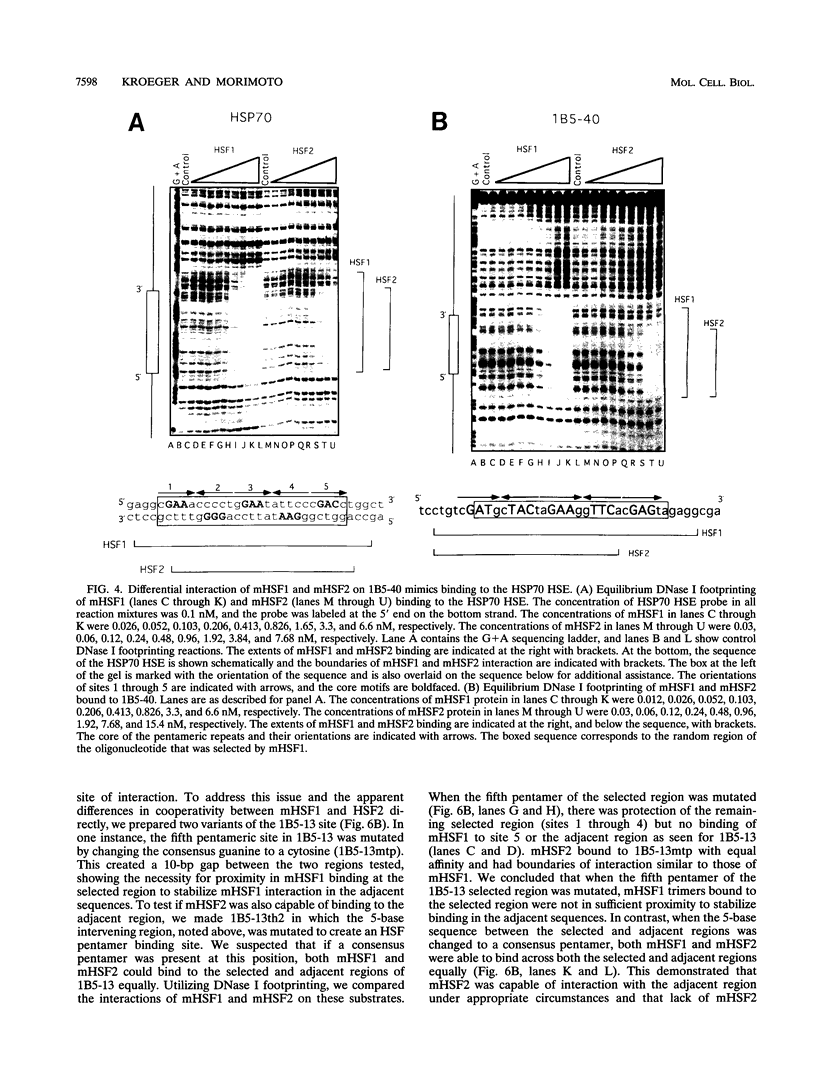
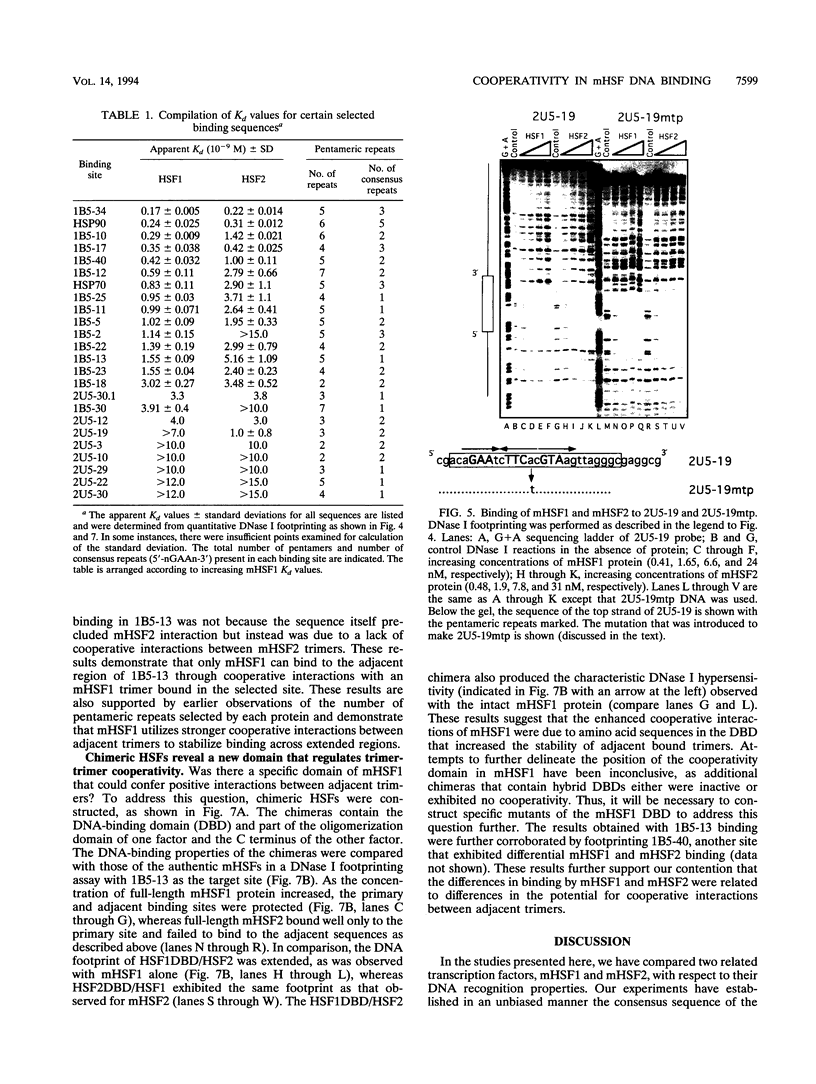
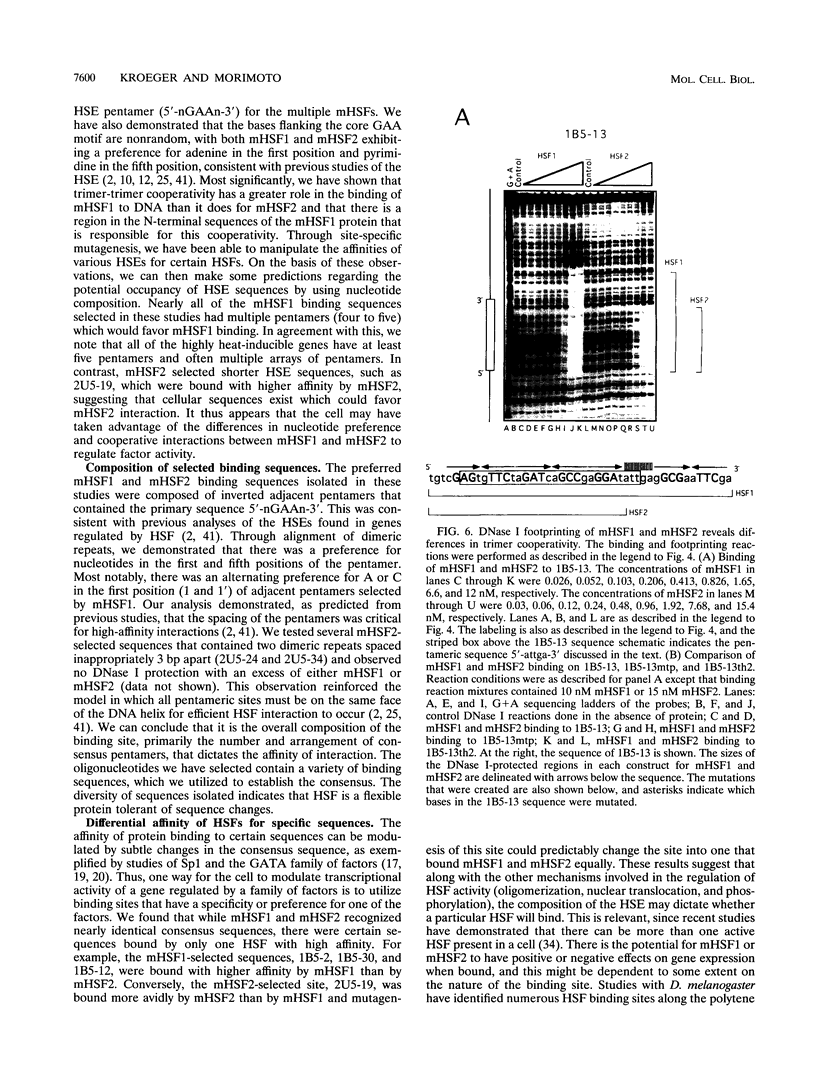
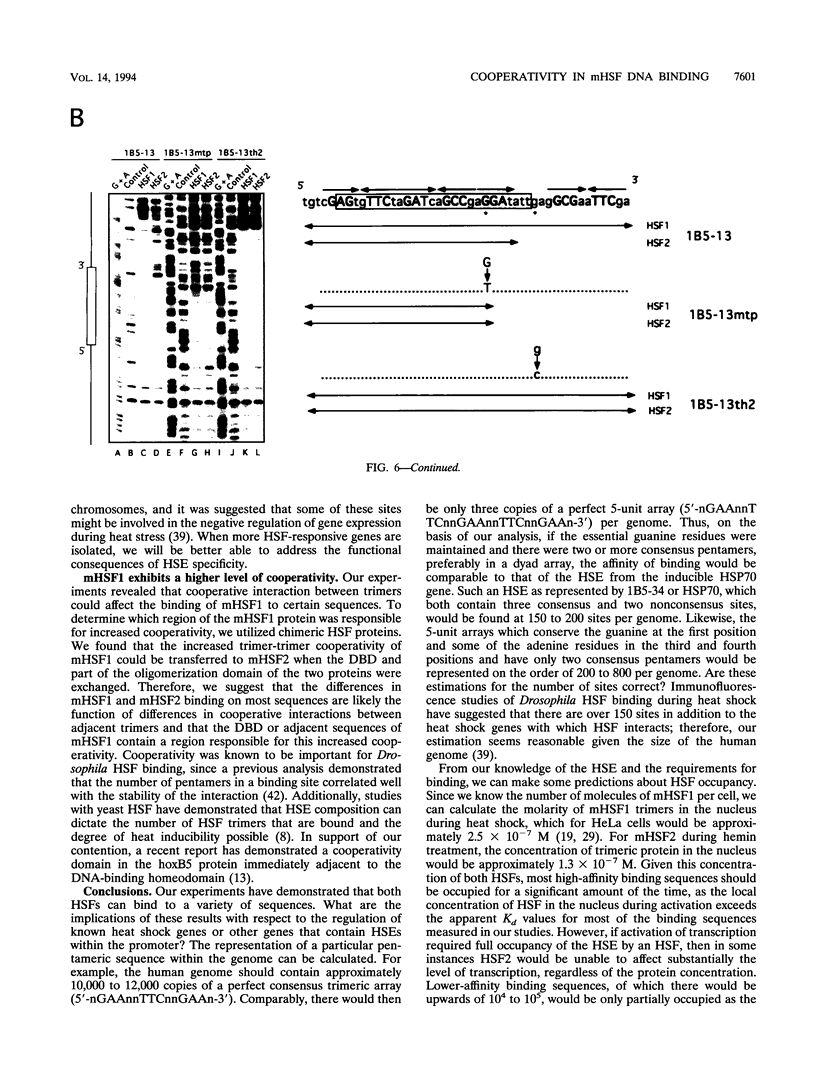
Abstract
Multiple heat shock transcription factors (HSFs) have been discovered in several higher eukaryotes, raising questions about their respective functions in the cellular stress response. Previously, we had demonstrated that the two mouse HSFs (mHSF1 and mHSF2) interacted differently with the HSP70 heat shock element (HSE). To further address the issues of cooperativity and the interaction of multiple HSFs with the HSE, we selected new mHSF1 and mHSF2 DNA-binding sites through protein binding and PCR amplification. The selected sequences, isolated from a random population, were composed primarily of alternating inverted arrays of the pentameric consensus 5'-nGAAn-3', and the nucleotides flanking the core GAA motif were nonrandom. The average number of pentamers selected in each binding site was four to five for mHSF1 and two to three for mHSF2, suggesting differences in the potential for cooperative interactions between adjacent trimers. Our comparison of mHSF1 and mHSF2 binding to selected sequences further substantiated these differences in cooperativity as mHSF1, unlike mHSF2, was able to bind to extended HSE sequences, confirming previous observations on the HSP70 HSE. Certain selected sequences that exhibited preferential binding of mHSF1 or mHSF2 were mutagenized, and these studies demonstrated that the affinity of an HSE for a particular HSF and the extent of HSF interaction could be altered by single base substitutions. The domain of mHSF1 utilized for cooperative interactions was transferable, as chimeric mHSF1/mHSF2 proteins demonstrated