours after HGF treatment. Total MET, total ERK1/2 and actin were used as loading controls.
Mutant p53 drives MET-dependent invasion
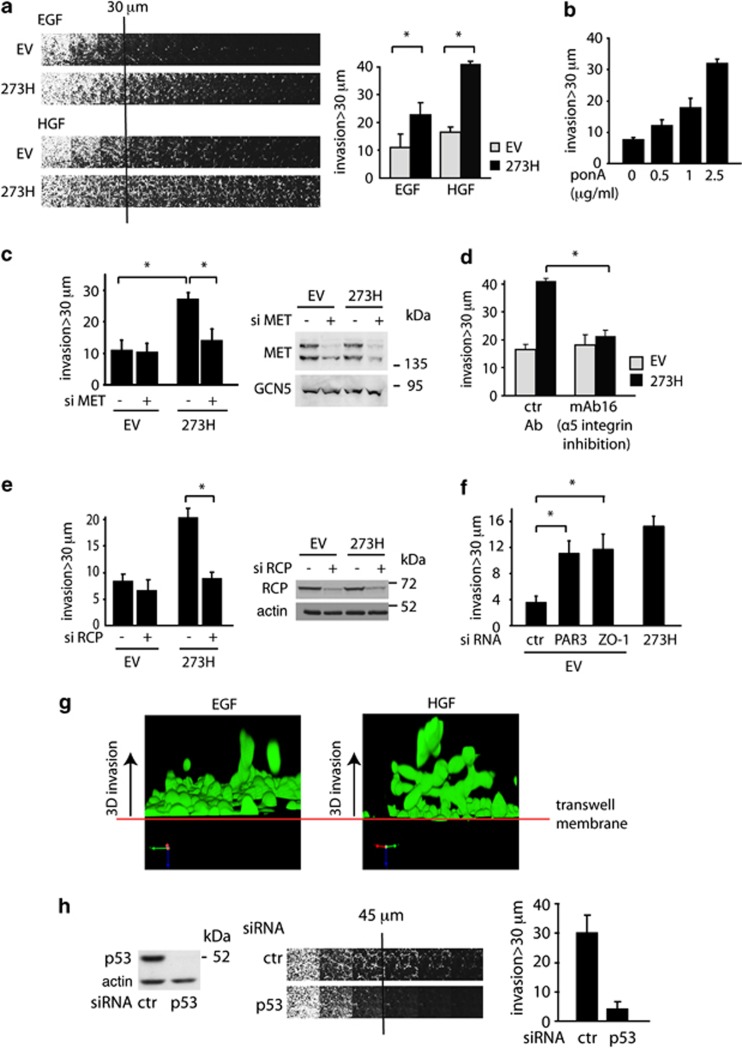
Previously we showed that mutant p53 promotes the ability of cells to invade through Matrigel towards EGF by promoting RCP-dependent increased recycling of α5β1 integrin and EGFR.20 As both EGFR and MET signalling have been shown to contribute to invasion,33 we examined the ability of mutant p53 expression to influence the invasive behaviour of cells in Matrigel using HGF as an attractant. These assays showed increased invasion of mutant p53 cells towards either EGF or HGF (Figure 4a) and induction of mutant p53 in the ponA-inducible cell line also led to increased invasion towards HGF (Figure 4b and Supplementary Figure S3a). IGF-1 (insulin-like growth factor) or PDGF-β (platelet-derived growth factor-β) were unable to enhance the invasive behaviour of mutant p53-expressing cells (Supplementary Figure S3b), although the cells were responsive to both growth factors as demonstrated by AKT phosphorylation (Supplementary Figure S3c). Interestingly, the IGF-1-R and PDGF-β-R were not detected as RCP-binding partners in our SILAC analysis, suggesting some specificity in the receptor tyrosine kinases that are influenced by RCP and therefore targets for mutant p53. The increased invasion of mutant p53 cells towards HGF was dependent on MET (Figure 4c) and α5β1 integrin (Figure 4d) function. Importantly, the increased invasion (although not basal invasion) seen in mutant p53-expressing cells was also lost following RCP depletion (Figure 4e). These data indicate that RCP-mediated recycling of integrin and MET is required for the enhanced invasiveness of mutant p53 cells. Notably, loss of ZO-1 or PAR3 (which led to cell scattering, Figure 2f) also resulted in enhanced invasion of empty vector (EV) cells, suggesting that loss of cell–cell junctions can at least partially contribute to the invasion of mutant p53 cells (Figure 4f).
Figure 4.
Mutant p53 promotes MET-dependent invasion towards HGF. (a) Mutant p53 (273H) or control (EV) H1299 cells were analysed for invasion capacity in fibronectin-supplemented Matrigel using EGF or HGF as a chemo-attractant (left panel). Invasion was quantified as described in the Materials and methods section and values are means±s.e.m. of six replicates from each of three independent experiments (right panel). *indicates statistical significant changes (P<0.05) as determined by a T-test. (b) EI 175H H1299 cells were analysed for invasion capacity in fibronectin-supplemented Matrigel using HGF as a chemo-attractant after induction with increasing doses of ponA. Invasion was quantified as described in the Materials and methods section and values are means±s.e.m. of six replicates from each of three independent experiments (right panel). * indicates statistical significant changes (P<0.05) as determined by a T-test. (c) Mutant p53 (273H) or control (EV) H1299 cells were analysed for invasion after knockdown of MET using HGF as a chemo-attractant. * indicates statistical significant changes (P<0.05) as determined by a T-test. Knockdown of MET was verified by western blot (right panel), using GCN5 as a loading control. (d) Mutant p53 (273H) or control (EV) H1299 cells were analysed for invasion capacity in fibronectin-supplemented Matrigel with monoclonal antibodies against α5 integrin, using HGF as a chemo-attractant. * indicates statistical significant changes (P<0.05) as determined by a T-test. (e) Mutant p53 (273H) or control (EV) H1299 cells were transfected with RCP siRNA and analysed for invasion capacity in fibronectin-supplemented Matrigel using HGF as a chemo-attractant. Knockdown of RCP was verified by western blot using actin as loading control (right panel). * indicates statistical significant changes (P<0.05) as determined by a T-test. (f) H1299 EV cells were transfected with siRNA against ZO-1 or PAR3 and compared with 273H cells for invasion capacity in fibronectin-supplemented Matrigel using HGF as chemo-attractant. * indicates statistical significant changes (P<0.05) as determined by a T-test. (g) 3D reconstructive images of 273H H1299 cells invading towards EGF or HGF. (h) MDA MB231 cells were transfected with siRNA-targeting (mutant) p53 and analysed for invasion capacity into fibronectin-supplemented Matrigel using HGF as chemo-attractant (middle panel). Knockdown was verified by western blot using actin as loading control (left panel). Quantified invasion is shown in the right panel.
