Abstract
Troponin T (TnT) is known to mediate the interaction between Tn complex and tropomyosin (Tm), which is essential for calcium-activated striated muscle contraction. This regulatory function takes place in the myoplasm, where TnT binds Tm. However, recent findings of troponin I and Tm nuclear translocation in Drosophila and mammalian cells imply other roles for the Tn–Tm complex. We hypothesized that TnT plays a nonclassical role through nuclear translocation. Immunoblotting with different antibodies targeting the NH2- or COOH-terminal region uncovered a pool of fast skeletal muscle TnT3 localized in the nuclear fraction of mouse skeletal muscle as either an intact or fragmented protein. Construction of TnT3–DsRed fusion proteins led to the further observation that TnT3 fragments are closely related to nucleolus and RNA polymerase activity, suggesting a role for TnT3 in regulating transcription. Functionally, overexpression of TnT3 fragments produced significant defects in nuclear shape and caused high levels of apoptosis. Interestingly, nuclear TnT3 and its fragments were highly regulated by aging, thus creating a possible link between the deleterious effects of TnT3 and sarcopenia. We propose that changes in nuclear TnT3 and its fragments cause the number of myonuclei to decrease with age, contributing to muscle damage and wasting.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-011-9368-4) contains supplementary material, which is available to authorized users.
Keywords: Troponin T, Nuclear localization, Skeletal muscle, Aging, Apoptosis, Nucleolus, RNA polymerase
Introduction
The thin filament regulatory proteins troponin (Tn) and tropomyosin (Tm) are essential for contraction of striated muscle (skeletal and cardiac), which is regulated by the concentration of intracellular calcium (Tobacman 1996; Gordon et al. 2000; Szczesna and Potter 2002). The Tn complex is composed of three subunits: calcium-binding troponin C (TnC), inhibitory troponin I (TnI), and Tm-binding troponin T (TnT). Muscle fiber depolarization leads to sarcoplasmic reticulum calcium release and contraction (Jimenez-Moreno et al. 2008, 2010). Calcium binds to TnC, causing conformational changes in the Tn complex, exposing binding sites for myosin on the actin filaments, and initiating interaction between myosin and actin, with consequent muscle contraction (Geeves and Holmes 1999).
Vertebrates have various TnT isoforms: cardiac TnT1, slow skeletal muscle TnT2, and fast skeletal muscle TnT3, with variable sizes (233–305 amino acids) and charges resulting from multiple regulatory mechanisms, including alternative RNA splicing and posttranslational modifications. The TnT NH2-terminal region is hypervariable in contrast to the COOH-terminal and middle regions, which are highly conserved across species (Jin et al. 2008). Functionally, the NH2-terminal region of TnT does not bind any known myofilament proteins and can be selectively removed during cardiac ischemia reperfusion through calpain cleavage (Zhang et al. 2006). In contrast, the COOH-terminal and middle regions of TnT carry the binding sites for TnI, TnC, and Tm (Perry 1998) and play a central role, activating actomyosin even when the NH2-terminal variable region is deleted (Pan et al. 1991). Therefore, the COOH-terminal and middle regions are the core TnT domain, making TnT the key mediator of the Tn complex self-organization and interaction with the muscle thin filament (Tobacman 1996; Perry 1998).
Still, evolutionarily and developmentally, the hypervariable N-terminus of TnT seems to be required for the fine functional regulation of the core domain of each TnT isoform. The NH2-terminal structure modulates TnT conformation and middle and C-terminal region interaction with other thin filament proteins (Ogut and Jin 1996; Wang and Jin 1998; Biesiadecki et al. 2007; Jin and Root 2000). Deletion of the N-terminal variable region of cardiac TnT increases heart efficiency (Zhang et al. 2006; Feng et al. 2008). TnT N-terminal variation alters thin filament Ca2+ sensitivity and force production (Reiser et al. 1992; Gomes et al. 2002; Biesiadecki and Jin 2002; Chandra et al. 1999), and the negative charges of the N-terminal region determine the overall TnT charge (Wang and Jin 1997), while the TnT N-terminal acidic residues provide binding capacity for Ca2+ (Zhang et al. 2004).