that sequences within or adjacent to the mHSF1 DNA-binding domain were responsible. We have demonstrated that HSEs can have a greater affinity for a specific HSF and that in mice, mHSF1 utilizes a higher degree of cooperativity in DNA binding. This suggests two ways in which cells have developed to regulate the activity of closely related transcription factors: developing the ability to fully occupy the target binding site and alteration of the target site to favor interaction with a specific factor.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abravaya K., Phillips B., Morimoto R. I. Heat shock-induced interactions of heat shock transcription factor and the human hsp70 promoter examined by in vivo footprinting. Mol Cell Biol. 1991 Jan;11(1):586–592. doi: 10.1128/mcb.11.1.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin J., Ananthan J., Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988 Sep;8(9):3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin J., Fernandez M., Ananthan J., Lis J. T., Voellmy R. Cooperative binding of heat shock transcription factor to the Hsp70 promoter in vivo and in vitro. J Biol Chem. 1994 Feb 18;269(7):4804–4811. [PubMed] [Google Scholar]
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. V. Responses to environmental treatments. Chromosoma. 1970;31(3):356–376. doi: 10.1007/BF00321231. [DOI] [PubMed] [Google Scholar]
- Baler R., Dahl G., Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol. 1993 Apr;13(4):2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P. B., Rabindran S. K., Wu C. Heat shock-regulated transcription in vitro from a reconstituted chromatin template. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4109–4113. doi: 10.1073/pnas.88.10.