MET and EGFR share several downstream signalling targets and crosstalk between these two receptors has been demonstrated to enhance proliferation, invasion and scattering.34, 35, 36 This prompted us to investigate whether mutant p53 specifically increased signalling via MET to enhance invasion towards HGF, or whether this reflected concurrent activation of the EGFR. Treatment of cells with the EGFR inhibitor PD153035 resulted in an overall decrease in cell invasion towards HGF, but the difference in invasion between mutant p53 and control cells was unaffected (Supplementary Figure S3d). Therefore, although EGFR signalling contributes to the basal invasive activity of these cells, enhanced HGF-driven invasion promoted by mutant p53 requires MET, but not EGFR signalling. Interestingly, a clear difference in the invasion of mutant p53 cells promoted by the two growth factors was evident in 3D images of invading cells, with HGF promoting more extensive invasion and also more branched invasive structures (Figure 4g and Supplementary Movies S1a and S1b). To confirm that endogenous mutant p53 also enhanced invasion towards HGF, we repeated the assay using MDA MB231 cells (expressing R280K p53). These cells invaded efficiently towards HGF, an activity that was strongly inhibited following depletion of the endogenous mutant p53 (Figure 4h).
Endogenous mutant p53 can modulate MET-dependent scattering and invasion
To address the role of endogenous mutant p53 in regulating the scatter process, we used HT29 cells and A431 cells (both of which express endogenous p53 273H). These cells express E-cadherin37, 38 and form tight colonies under normal growth conditions. In response to high levels of HGF, the cells scatter30, 39 (Figures 5a and c) and E-cadherin is lost from the cell junctions (Supplementary Figures S4a and S4b). siRNA-mediated depletion of the endogenous mutant p53 resulted in a decrease in the ability of these cells to scatter in response to HGF (Figures 5a and c) and the retention of E-cadherin and ZO-1 at the cell junctions after prolonged HGF treatment (Supplementary Figures S4a and S4b). To quantify this effect, we made Z-stacks of A431 and HT29 colonies and measured the height of the E-cadherin staining at the junctions in cells that were treated with HGF. It is important to note that for these experiments we looked at a time point prior to cell scattering. At these early times following HGF treatment, control cells showed a clear reduction in the height of E-cadherin staining in response to HGF, which was impaired by depleting mutant p53 (Figures 5b and d). Furthermore, A431 cells depleted of endogenous mutant p53 showed decreased MET phosphorylation in response to HGF (Figure 5e), which is consistent with our results in H1299 cells. Consistent with these cell models, cells derived from mutant p53-expressing PDAC tumours scattered better after HGF exposure compared with cells from p53 null tumours (Figure 5f). Expression of the human mutant p53 constructs 175H and 273H in the cells derived from the p53 −/− PDAC tumours also enhanced scattering (Figure 5g), invasion into organotypic assays (Figure 5h) and phosphorylation of ERK1/2 and MET in response to HGF (Figure 5i), indicating that our observations are not the result of heterogeneity of the pancreatic tumours or clonal selection of PDAC cells.
Figure 5.
Endogenous mutant p53 also promotes scattering. (a and c) Scattering images of HT29 cells (a) or A431 (c) after transfection with control siRNA or p53 siRNA in the presence of 30 ng/ml HGF for 48 h (upper panels). Scale bars indicate 50 μm. Knockdown of p53 was verified by western blot using actin as loading control (bottom panels). (b and d) Ortho-images of Z-stacks of overlay immune fluorescence of HT29 (b) and A431 (d) cells, with views of XY (centre image), XZ (above centre image) or YZ (right of centre image) axes. Cells were transfected with control siRNA or p53 siRNA, then treated or not with 30 ng/ml HGF for 16 h and the height of E-Cadherin staining in cell junctions is shown (right panels). p53 (red), E-cadherin (green), DAPI (blue) and scale bars indicate 10 μm. * indicates statistical significant changes (P<0.05) as determined by a T-test. (e) Immunoblot analysis showing MET phosphorylation in A431 cells transfected with siRNA-targeting (mutant) p53 and incubated with HGF for indicated times. Actin and total MET expression were used as loading controls. (f) Scattering of cell lines established from the p53 null and mutant p53 (172H) pancreatic tumours after HGF treatment for 48 h. Scale bars indicate 50 μm. Scattering (g), invasion in organotypic assays (h) and MET/ ERK1/2 phosphorylation (i) in response to HGF of cell lines established from the p53 null pancreatic tumours that were stably transfected with human mutant p53 175H and 273H. Scale bars indicate 50 μm. Total MET, total ERK1/2 and actin were used as loading controls.
The role of TAp63 in mediating mutant p53 activities
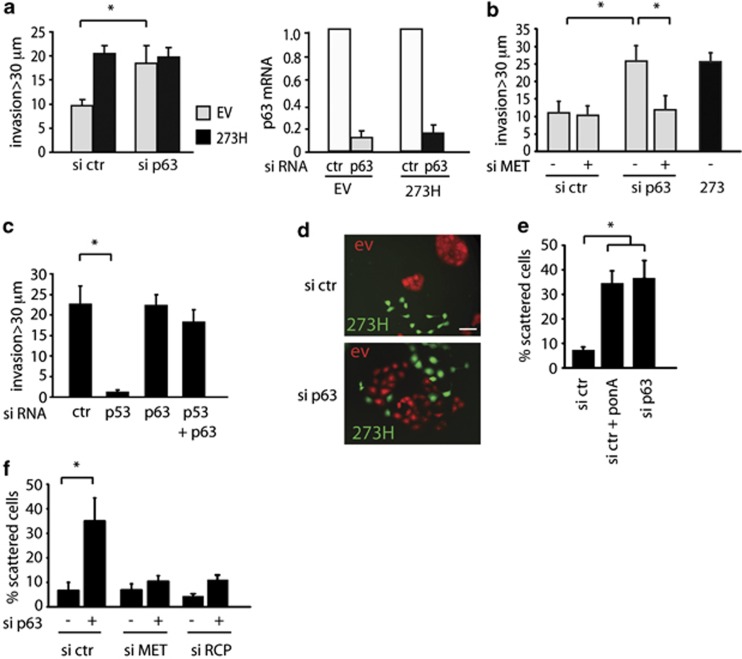
Several functions of mutant p53 have been attributed to the ability to bind and inhibit the activity of p53 family members p63 and p73. Indeed, loss of the TA isoform of p63 has been shown to drive invasion and metastasis in a manner very similar to that seen in response to mutant p53 expression.16, 40 Our previous studies showed that depletion of p63, but not p73, promoted invasion of H1299 cells towards EGF, and we found a similar effect of p63 depletion on the ability of these cells to invade towards HGF (Figure 6a). Neither mutant p53 expression nor loss of p63 promoted invasion in general towards medium without additional growth factors (Supplementary Figure S5a). The invasion promoted by p63 depletion was also dependent on MET expression (Figure 6b). In MDA MB231 cells, where depletion of the endogenous mutant p53 inhibited invasion towards HGF, depletion of p63 had no effect on overall invasion (Figure 6c). However, loss of p63 rescued the invasive behaviour in cells depleted of mutant p53 (Figure 6c). Turning to cell scattering in H1299 cells, we observed scattering in Cherry-tagged control cells after knockdown of p63, similar to that seen in the GFP-tagged mutant p53 cells (Figure 6d). Consistently, reduced expression of p63 in control cells using a pool of siRNA (Figure 6e) or single siRNA (Supplementary Figure S5b) resulted in scattering similar to that observed following mutant p53 induction (Figure 6e). The scattering induced by loss of p63 in these cells was also MET and RCP dependent (Figure 6f). Taken together, the results support a model in which mutant p53 functions to inhibit the activity of p63, and that either expression of mutant p53 or depletion of p63 promotes invasion and cell scattering through an RCP-dependent mechanism.