Age-related loss of muscle mass, muscle function, and muscle quality, termed sarcopenia (Zinna and Yarasheski 2003), is characterized by muscle weakness (Degens and Alway 2003, 2006; Onambele et al. 2006; Morse et al. 2004, 2005; Klitgaard et al. 1990; Larsson 1978; Larsson and Ansved 1995), reduced maximal shortening velocity, and slow contraction and relaxation (Klitgaard et al. 1990; Larsson 1978; Larsson and Ansved 1995; Narici and Maganaris 2006) with a consequent decrease in force-generating capacity (Barbieri et al. 2003; Brooks and Faulkner 1991; Runge et al. 2004; Gonzalez and Delbono 2001; Gonzalez et al. 2000; for a review see, Delbono 2011). Sedentary lifestyle and reduced levels and/or responsiveness to trophic hormones and factors contribute to muscle atrophy (Thomas 2010). In addition, age-related decrease in muscle mass has been associated with myonuclei loss (Marzetti et al. 2010; Buford et al. 2010), fewer stem cells, and diminished regenerative capacity (Snijders et al. 2009; Carlson and Conboy 2007; Day et al. 2010; Shefer et al. 2010). Loss in fiber type II (Larsson 1978; Tomlinson et al. 1973) and fewer satellite cells (Verdijk et al. 2007) also contribute to the predominant atrophy of fast-twitch fibers with aging. A rigorous analysis of the involvement of Tn isoforms in muscle fiber atrophy is needed.
Whether Tn plays another role than the classically described regulation of striated muscle contraction is unknown. Here, for the first time, we show that fast skeletal muscle TnT3 and its fragments translocate into the skeletal myofiber nuclei in vivo. We further characterized their subnuclear localization, proved that they are closely related to the nucleolus and RNA polymerase activity, and examined changes in full-length TnT3 and TnT COOH-terminal fragment myonuclei expression with aging. Finally, we examined the cytotoxicity of TnT3 fragments. We propose that nuclear TnT3 and its fragments cause myonuclear decrease in aging muscle and mediate muscle damage and disease. TnT3 may prove an effective therapeutic target to improve muscle quality in health and disease.
Methods
Cell culture and transfection
The mouse skeletal muscle cell line C2C12 was cultured as described previously (Zhang et al. 2009). Only undifferentiated C2C12 myoblasts, which do not exhibit endogenous myofilaments, were used for transient exogenous protein expression. Briefly, undifferentiated C2C12 myoblasts were plated on tissue culture dishes or glass coverslips coated with 0.5% gelatin in growth medium consisting of Dulbecco’s modified Eagle medium (1 g/l glucose), containing 10% FBS (Atlanta Biologicals, Atlanta, GA, USA) and 2 mM Glutamax (Invitrogen, Carlsbad, CA, USA). The mouse NIH3T3 fibroblasts were cultured in the same medium used for C2C12. Lipofactamine 2000 (Invitrogen) was used for cell transfection.
TnT3 cDNA construct and primers
The full-length cDNA of TnT3 was amplified by PCR using a TnT3 cDNA fragment subcloned in the pGADT7 vector as a template (obtained from a Yeast Two Hybrid assay from our lab, unpublished data) and into the NH2-terminal region of pDsRed2-N1 vector (Clontech, Mountain View, CA, USA) between HindIII and SacII restriction enzyme digestion sites. The other three TnT3 fragments encoding cDNA were further cloned by PCR using the same strategy. Primer sequences are listed in Fig. S1. Sequencing confirmed all constructs (DNA sequencing laboratory, Wake Forest University School of Medicine [WFUSM]).
Transcription inhibition
Three hours before fixation, C2C12 cells transfected with different TnT3/DsRed constructs were treated with 0.05 or 20 μg/ml actinomycin D (ActD) (Sigma-Aldrich, St. Louis, MO, USA) to inhibit RNA polymerase I (Pol I) or Pol I and polymerase II (Pol II), respectively (Dousset et al. 2000; Pinol-Roma and Dreyfuss 1992) or treated with 25 μg/ml 5,6-dichlorobenimidazole ribose (DRB) (Sigma-Aldrich) to inhibit RNA Pol II specifically (van Koningsbruggen et al. 2004).