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Bonner J. J., Ballou C., Fackenthal D. L. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol. 1994 Jan;14(1):501–508. doi: 10.1128/mcb.14.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila M. P., Garcia-Bellido A. Development and genetic analysis of bithorax phenocopies in Drosophila. Nature. 1974 Aug 9;250(5466):500–502. doi: 10.1038/250500a0. [DOI] [PubMed] [Google Scholar]
- Cunniff N. F., Morgan W. D. Analysis of heat shock element recognition by saturation mutagenesis of the human HSP70.1 gene promoter. J Biol Chem. 1993 Apr 15;268(11):8317–8324. [PubMed] [Google Scholar]
- Fernandes M., Xiao H., Lis J. T. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor--heat shock element interactions. Nucleic Acids Res. 1994 Jan 25;22(2):167–173. doi: 10.1093/nar/22.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galang C. K., Hauser C. A. Cooperative DNA binding of the human HoxB5 (Hox-2.1) protein is under redox regulation in vitro. Mol Cell Biol. 1993 Aug;13(8):4609–4617. doi: 10.1128/mcb.13.8.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey E., Brandon S. E., Smale G., Lloyd D., Weber L. A. Sequence and regulation of a gene encoding a human 89-kilodalton heat shock protein. Mol Cell Biol. 1989 Jun;9(6):2615–2626. doi: 10.1128/mcb.9.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R. E., Schuetz T. J., Larin Z. Heat-inducible human factor that binds to a human hsp70 promoter. Mol Cell Biol. 1987 Apr;7(4):1530–1534. doi: 10.1128/mcb.7.4.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L. J., Engel J. D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993 Jul;13(7):4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