Figure 6.
Effect of p63 depletion on cell scattering and invasion. (a) Mutant p53 (273H) or control (EV) H1299 cells were transfected with p63 siRNA and analysed for their invasion capacity to HGF as chemo-attractant. To verify knockdown of p63, relative p63 mRNA expression levels are shown in the right panel. * indicates statistical significant changes (P<0.05) as determined by a T-test. (b) Analysis of invasion of EV cells transfected with a combination of p63 and MET siRNA compared with 273H cells using HGF as chemo-attractant. (c) MDA MB231 cells were analysed for invasion capacity in fibronectin-supplemented Matrigel after knockdown of (mutant) p53 and/or p63, using HGF as chemo-attractant. * indicates statistical significant changes (P<0.05) as determined by a T-test. (d) Fluorescence images of co-cultures of H1299 cells expressing mutant p53 273H and GFP with empty vector (EV) cells expressing Cherry, following transfection of p63 siRNA. Scale bars indicate 50 μm. (e) EI 175H cells were transfected with p63 or ctr siRNA and incubated in ponA (2.5 μg/ml) as indicated. After 48 h, scattering was quantified. * indicates statistical significant changes (P<0.05) as determined by a T-test. Scale bars indicate 50 μm. (f) H1299 cells were transfected with MET or RCP siRNA in combination with p63 siRNA and quantified for scattering 48 h later. * indicates statistical significant changes (P<0.05) as determined by a T-test.
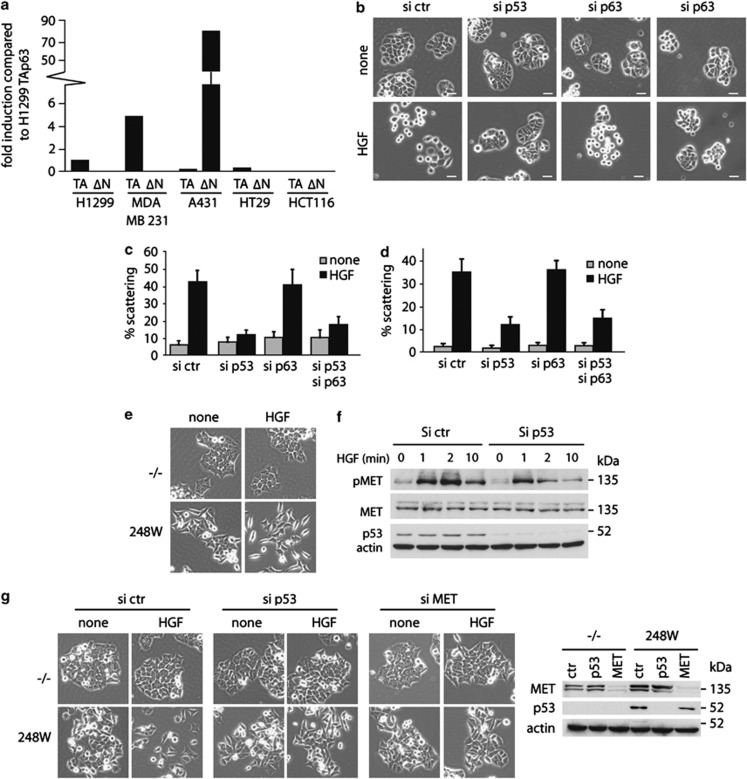
To examine the relationship between mutant p53-induced scattering and the inhibition of p63 more closely, we tested a number of cell lines for patterns of p63 expression. There is some lack of consensus in the literature concerning which isoforms of p63 are expressed in which cell lines, inconsistencies that may reflect drift of different clones of these established cells lines. In our hands (Figure 7a), both H1299 and MDA MB231 cells expressed the TA isoforms of p63, with no detectable expression of ΔNp63 (TAp63 expression in H1299 cells has been confirmed by others).41 By contrast, we found that A431 and HT29 cells expressed very low levels of TAp63, with high levels of ΔNp63 expression in A431 cells (as previously described42 (Figure 7a). However, despite this very low level of TAp63 expression, both A431 and HT29 cells showed mutant p53-dependent scattering (Figure 4). Depletion of p63 did not alter the scattering of these cells in response to HGF (Figure 7b) but, surprisingly, depletion of p63 did not reverse the inhibition of scattering following depletion of mutant p53 in either cell line (Figure 7b), suggesting that mutant p53 can function independently of p63. We therefore turned to the colorectal carcinoma HCT116 cell lines, in which we were unable to detect expression of any p63 isoform (Figure 7a). Lack of p63 expression in these cells has been described by others, although TAp63 expression can be induced by treatment with the HDAC inhibitor TSA.43 Mutant p53-dependent scattering was also observed in these cells, where we compared cells null for p53 with those engineered to express mutant p53 (Figure 7e). Although the null cells formed very tight colonies that were not responsive to HGF, the mutant p53 cells formed looser colonies with many elongated cells, which scattered in response to HGF (Figure 7e). Consistent with our observation in other cells lines, these cells showed a mutant p53-dependent enhanced activation of MET in response to HGF (Figure 7f). Finally, siRNA-mediated depletion of mutant p53 and MET in the mutant p53-expressing HCT116 cells inhibited scattering in response to HGF, confirming the specificity of mutant p53 driving this scattering via MET (Figure 7g).