RNA extraction, reverse transcription, and qPCR
Total RNA was extracted from young and old mouse flexor digitorum brevis (FDB) muscles using Trizol reagent (Invitrogen) following the user manual. Gene expression was analyzed by quantitative real-time PCR (qPCR) using Stratagene Mx3000 (Stratagene, La Jolla, CA, USA). qPCR master mix and TaqMan primer/probes for TnT3 and GAPDH genes were purchased from Applied Biosystems (Foster City, CA, USA). To determine TnT3 or GAPDH tissue expression levels, 10 ng of total RNA was added to the PCR reaction tube with a mixture of qPCR master mix, Taqman primer/probes, superscript III reverse transcriptase (Invitrogen), and RNAse inhibitor (Promega, Fitchburg, WI, USA) in a total reaction volume of 25 μl. The PCR parameters were 48°C, 45 min × 1 cycle; 95°C, 10 min × 1 cycle; 95°C, 15 s, and 60°C, 1 min × 40 cycles.
Animal and tissue lysates
Tibialis anterior (TA) muscles were dissected from 3- to 28-month-old FVB (Friend Virus B, our colony) mice, which have been used as a model of aging skeletal muscle in our laboratory (Renganathan et al. 1998; Payne et al. 2004). Animals were housed at WFUSM and killed by cervical dislocation. Handling and procedures were approved by the WFUSM Animal Care and Use Committee.
To extract cytosolic and nuclear protein fractions from the mouse TA, we used a procedure reported before (Siu et al. 2006) as shown in Fig. S2. In brief, 50 mg of muscle was ground in liquid nitrogen and homogenized on ice in 1 ml lysis buffer (10 mM NaCl, 1.5 mM MgCl2, 20 mM HEPES, pH 7.4, 20% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol) supplemented with a protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Then the samples were filtered through a 40-μm cell strainer (BD Biosciences, Bedford, MA, USA) to remove tissue debris and centrifuged (Heraeus’ Biofuge Fresco, Thermo Fisher Scientific, Pittsburgh, PA, USA) at 5,000 rpm for 5 min at 4°C to pellet the nuclei and cell debris. Supernatants were collected and centrifuged three more times at 6,000 rpm for 5 min at 4°C to remove residual nuclei. The final collected supernatants were stored as nuclei-free total cytosolic protein fractions. The remaining pellet was resuspended in 360 μl of lysis buffer in the presence of 49.8 μl of 5 M NaCl and protease inhibitor cocktail and rotated for 2 h at 4°C to release soluble nuclear protein. Following a spin at 13,000 rpm for 15 min at 4°C, the supernatants were collected and stored as cytosolic-free nuclear protein fractions. The final pellet, mainly containing the myofibrillar proteins and insoluble nuclear matrix, was resuspended in Laemmli SDS-PAGE sample buffer containing 2% SDS. Protein concentration was determined by Bio-Rad DC assay (Hercules, CA, USA). For immunoblots, 10 μg protein from each preparation was used.
To prepare whole tissue lysates, muscles were homogenized in Laemmli SDS-PAGE sample buffer containing 2% SDS and heated at 95°C for 5 min. Insoluble material was removed by centrifugation, and the soluble supernatants saved as whole cell lysis.
Muscle electroporation and single intact fiber preparation
Two mouse strains, C57B16 and FVB, were used for intramuscular plasmid injection and electroporation according to DiFranco et al. (2006). Briefly, FDB muscles were injected with 5 μl of 2 mg/ml hyaluronidase and injected 1 h later with 20 μg DsRed-conjugated TnT3 cDNAs or control DsRed cDNA. Ten minutes later, two sterile, gold-plated acupuncture needles were placed under the skin on adjacent sides of the muscle. Twenty 100 V/cm, 20-ms square-wave pulses of 1-Hz frequency were applied to the muscle for 1 s each using a Grass stimulator (Grass S48; W. Warwick, RI, USA).
The technique for single intact fiber dissection followed the procedures previously described (Gonzalez et al. 2000; Lannergren and Westerblad 1987; Birbrair et al. 2011). After isolation, suspended fibers were pelleted for 20 min and fixed with 4% paraformaldehyde (15 min at room temperature), then stained with Hoechst 33342 and mounted for microscopic analysis.
Microscopy and image analysis
Cells cultured on coverslips were fixed with 4% paraformaldehyde (15 min at room temperature), and the cell membrane was permeabilized for 5 min at room temperature with 0.5% Triton X-100 in PBS buffer after fixation. After three washes with PBS, cells were incubated for 1 h at room temperature in blocking buffer (PBS with 10% normal goat serum [Sigma]) and labeled at room temperature with primary and secondary antibodies for 2 and 1 h, respectively. Cells were counterstained with Hoechst 33342 (Invitrogen) and mounted in DAKO fluorescent mounting medium (DAKO, Carpinteria, CA, USA).