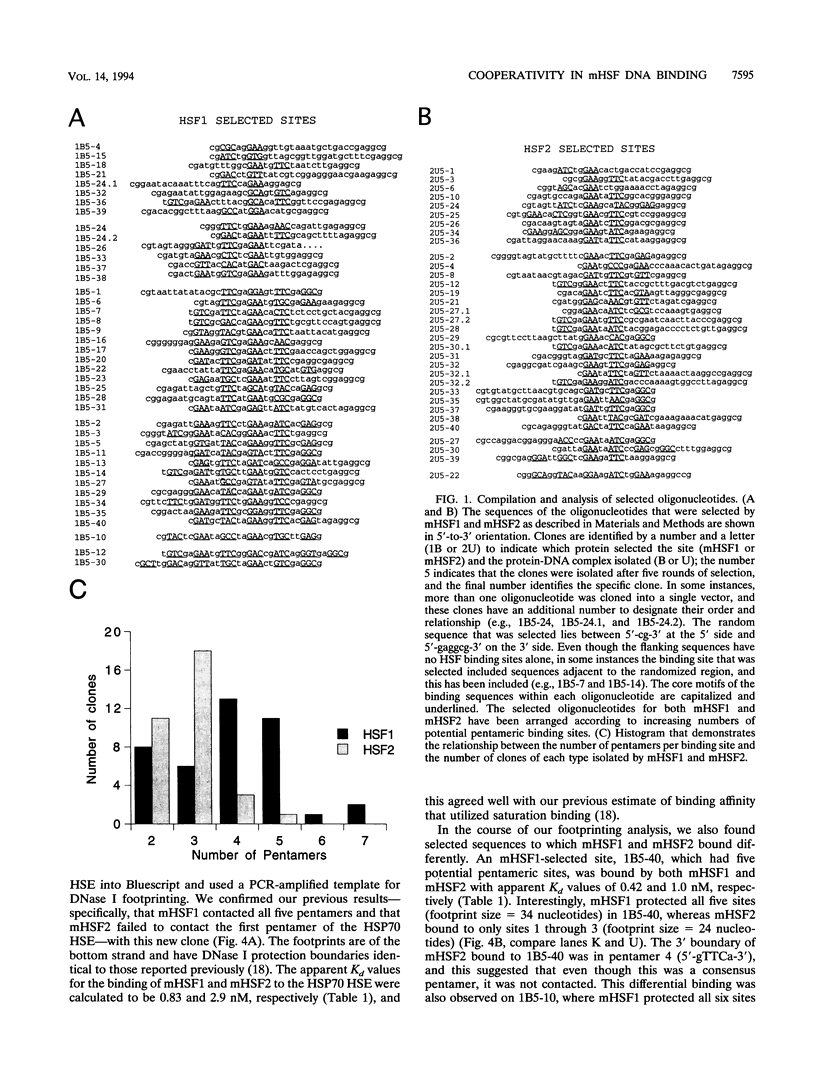
- Kroeger P. E., Sarge K. D., Morimoto R. I. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol Cell Biol. 1993 Jun;13(6):3370–3383. doi: 10.1128/mcb.13.6.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letovsky J., Dynan W. S. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989 Apr 11;17(7):2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M., Orkin S. H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993 Jul;13(7):3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R. I., Sarge K. D., Abravaya K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J Biol Chem. 1992 Nov 5;267(31):21987–21990. [PubMed] [Google Scholar]
- Mosser D. D., Theodorakis N. G., Morimoto R. I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol Cell Biol. 1988 Nov;8(11):4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A., Morimoto R. I. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Mol Cell Biol. 1993 Apr;13(4):1983–1997. doi: 10.1128/mcb.13.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O., Xiao H., Lis J. T. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989 Dec 1;59(5):797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran S. K., Giorgi G., Clos J., Wu C. Molecular cloning and expression of a human heat shock factor, HSF1. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge K. D., Murphy S. P., Morimoto R. I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993 Mar;13(3):1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge K. D., Zimarino V., Holm K., Wu C., Morimoto R. I. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991 Oct;5(10):1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- Scharf K. D., Rose S., Zott W., Schöffl F., Nover L., Schöff F. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990 Dec;9(13):4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz T. J., Gallo G. J., Sheldon L., Tempst P., Kingston R. E. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuey D. J., Parker C. S. Binding of Drosophila heat-shock gene transcription factor to the hsp 70 promoter. Evidence for symmetric and dynamic interactions. J Biol Chem. 1986 Jun 15;261(17):7934–7940. [PubMed] [Google Scholar]
- Sistonen L., Sarge K. D., Morimoto R. I. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994 Mar;14(3):2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L., Sarge K. D., Phillips B., Abravaya K., Morimoto R. I. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992 Sep;12(9):4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988 Sep 9;54(6):855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Sorger P. K. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990 Aug 24;62(4):793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Westwood J. T., Clos J., Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991 Oct 31;353(6347):822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wilson S., Walker B., Dawid I., Paisley T., Zimarino V., Ueda H. Purification and properties of Drosophila heat shock activator protein. Science. 1987 Nov 27;238(4831):1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- Xiao H., Lis J. T. Germline transformation used to define key features of heat-shock response elements. Science. 1988 Mar 4;239(4844):1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- Xiao H., Perisic O., Lis J. T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991 Feb 8;64(3):585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]