Figure 7.
Mutant p53 regulates scattering via p63-dependent and -independent mechanisms. (a) p63 isoform expression in H1299, MDA MB231, A431, HT29 and HCT116 cells using isoform-specific oligos and quantitative RT–PCR. Expression is compared with expression of the TAp63 isoform in H1299 cells and corrected for GAPDH expression in each cell line. (b) Images of scattering of HT29 cells that were transfected with siRNA against (mutant) p53 and/or p63 and after 16 h incubated in HGF for 48 h. (c) Quantification of scattering of HT29 cells that were transfected with siRNA against (mutant) p53 and/or p63 and after 16 h incubated in HGF for 48 h. (d) Quantification of scattering of HT29 cells that were transfected with siRNA against (mutant) p53 and/or p63 for 16 h and incubated in HGF for 48 h. (e) HCT 116 −/− or HCT 248W/− cells were analysed for scattering after HGF treatment for 48 h. (f) HCT116 248W/− cells were transfected with p53 siRNA, serum starved for 1 h and incubated with HGF as indicated. pMET and p53 levels were analysed by western blot, total MET and actin were used as loading control. (g) HCT116 −/− or HCT248 W/− cells were transfected with siRNA against p53 or MET for 16 h and analysed for scattering after HGF treatment for 48 h (left panels). Knockdown of MET and p53 was verified by western blot as indicated in the right panel. Actin was used as loading control.
Taken together, these results indicate that whereas mutant p53 can function through the inhibition of p63 in cells expressing TAp63, the effects of mutant p53 on cell invasion and scattering can also be mediated by p63-independent mechanisms.
Discussion
The activation of MET has been particularly associated with metastatic behaviour of tumours44 and silencing of MET in established metastases using lentiviral RNAi approaches has been shown to lead to tumour regression in mice.45 MET is a receptor tyrosine kinase (RTK) that has a role in various cellular processes, including proliferation, invasion and cell scattering. The ability of MET signalling to induce cell scattering has been well established in the MDCK canine cell line46 and we show here that several human cell lines derived from various tumour types scatter in response to HGF in a mutant p53-dependent manner. Furthermore, we show that whereas cell lines derived from invasive pancreatic tumours that harbour a p53 mutation scatter in response to HGF, non-invasive p53 null cancers do not show this phenotype. These results suggest the scattering process induced in response to mutant p53 expression might have a role in cancer metastasis in vivo. The enhanced recycling of the MET receptor has previously been shown to promote ERK activation and cell migration47 and oncogenic mutant MET shows enhanced trafficking and signalling.22 Our data reveal another mechanism for oncogenic MET activation in the absence of mutations in MET, which occurs as a response to mutant p53 expression.
The intersection of p53 and MET signalling has been proposed by several previous studies that demonstrated an ability of wild-type p53 function to control MET expression. Whereas MET has been reported as target for transcriptional activation by p53,28 other studies have shown that wild-type p53 suppresses expression of MET, both through direct repression of the MET promoter and indirectly through enhanced expression of miR-34.48 Although mutant p53 proteins were shown to retain some ability to repress MET expression in ovarian surface epithelial cells,27 we were unable to detect a clear effect of mutant p53 in altering the overall levels of MET in cells lacking wild-type p53. These observations may reveal dissociation between MET mRNA and protein levels. Indeed, it is possible that in the longer term the effects of p53 on MET recycling may lead to its accumulation (and other similarly regulated receptors such as the EGFR) by interfering with the normal internalization and lysosomal degradation of the activated receptors. Taken together, the data suggest that in most cell types loss of wild-type p53 results in increased MET expression, whereas expression of mutant p53 further sensitizes cells to enhance MET signalling by promoting its RCP-dependent recycling. Importantly, our results show the mutant p53-driven recycling of MET results in enhanced sensitivity to HGF either in basal medium (where the only HGF available is that secreted by the cells) or in response to addition of HGF to the medium. Although we see enhanced activation of ERK1/2 in mutant p53-expressing cells, it is not yet clear whether this RCP-dependent signalling is qualitatively the same as the response of cells to high levels of HGF in all respects. We previously described differential phosphorylation of EGFR in mutant p53 cells that distinguished this signal from that seen in response to EGF.20 RTK activation consists of a series of events that include (auto) phosphorylation, dimerization, decreased dephosphorylation and decreased degradation, followed by the concerted action of numerous different signalling molecules that bind RTKs and subsequently determine the cellular response.49 Differential modification and activation of downstream signalling cascades in response to different stimuli has been described for these receptors, and the exact consequences of mutant p53 on further downstream events remain to be determined.
Although we have shown that recycling and signalling from both the EGFR and MET are enhanced by mutant p53, mutant p53-expressing cells did not show enhanced invasion in response to PDGF-β or IGF-1. These results illustrate some degree of selectivity in either mutant p53- or RCP-driven receptor recycling, although it seems possible that other RTKs will be regulated in the same way as both the EGFR and MET, and so contribute to the response to mutant p53. We are presently analysing the contribution to enhanced invasion and tumorigenesis that is made by several additional RTKs that were found to be associated with RCP in our mass spectrometry analysis. As knockdown of RCP can prevent invasion and scattering, future work will focus on understanding this process and the possibilities of inhibiting RCP as a means of therapy.
Mutant p53s function through a number of mechanisms, including control of transcription by direct binding to regulatory regions of target genes, indirect modulation of gene expression through interaction with other transcription factors and transcriptionally independent activities that reflect the interaction of p53 with other proteins.50, 51 Through these mechanisms, mutant p53 controls numerous responses, including cell survival, migration, invasion, autophagy and metabolism. The ability of mutant p53 to inhibit the activity of TAp63 is appealing as a mechanism to promote invasion, as depletion or deletion of TAp63 promotes many of the invasive phenotypes also seen in response to mutant p53.17, 20, 40 We show here that regulation of TAp63 by mutant p53 contributes to the cell's scattering response in those cells that retain TAp63 expression. However, the scattering activity of mutant p53 in cells that have low or no detectable TAp63 is mediated by functions that are distinct from the ability to inhibit p63. This may reflect any of the other reported activities of mutant p53 and highlights the recognition of numerous, potentially independent, activities of mutant p53 that contribute to phenotypes such as enhanced invasion and metastasis. It is clear that the final cell response to mutant p53 expression will reflect the integration of a plethora of signals that are both p63 dependent and independent.