Widefield immunofluorescence images were taken on an inverted motorized fluorescent microscope (Olympus, IX81, Tokyo, Japan), and an Orca-R2 Hamamatsu CCD camera (Hamamatsu, Japan) was used to acquire images. Camera driver and image acquisition were controlled with a MetaMorph Imaging System (Olympus). Digital image files were transferred to Photoshop 7.0 to assemble montages. Images are representative examples from three independent experiments.
Annexin V and 7-AAD staining and flow cytometry
The C2C12 or NIH3T3 cells were transfected with different constructs (TnFL/DsRed, TnNT/DsRed, TnM/DsRed, TnCT/DsRed, and DsRed) (Fig. S1), cultured for 48 h, and stained with FITC Annexin V and 7-amino actinomycin D (7-AAD) (BD Bioscience, San Jose, CA, USA), following the manufacturer’s protocol. Briefly, the cells were harvested by trypsinization, washed twice with PBS, and stained with 2.5 μg/ml FITC Annexin V and 7-AAD in 1× binding buffer (10 mM HEPES (pH 7.4), 140 mM NaCl, 2.5 mM CaCl2) at room temperature for 15 min. This method allows us to discriminate viable (AnnexinV−/7-AAD−), apoptotic (Annexin V+/7-AAD−), and necrotic (Annexin V+/7-AAD+; Annexin V−/7-AAD+) cells. Flow cytometry analysis was carried out using a FACSCalibur flow cytometer (FACScan; Becton Dickinson Immunocytometry Systems, San Jose, CA, USA). The samples were acquired immediately after staining, and the value in the dot plot indicates the percent of each gated region that fell into the quadrant. DsRed fluorescence was detected with the FL2 channel, Annexin V FITC fluorescence with the FL1 channel, and 7-AAD fluorescence with the FL3 channel. Background fluorescence of (1) unstained nontransfected cells, (2) nontransfected cells stained for FITC Annexin V or 7-AAD, and (3) transfected DsRed-positive cells was used to set up gates, photomultiplier tube voltages, and amperage gain for each channel. The gate was set up to quantify only the transfected DsRed-positive cells for 7-AAD and Annexin V staining. Data were collected on at least 100,000 freshly stained cells and analyzed using CellQuest software (BD).
Electrophoresis and immunoblotting
SDS-PAGE was conducted using a 4.5% stacking gel with a 10% resolving gel in a Mini-Protean gel system (BioRad Laboratories, Hemel-Hemptstead, Herts, UK) as described (Taylor et al. 2009). Gels were transferred to PVDF membranes (Amersham Health, Little Chalfont, Buckinghamshire, UK) and held overnight at 4°C. Blots were blocked in 5% nonfat dry milk with 0.1% Tween in TBS for primary antibody incubation. Horseradish peroxidase-conjugated secondary antibodies were used at a 1:5,000 dilution at room temperature for 1 h. Band intensity was measured using the Kodak Gel Doc imaging system (Carestream Health, Inc, Rochester, NY, USA). Peroxidase activity was revealed with the Amersham ECL plus western blot detection reagents (GE Healthcare, Piscataway, NJ, USA). Data are representative of three independent experiments.
Reagents and antibodies
Rabbit anti-TnT3 polyclonal antibody was purchased from Aviva Systems Biology (San Diego, CA, USA), mouse anti-TnT3 monoclonal antibody from NOVUS Biologicals (Littleton, CO, USA), rabbit anti-histone H3 from Cell Signaling Technology (Beverly, MA, USA), mouse anti-myosin heavy chain (MHC) MF20 from Developmental Studies Hybridoma Bank (University of Iowa), mouse anti-α-tubulin from Sigma, mouse anti-actin from Millipore (Billerica, MA, USA), mouse anti-fibrillarin from Abcam (Cambridge, MA, USA), mouse anti-PRA194 (C-1) monoclonal antibody from Santa Cruz (Santa Cruz, CA, USA), and mouse anti-RNA polymerase II 8GW16 monoclonal antibody from Covance (Emeryville, CA, USA).
Alexa 488- or 568-conjugated anti-mouse or anti-rabbit IgG was purchased from Invitrogen. NA931V goat anti-mouse (Amersham Health) was used as a secondary antibody for immunoblotting.