From a clinical perspective, the ability of mutant p53 to promote both EGFR and MET trafficking and signalling is an interesting finding, because both receptors have been reported to be activated in various cancers, and combination strategies in vivo and in vitro to inhibit both receptors have been found to be more effective than inhibition of only one.52, 53, 54, 55 Thus, the identification of RTKs that are activated by mutant p53 may highlight combinations of inhibitors that would be particularly effective in the treatment of mutant p53-expressing cancers.
Materials and methods
Cell culture, generation of PDAC cell lines and constructs
H1299 cells, HT29, MDA-MB-231 and A431 cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Paisley, UK) supplemented with 10% FCS (fetal calf serum) and 1% glutamine at 37 °C and 5% CO2. HCT116 p53 null and HCT116 248W cells were described previously.56 The EI system was previously described57 and cells were established as described in Noll et al.13 Stable cell lines and mutant p53 constructs were generated as described previously.20 GFP and Cherry constructs were purchased from BD biosciences, Clontech (Oxford, UK). Cells were authenticated by morphology and Mycoplasma detection in accordance with the ATCC cell line verification test recommendations.
Primary mouse PDACs were derived from tumours harvested from Pdx1-Cre-GFP, LSL-KRasG12D/+, Trp53LoxP/+ mice.32 p53 null PDAC cells were subsequently transfected with empty vector, mutant p53R175H or mutant p53R273H using Polyfect as described by manufacturer's protocol (Qiagen, Crawley, UK). Cells were selected using 0.6 mg/ml G418 and stable pools generated using standard procedures. Cell lines were cultured in DMEM (Invitrogen) supplemented with 10% FBS (fetal bovine serum) and 2 mℳℒ-glutamine (Invitrogen).
Cell transfections
The following oligos were used for siRNA experiments: control siRNA (1) 5′-GCAACGGCAUUCCACCUUU(TT)-3′, ctr siRNA (2) control pool Dharmacon (D-001810-10-20), RCP (smartpool Dharmacon, L-015968-00-0005), EGFR smartpool Dharmacon (L-003114-00-0010), p53 5′-GACUCCAGUGGUAAUCUAC(TT)-3′, p63 (1) 5′-UGAACAGCAUGAACAAGCU(TT)-3′ and p63 (2) 5′-UGACUUCAACUUUGACAUG(TT)-3′, MET (smartpool Dharmacon, L-003156-00-0005). ZO-1 (1) 5′-GGAAACAGCUAUAUGGGAA(TT)-3′, ZO-1 (2) 5′-GCCUGUGUAUGCCCAAGUU(TT)-3′, PAR3 (1) 5′-CCAGGGAAUUUCUGACAUU(TT), PAR3 (2) 5′-GCGUGACUAUGCUGAAAUU(TT)-3′. Cells were transfected with siRNA using Hiperfect (Qiagen) or nucleofection (Lonza, Slough, UK). For hiperfect, 30 pmol of a pool of two siRNA oligos or smartpool siRNA from Dharmacon was combined with 7.5 μl Hiperfect in 800 μl of serum-free DMEM for six-well plates. This mixture was vortexed and incubated for 10 min before adding to cells grown for 24 h in six-well dishes in normal medium. All siRNAs were tested separately for efficient knockdown in immuno blot analysis or quantitative RT–PCR and potential off-target effects. For Amaxa nucleofection, 120 pmol siRNA was transfected per 1 × 107 cells using solution T and program X-001 for H1299 and A431 cells and X-023 for HT-29 cells (Lonza).
For overexpression, lipofectamine (Invitrogen), effectene (Qiagen) or Genejuice (Merck Biosciences, Nottingham, UK) were used according to the manufacturer's protocols.
SILAC-based mass spectrometry
H1299 EV and H1299 273H cells were grown in SILAC DMEM (PAA) 10% FBS (10 KDa dialysed, PAA), supplemented with light — ℒ-arginine (Arg0) and ℒ-lysine (Lys0) — medium — ℒ-arginine-U-13C6 (Arg6) and ℒ-lysine 2H4 (Lys4) — or heavy — ℒ-arginine-U-13C615N4 (Arg10) and ℒ-lysine-U-13C6-15N2 (Lys8) — amino acids (Cambridge Isotope Laboratories, Cambridge, UK). EV cells were labelled in light or medium DMEM. Mutant p53 273H cells were labelled in heavy. Cells were cultured for >8 passages until an incorporation of >97% of medium and heavy amino acids was measured. Cells were subsequently transfected with GFP or GFP-RCP and lysed.
Immunoprecipitations were performed as described previously,58 with the addition that antibodies were cross-linked to the beads. Briefly, magnetic beads conjugated to sheep anti-mouse IgG (Invitrogen) were bound to mouse-anti-GFP (Abcam, Cambridge, UK). Antibody-coated beads were washed twice (0.2 ℳ Sodium Borate, 0.1% NP-40, pH 9.0) prior to cross-linking with 25 mℳ DMP (dimethyl pimelimidate dihydrochloride) (in 0.2 ℳ Sodium Borate, 0.1% NP-40, pH 9.0) for 45 min at room temperature. Beads were washed once (0.2 ℳ Sodium Borate, 0.1% NP-40, pH 9.0) and primary amines were blocked with 0.2 ℳ ethanolamine (phosphate-buffered saline (PBS), pH 8.0 and 0.1% NP-40) for 1 h at room temperature. Beads were washed briefly with 100 mℳ glycine pH 2.5 and neutralized with 50 mℳ Tris–Hcl pH 7.4, 0.1% NP-40. Lysates were pre-cleared with magnetic beads conjugated to sheep anti-mouse IgG, and antibody-coated beads were incubated with 8 mg lysate per condition for 2 h at 4 °C with constant rotation. Unbound proteins were removed by extensive washing in lysis buffer, and specifically associated proteins were eluted from the beads by boiling for 10 min in Laemmli sample buffer. Proteins were resolved by SDS–PAGE and analysed by western blotting as described previously.59 For mass spectrometry analysis, proteins were gel-digested60 and peptides were analysed on a LTQ-Orbitrap Velos equipped with nanoelectrospray source (Thermo Fisher Scientific, Basingstoke, UK). Peptides and proteins were identified and quantified using the MaxQuant software61 and searched with the Andromeda search engine62 against the human IPI database 3.68.62
Organotypic assays
Organotypic assays were done as described previously.63 Briefly, ∼7.5 × 104/ml primary human fibroblasts were embedded in a 3D (three-dimensional) matrix of rat tail collagen I. Rat tail tendon collagen solution was prepared by the extraction of tendons with 0.5 ℳ acetic acid to a concentration of ∼2 mg/ml. Detached, polymerized matrix (2.5 ml) in 35 mm petri dishes was allowed to contract for approximately 6 days in complete media (DMEM, supplemented with 10% FCS, Invitrogen) until the fibroblasts had contracted the matrix to ∼1.5 cm diameter. Subsequently, 4 × 104 PDAC cells were plated on top of the matrix in complete media and allowed to grow to confluence for 5 days. The matrix was then mounted on a metal grid and raised to the air/liquid interface resulting in the matrix being fed from below with complete media that was changed every 2 days. After 8–12 days, the cultures were fixed using 4% paraformaldehyde and processed by standard methods for hematoxylin and eosin staining. Representative images of at least three independent experiments are shown.
Scattering assays
For live cell imaging, 2500 cells were seeded in six-well plates and allowed to settle for 32 h. Cells were incubated in the indicated chemicals and scattering was monitored using a phase-contrast × 20 objective of an inverted microscope (Nikon TE2000, Nikon, Surrey, UK) capturing pictures every 10 min for 16 h.
For knockdown experiments, H1299 cells or HCT116 cells were transfected with siRNA using hiperfect 24 h after seeding, washed 5 h later and incubated in 2.5 μg/ml ponA (Invitrogen), 10 μℳ U1026 (Sigma, Gllingham, UK), 10 ng/ml HGF (Sigma) or 25 ng/ml EGF (Sigma) for 48 h as indicated. For A431 cells and HT-29 cells, nucleofection was used to knockdown mutant p53 and cells were subsequently plated sparsely in six-well dishes and incubated with 20 ng/ml HGF for 48 h. Phase-contrast images were taken using an Olympus CKX41 microscope, Olympus, East Grinstead, UK and lysates were harvested for immunoblot analysis to verify knockdown after scattering. For each condition, >1000 cells from three independent experiments were counted and tagged as ‘scattered' or ‘non-scattered' in the ImageJ64 plugin ‘cell counter' to calculate the proportion of scattered cells. Scattered cells were defined as cells that were growing in colonies of four cells or less or cells that were touching only one other cell.
Immunohistochemistry
pERK1/2 (p44/42) MAPK ab (9101, Cell Signaling, Danvers, MA, USA; 1:200) was used to detect mouse pERK1/2 levels and p53 CM-5 (Vector Labs, Peterborough, UK, VP-P956) was used to detect mouse p53 levels. Both antibodies underwent pH 6 Na Citrate heat-induced epitope retrieval. The immunohistochemistryIHC was carried out on paraffin-embedded tissue(obtained as described previously20 using an EnVision kit (Dako, Hamburg, Germany) according to manufacturer's instructions.
Inverted Matrigel invasion assay
Matrigel assays were performed as previously described.58 In short, Matrigel (BD Biosciences, Oxford, UK), supplemented with 25 μg/ml fibronectin (FN, Sigma) was polymerized in transwell inserts (Corning Life Sciences, High Wycombe, UK) for 45 min at 37 °C. The inserts were inverted and 2.5 × 105 cells were seeded at the base of the insert. The remaining cells were seeded in new dishes and lysates were taken to verify knockdown. The inserts were placed in serum-free medium and the upper chamber was filled with 10% FCS and 25 ng/ml EGF, 10 ng/ml HGF, 5 ng/ml IGF-1 (Sigma) or 10 ng/ml PDGF-β (Sigma). Where necessary, 0.1 nℳ EGF inhibitor (PD153035, Calbiochem, Nottingham, UK) and 0.5 μg/ml of integrin-blocking antibodies (Mab16) were added to the Matrigel before polymerization were necessary and to the medium in all compartments. Migrating cells were stained in medium containing 4 nℳ Calcein (Invitrogen) and were visualized by confocal microscopy (Leica2) in serial sections of 10 μm (or 15 μm for MDA-MB-231 cells) intervals. Quantification of Matrigel images was performed in ImageJ, in which the intensity of each slide was measured. Invasion was measured by dividing the sum of the intensity of all slides beyond 30 μm (or 45 μm for MDAMB231 cells) by the sum of the intensity of all slides.
3D reconstruction images and movies of invading 273H H1299 cells were made by taking pictures in serial sections of 1 μm on the Fluoview FV1000 confocal microscope (Olympus) using a × 40 water lens. The software program Velocity (Improvision, Coventry, UK) was used to reconstruct a 3D image and generate movies of invading cells.
AKT/ ERK1/2 activation, immunoprecipitation, ELISA and immunoblot analyses
Activation of ERK1/2, MET or AKT was determined using phospho-specific antibodies in western blot. For these assays, cells were serum starved overnight or 1 h and incubated for the indicated times in IGF, PDGF, HGF or EGF (all at 20 ng/ml), ponA (2.5 μg/ml) or 50 nℳ SU11274 (Selleck, Cambridge, UK). For transfection experiments, cells were transfected using hiperfect or nucleofection for knockdown and allowed to reach confluence in approximately 48 h. PonA was added 24 h before harvest when necessary and added throughout the starvation period. Cells were starved for 4 h followed by HGF (5 ng/ml) treatment for 10 or 30 min as indicated, harvested in cold RIPA lysis buffer and immediately processed for western blot analysis to detect ERK1/2 phosphorylation. MET phosphorylation was best detected within a time interval of 10 min of HGF exposure after 1 h of starvation, whereafter cells were harvested immediately in sample buffer. RCP co-immunoprecipitation experiments were performed as described.58 Antibodies to detect the following proteins were used in western blotting: p53 DO-1 (1:10 000, monoclonal65), p53 1801 (1:5000), pMET (Y1230-1234-1234 1:2000; Millipore), pMET (Y1234, Y1235; 1:1000 Cell Signaling), pMET (Y1003 1:1000 Abcam), actin (1:5000; Millipore), α5 integrin (1:1000; BD biosciences), RCP (1:3000; Invitrogen), AKT (1:1000, Cell Signaling), p-AKT (1:1000, Cell Signaling), ERK1/2 (1:1000, Cell Signaling), pERK1/2 (1:1000, Cell Signaling), MET (1:250, R&D systems, Abingdon, UK), MET (1:250, R&D systems, goat), MET (1:1000, Santa Cruz (Santa Cruz, CA, USA), mouse), ZO-1 (1:500, Invitrogen), PAR3 (1:500, Millipore). For measuring HGF in medium, an ELISA kit (Invitrogen) was used according to the manufacturer's instructions.