Statistical analysis
All graphs and statistical analyses were made with Prism 5.0a (GraphPad Software, Inc., La Jolla, CA, USA). Data are expressed as means±D. The unpaired Student’s t test was used to compare experimental groups.
Results
TnT3 is located in the myofiber nucleus and cytoplasm
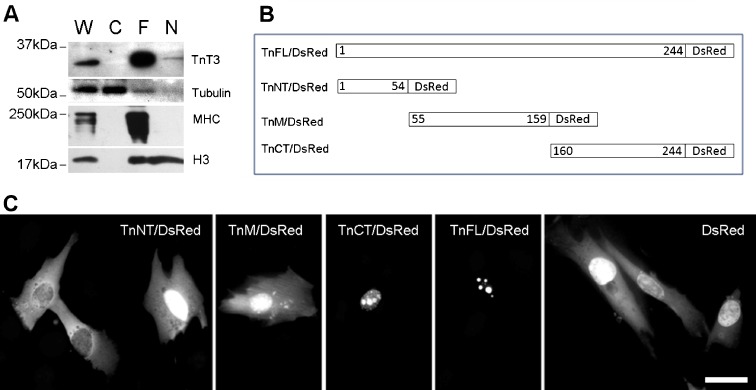
Western blot analysis of total (W), nuclear (N), myofibrillar (F), and cytoplasmic (C) extracts of untransfected mouse TA muscles showed that endogenous TnT3 localizes mainly in the myofibrillar pool. However, a small fraction localized in the nucleus (Fig. 1a). To verify the purity of the fractions, we looked for α-tubulin as a cytoplasmic marker and MHC as a myofibrillar marker. We detected α-tubulin predominantly in the total and cytoplasmic fractions and MHC only in the total and myofibrillar fractions, with the strongest bands in the myofibrillar pool. Histone H3 was consistently in both the total and nuclear fractions but not the cytoplasmic pool. H3 was also detected in the myofibrillar pool, indicating that a fraction of the nuclear proteins was not completely released (Fig. 1a).
Fig. 1.
Troponin T3 is localized in the muscle nucleus. Whole cell lysis protein extract (W), cytosolic (C), myofibrillar (F), and nuclear (N) pools were prepared from the TA muscle. a A 35-kDa band showed that full-length TnT3 (TnFL) was most abundant in the myofibrillar pool and the whole cell lysate. A small portion was present in the nuclear pool. Histone H3 was used as a nuclear marker and detected in the nuclear pool and the whole cell lysis and the myofibrillar pool. Consistently, tubulin was mainly detected in the whole cell lysis and the cytosolic pool. b Schematic drawings of different TnT3 cDNA constructs with DsRed conjugation to the C-terminus. c Immunofluorescence images of various TnT3/DsRed proteins transiently expressed in the C2C12 cells. TnNT/DsRed, TnM/DsRed, and control DsRed are distributed throughout the cell. A distribution along the stress fibers was also noticed for TnM/DsRed. In contrast, TnCT/DsRed and TnFL/DsRed are mainly found in the nucleus and showed a punctate distribution pattern. Scale bar, 20 μm
Subcellular localization of TnT3/DsRed fusion constructs in mouse C2C12 myoblasts
To investigate the subnuclear localization of TnT3 and its fragments and to determine the domain that contains the nuclear localization signal, we conjugated DsRed with intact TnT3 or each of its three fragments, representing the NH2-terminal variable region, the middle, conserved region, and the COOH-terminal conserved region (Figs. 1b and S1). All constructs were overexpressed in the mouse C2C12 myoblasts, and their distribution pattern was compared with control DsRed protein (Fig. 1c). Surprisingly, both the TnFL/DsRed and the TnCT/DsRed showed sharp, punctate nuclear localization, while TnCT/DsRed also showed diffuse distribution in the nucleoplasm. In contrast, both TnNT/DsRed and TnM/DsRed, similar to control DsRed protein, showed diffuse localization throughout the cytoplasm and nucleus. Note that the TnM/DsRed construct may have been distributed along the stress fibers in the cytosolic area. Similar subcellular localization patterns for all of these constructs were observed in NIH3T3 and COS7 cell lines (data not shown). GFP NH2-terminal-tagged, full-length TnT3 exhibited similar subcellular localization in C2C12 cells (Fig. S3). These data indicate that TnT3 is mainly localized in the nucleus when overexpressed in cells in the absence of myofilaments, and the nuclear localization signal resides in the TnT3 COOH-terminal region.
Nuclear TnT3 associates with dense nucleolar structures
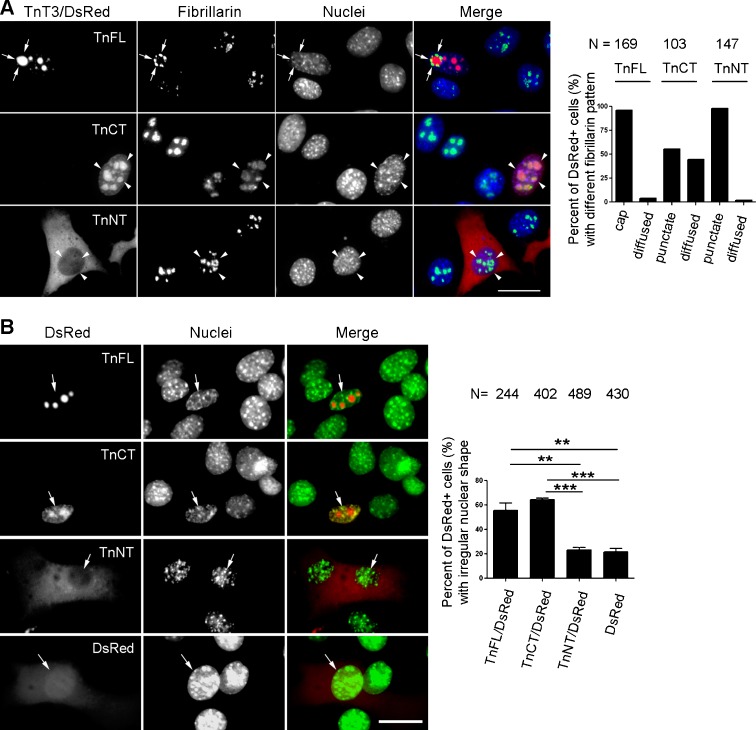
To further examine the subnuclear localization of TnT3, we performed immunofluorescence staining using an antibody against the nucleolar protein fibrillarin, a dense nucleolar marker. TnCT/DsRed co-localized with fibrillarin in large puncta, while TnFL/DsRed was surrounded mainly with fibrillarin “caps” (Fig. 2a). TnNT/DsRed, TnM, and control DsRed did not have any effect on the fibrillarin staining pattern (data not shown). The NH2-terminal region of TnT3 seems to affect TnCT’s subnuclear localization or its relationship with fibrillarin.
Fig. 2.
Transiently overexpressed TnFL/DsRed or TnCT/DsRed co-localized with fibrillarin and affected C2C12 nuclear morphology. TnT3/DsRed constructs were transiently expressed in C2C12 in growth medium for 2 days. Cells were then fixed, immunostained with fibrillarin antibody (nucleolar marker), Hoechst 33342, and examined by immunofluorescence microscopy. a Both TnFL/DsRed and TnCT/DsRed localized in the nucleolar area, yet TnFL/DsRed gathered fibrillarin around it and formed the nucleolar “cap” (arrows). TnCT/DsRed both co-localized with fibrillarin puncta and exhibited a diffuse nucleoplasmic distribution (arrow heads). In contrast, TnNT/DsRed showed no obvious effect on fibrillarin distribution (arrow heads). Data shown are representative of two independent experiments. b TnFL/DsRed and TnCT/DsRed were the only two constructs found in the nucleus, which changed from smooth and round or elliptic to irregular. In contrast, TnNT/DsRed showed the least effect on nuclear morphology, like control DsRed (arrows indicating nuclei from red fluorescence-positive cells). Scale bars, 25 μm. The graph quantifies the nuclei with irregular morphology for TnFL/DsRed-, TnCT/DsRed-, TnNT/DsRed-, and DsRed-transfected cells. The number of irregular nuclei is greater in TnFL/DsRed- and TnCT/DsRed- than TnNT/DsRed- and control DsRed-transfected cells (***P ≤ 0.001, n = 3; **P < 0.01, n = 3)
Since fibrillarin-depleted cells showed abnormal nuclear morphology (Amin et al. 2007), we used reported parameters to analyze the effects of TnT constructs on nuclear morphology and found that TnFL/DsRed and TnCT/DsRed, which both show close spatial association with fibrillarin, had the strongest effects on changes from normal (round or ellipse with smooth edge) to abnormal (indented, curved with uneven edge) when transiently overexpressed in C2C12 cells (Fig. 2b).