Immunofluorescence
For immunofluorescence, H1299 cells were grown on glass coverslips, transfected with siRNA using hiperfect and incubated with HGF or ponA as indicated. A431 cells or HT29 cells were transfected with nucleofection to knockdown mutant p53 and incubated in HGF where necessary. Cells were washed with PBS, fixed in 4% paraformaldehyde for 10 min in 4 °C and permeabilized with 0.5% Triton X-100 in PBS for 1 min. Cells were washed four times with PBS and blocked for 30 min in 1% BSA (bovine serum albumin) in PBS, followed by 1 h incubation with first antibodies 1% BSA-PBS. Antibodies were used in the following dilutions: PAR3 (1:75, Millipore), ZO-1 (1:100, Invitrogen), p53 DO-1 (1:300, monoclonal Cancer Research UK), E-cadherin (1:125, Cell Signaling), P-Cadherin (1:125, Cell Signaling), MET (1:50, R&D systems). After three washes with PBS, cells were incubated with Alexa 488-conjugated or Alexa 594-conjugated secondary antibodies (Invitrogen, 1:300), supplemented with DAPI (Sigma, 0.5 μg/ml) for 30 min. Cells were rinsed five times with PBS and mounted on microscope slides. Images were captured using an inverted confocal fluorescent microscope (Fluoview FV1000, Olympus). Z-stacks were generated with 0.2 μm intervals and E-cadherin in the Z-stacks was quantified by determining the number of slides in a stack over which a continuous staining of E-cadherin between two cells could be detected.
Quantitative RT–PCR
For quantitative RT–PCR analyses, RNA was isolated from cells using a Qiagen kit according to the manufacturer's instructions. Complementary DNA was generated using the first-strand cDNA kit from Invitrogen according to the manufacturer's instructions on 0.5 μg DNAse-treated total RNA. For the RT–PCR reaction, 5 μl of the 40 times diluted complementary DNA was used in combination with 10 μl Sybr green Mastermix (Thermo Scientific), 1 μl of each oligo (10 μℳ) and 3 μl H2O with the following PCR conditions: 94 °C annealing 2 min, 40 cycles of 30 s at 94 °C, 30 s at 60 °C and 1 min at 72 °C, followed by a 10-min 72 °C incubation. After finishing these cycles, a melting curve was generated to check for specificity. mRNA expression of GAPDH was used as a reference. Oligos: GAPDH fw 5′-GCAGAGATGATGACCCTTTTGGCT-3′ GAPDH rev 5′-TGAAGCTCGGAGTCAACGGATTTGGT-3′ total p63 H1299 cells fw 5′-TTCTTAGCGAGGTTGGGCTG-3′, total p63 H1299 cells rev 5′-GATCGCATGTCGAAATTGCTC-3′ TA p63 fw 5′-GGACTGTATCCGCATGCAG-3′ TA p63 rev 5′-GAGCTGGGCTGTGCGTAG-3′ dN p63 fw 5′-GAAGAAAGGACAGCAGCATTGA-3′ dN p63 rev 5′-GGGACTGGTGGACGAGGAG-3′.
Recycling assays
Measurement of integrin recycling has been described previously.59 This protocol was optimized for MET using 5 μg/ml of MET antibody (R&D systems) to detect levels of biotinylated MET in 50 μl lysate by capture enzyme-linked immunosorbent assays. The quality of the MET antibody to pulldown biotinylated MET in H1299 cells that were labelled with biotin was determined in western blots/IPs (immunoprecipitations) in which MET levels and biotinylated MET (using a streptavidin-HRP antibody to detect MET in the IP lysate) were determined. Competition experiments to measure the specificity of the MET antibody for pulldown of biotinylated MET were performed using a capture enzyme-linked immunosorbent assay.
Acknowledgments
We thank Cancer Research UK and the AICR for funding of work in KHV and JCN's laboratories. PAJM is a recipient of a Rubicon Fellowship from the Netherlands Organisation for Scientific Research. We thank Julin Wong from DPL's lab for the MET construct, Bert Vogelstein for the HCT116 p53 null and HCT116 248W cells, Karen Oien for help with quantifying the staining in the pancreatic tumours and Christine Gundry for help with optimizing the recycling assays.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- O'Farrell TJ, Ghosh P, Dobashi N, Sasaki CY, Longo DL. Comparison of the effect of mutant and wild-type p53 on global gene expression. Cancer Res. 2004;64:8199–8207. doi: 10.1158/0008-5472.CAN-03-3639. [DOI] [PubMed] [Google Scholar]
- Tepper CG, Gregg JP, Shi XB, Vinall RL, Baron CA, Ryan PE, et al. Profiling of gene expression changes caused by p53 gain-of-function mutant alleles in prostate cancer cells. Prostate. 2005;65:375–389. doi: 10.1002/pros.20308. [DOI] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Weisz L, Zalcenstein A, Stambolsky P, Cohen Y, Goldfinger N, Oren M, et al. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 2004;64:8318–8327. doi: 10.1158/0008-5472.CAN-04-1145. [DOI] [PubMed] [Google Scholar]
- Zalcenstein A, Weisz L, Stambolsky P, Bar J, Rotter V, Oren M. Repression of the MSP/MST-1 gene contributes to the antiapoptotic gain of function of mutant p53. Oncogene. 2006;25:359–369. doi: 10.1038/sj.onc.1209061. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A, et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;277:18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- Stambolsky P, Tabach Y, Fontemaggi G, Weisz L, Maor-Aloni R, Siegfried Z, et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 2010;17:273–285. doi: 10.1016/j.ccr.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Noll JE, Jeffery J, Al-Ejeh F, Kumar R, Khanna KK, Callen DF, et al. Mutant p53 drives multinucleation and invasion through a process that is suppressed by ANKRD11. Oncogene. 2011. [DOI] [PubMed]
- Liu K, Ling S, Lin WC. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol. 2011;31:4464–4481. doi: 10.1128/MCB.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin Y, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–991. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Lim LY, Vidnovic N, Ellisen LW, Leong CO. Mutant p53 mediates survival of breast cancer cells. Br J Cancer. 2009;101:1606–1612. doi: 10.1038/sj.bjc.6605335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan W, Chen X. Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J Biol Chem. 2011;286:16218–16228. doi: 10.1074/jbc.M110.214585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, et al. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest. 2009;119:2171–2183. doi: 10.1172/JCI37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre C, Barrow R, Menard L, Calleja V, Hart IR, Kermorgant S. A direct role for Met endocytosis in tumorigenesis. Nat Cell Biol. 2011;13:827–837. doi: 10.1038/ncb2257. [DOI] [PubMed] [Google Scholar]
- Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- Naldini L, Vigna E, Narsimhan RP, Gaudino G, Zarnegar R, Michalopoulos GK, et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene. 1991;6:501–504. [PubMed] [Google Scholar]