TnT3 is associated with RNA polymerase I and II and their related transcription activity
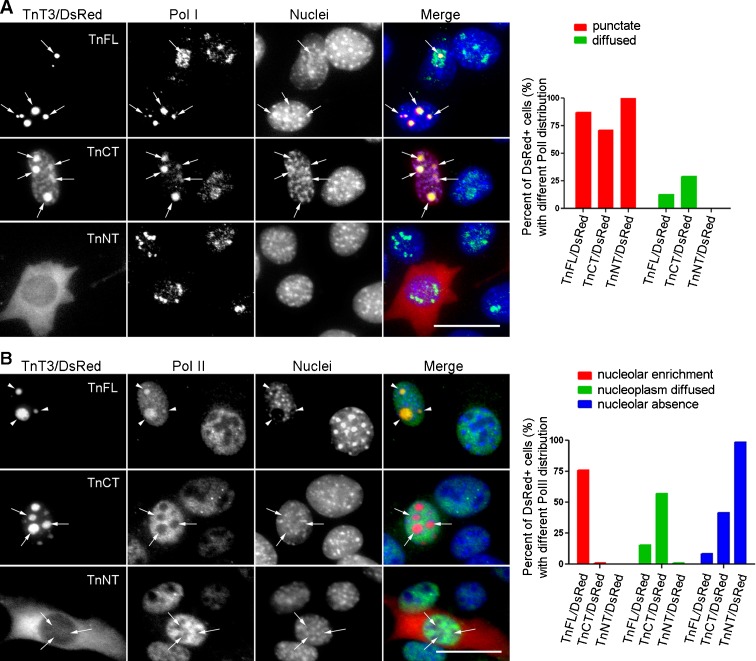
The nucleoli are the major nuclear structures where RNA polymerase I synthesizes ribosomal RNA (Sirri et al. 2008). We further stained C2C12 cells transfected with TnT3 constructs with either RNA Pol I antibody (PRA194(C-1)) or RNA Pol II antibody (8WG16). Pol I staining co-localized with both TnFL/DsRed and TnCT/DsRed in the nucleolar area, but interestingly, only TnFL/DsRed recruited Pol II, which normally localizes in the nucleoplasm, to the nucleolar area. As a control, TnNT/DsRed did not affect either Pol I or Pol II subnuclear localization (Fig. 3).
Fig. 3.
TnFL/DsRed and TnCT/DsRed showed different relationships with RNA polymerase I and II in C2C12 cells. C2C12 cells transiently expressing TnFL/DsRed, TnCT/DsRed, or TnNT/DsRed were immunostained with RNA polymerase I antibody (PRA194 [C-1], Pol I) or RNA polymerase II antibody (8WG16, Pol II), respectively. a Both TnFL/DsRed and TnCT/DsRed staining co-localized with Pol I in the nucleolar area (arrows). Unlike TnFL/DsRed, which showed mainly punctate nuclear distribution, TnCT/DsRed co-localized with Pol I in puncta and exhibited an additional diffused nucleoplasmic distribution. In contrast, TnNT/DsRed did not show any effect on RNA Pol I distribution. Data are representative of two independent experiments, and the numbers of cells analyzed are 94, 114, and 178 for TnFL/DsRed-, TnCT/DsRed-, and TnNT/DsRed-transfected cells, respectively. b TnFL/DsRed had a much stronger effect on Pol II enrichment in the nucleolar area (arrow heads). In contrast, most TnCT/DsRed or TnNT/DsRed showed no Pol II in the nucleolar area (arrows). Data are representative of two independent experiments, and the numbers of cells analyzed are 116, 110, and 175 for TnFL/DsRed-, TnCT/DsRed-, and TnNT/DsRed-transfected cells, respectively. Scale bars, 25 μm
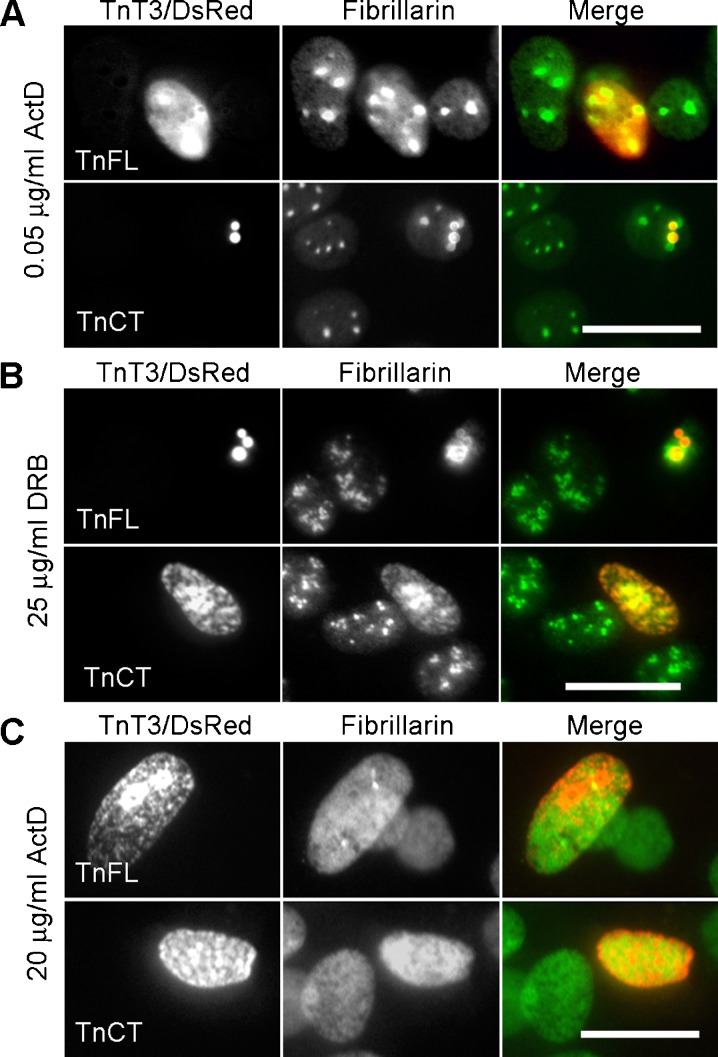