- Hwang CI, Matoso A, Corney DC, Flesken-Nikitin A, Korner S, Wang W, et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci USA. 2011;108:14240–14245. doi: 10.1073/pnas.1017536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol DW, Chen Q, Smith ML, Zarnegar R. Regulation of the c-met proto-oncogene promoter by p53. J Biol Chem. 1999;274:3565–3572. doi: 10.1074/jbc.274.6.3565. [DOI] [PubMed] [Google Scholar]
- Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J. 2006;25:3534–3545. doi: 10.1038/sj.emboj.7601213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera R. Modulation of hepatocyte growth factor-induced scattering of HT29 colon carcinoma cells. Involvement of the MAPK pathway. J Cell Sci. 1998;111:1039–1049. doi: 10.1242/jcs.111.8.1039. [DOI] [PubMed] [Google Scholar]
- Cirit M, Wang CC, Haugh JM. Systematic quantification of negative feedback mechanisms in the extracellular signal-regulated kinase (ERK) signaling network. J Biol Chem. 2010;285:36736–36744. doi: 10.1074/jbc.M110.148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti S, Comoglio PM. The MET receptor tyrosine kinase in invasion and metastasis. J Cell Physiol. 2007;213:316–325. doi: 10.1002/jcp.21183. [DOI] [PubMed] [Google Scholar]
- Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem. 2000;275:8806–8811. doi: 10.1074/jbc.275.12.8806. [DOI] [PubMed] [Google Scholar]
- Accornero P, Miretti S, Cucuzza LS, Martignani E, Baratta M. Epidermal growth factor and hepatocyte growth factor cooperate to enhance cell proliferation, scatter, and invasion in murine mammary epithelial cells. J Mol Endocrinol. 2010;44:115–125. doi: 10.1677/JME-09-0035. [DOI] [PubMed] [Google Scholar]
- Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama R, Shimizu N. Detection of epidermal growth factor receptors and E-cadherins in the basolateral membrane of A431 cells by laser scanning fluorescence microscopy. Jpn J Cancer Res. 1991;82:8–11. doi: 10.1111/j.1349-7006.1991.tb01737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan A, Wang ZD, Spencer J. E-cadherin expression in intestinal epithelium. J Clin Pathol. 1995;48:143–146. doi: 10.1136/jcp.48.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H, Matsumoto K, Nakamura T. Regulation of cell growth and motility by hepatocyte growth factor and receptor expression in various cell species. Exp Cell Res. 1992;202:423–431. doi: 10.1016/0014-4827(92)90095-p. [DOI] [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin M, Zhang C, Wang D, Feng Z, Hu W. TAp63gamma enhances nucleotide excision repair through transcriptional regulation of DNA repair genes. DNA Repair. 2012;11:167–176. doi: 10.1016/j.dnarep.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard MK, Kommagani R, Payal V, Mayo LD, Shamma HN, Kadakia MP. DeltaNp63alpha regulates keratinocyte proliferation by controlling PTEN expression and localization. Cell Death Differ. 2011;18:1924–1933. doi: 10.1038/cdd.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayan BS, Sayan AE, Yang AL, Aqeilan RI, Candi E, Cohen GM, et al. Cleavage of the transactivation-inhibitory domain of p63 by caspases enhances apoptosis. Proc Natl Acad Sci USA. 2007;104:10871–10876. doi: 10.1073/pnas.0700761104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Corso S, Migliore C, Ghiso E, De Rosa G, Comoglio PM, Giordano S. Silencing the MET oncogene leads to regression of experimental tumors and metastases. Oncogene. 2008;27:684–693. doi: 10.1038/sj.onc.1210697. [DOI] [PubMed] [Google Scholar]
- Stoker M, Perryman M. An epithelial scatter factor released by embryo fibroblasts. J Cell Sci. 1985;77:209–223. doi: 10.1242/jcs.77.1.209. [DOI] [PubMed] [Google Scholar]
- Parachoniak CA, Luo Y, Abella JV, Keen JH, Park M. GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev Cell. 2011;20:751–763. doi: 10.1016/j.devcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MY, Yu Y, Walsh WR, Yang JL. microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review) Int J Oncol. 2011;38:1189–1195. doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
- Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Xu H, Stabile LP, Gubish CT, Gooding WE, Grandis JR, Siegfried JM. Dual blockade of EGFR and c-Met abrogates redundant signaling and proliferation in head and neck carcinoma cells. Clin Cancer Res. 2011;17:4425–4438. doi: 10.1158/1078-0432.CCR-10-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Brevet M, Shimizu S, Bott MJ, Shukla N, Zhou Q, Olshen AB, et al. Coactivation of receptor tyrosine kinases in malignant mesothelioma as a rationale for combination targeted therapy. J Thorac Oncol. 2011;6:864–874. doi: 10.1097/jto.0b013e318215a07d. [DOI] [PubMed] [Google Scholar]
- Kawaguchi K, Murakami H, Taniguchi T, Fujii M, Kawata S, Fukui T, et al. Combined inhibition of MET and EGFR suppresses proliferation of malignant mesothelioma cells. Carcinogenesis. 2009;30:1097–1105. doi: 10.1093/carcin/bgp097. [DOI] [PubMed] [Google Scholar]
- Sur S, Pagliarini R, Bunz F, Rago C, Diaz LA, Kinzler KW, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci USA. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolarov J, Chang K, Reiner A, Rodgers L, Hannon GJ, Wigler MH, et al. Design of a retroviral-mediated ecdysone-inducible system and its application to the expression profiling of the PTEN tumor suppressor. Proc Natl Acad Sci USA. 2001;98:13043–13048. doi: 10.1073/pnas.221450598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–155. doi: 10.1083/jcb.200804140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M, Barry S, Woods A, van der Sluijs P, Norman J. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol. 2001;11:1392–1402. doi: 10.1016/s0960-9822(01)00442-0. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Edward M, Gillan C, Micha D, Tammi RH. Tumour regulation of fibroblast hyaluronan expression: a mechanism to facilitate tumour growth and invasion. Carcinogenesis. 2005;26:1215–1223. doi: 10.1093/carcin/bgi064. [DOI] [PubMed] [Google Scholar]
- Collins TJ. Image J for microscopy. Biotechniques. 2007;43 1 Suppl:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- Vojtesek B, Bartek J, Midgley CA, Lane DP. An immunochemical analysis of the human nuclear phosphoprotein p53. New monoclonal antibodies and epitope mapping using recombinant p53. J Immunol Methods. 1992;151:237–244. doi: 10.1016/0022-1759(92)90122-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.