To study the role of TnT3 in RNA processing, we performed transcription inhibition experiments in C2C12 cells transfected with TnFL/DsRed or TnCT/DsRed. A low ActD concentration (0.05 μg/ml), which specifically inhibits Pol I, resulted in TnFL/DsRed redistribution into the nucleoplasm, while nucleoplasmic TnCT/DsRed disappeared, leaving only nucleolar puncta (Fig. 4a). The Pol II-specific inhibitor DRB (25 μg/ml) showed effects mainly on TnCT/DsRed redistribution (Fig. 4b). When both Pol I and Pol II were inhibited with a higher ActD concentration (20 μg/ml), fibrillarin puncta disappeared almost completely and redistributed into the nucleoplasm. As a result, both TnFL/DsRed and TnCT/DsRed were also redistributed into the nucleoplasm (Fig. 4c). This transcription inhibition experiment and Pol I and/or Pol II co-localization with TnT3 constructs suggest a role for TnT3 in RNA biogenesis.
Fig. 4.
Transcription inhibition with actinomycin D (ActD) or DRB in TnFL/DsRed or TnCT/DsRed transiently transfected C2C12 cells. a Incubating transfected cells in 0.05 μg/ml ActD for 3 h did not seem to affect fibrillarin distribution, yet TnFL/DsRed was dispersed into the nucleoplasm. In contrast, TnCT/DsRed remained punctate and co-localized with some fibrillarin puncta. Notably, the weak diffusive distribution of TnCT/DsRed in the nucleoplasm disappeared. b When only RNA Pol II was inhibited with 25 μg/ml DRB for 3 h, fibrillarin adopted a large punctate pattern in both TnFL/DsRed- and TnCT/DsRed-transfected cells. In contrast, TnCT/DsRed showed a diffuse nucleoplasmic distribution while TnFL/DsRed remained mainly punctate. c RNA Pol I and Pol II inhibition by 20 μg/ml ActD for 3 h resulted in diffuse fibrillarin dispersion into the nucleoplasm. Consistently, both TnFL/DsRed and TnCT/DsRed showed mainly a diffuse nuclear distribution pattern. Data are representative of two independent experiments, and at least 50 cells were analyzed per group. Scale bars, 25 μm
Subcellular localization of TnT3/DsRed fusion proteins in mouse FDB fibers
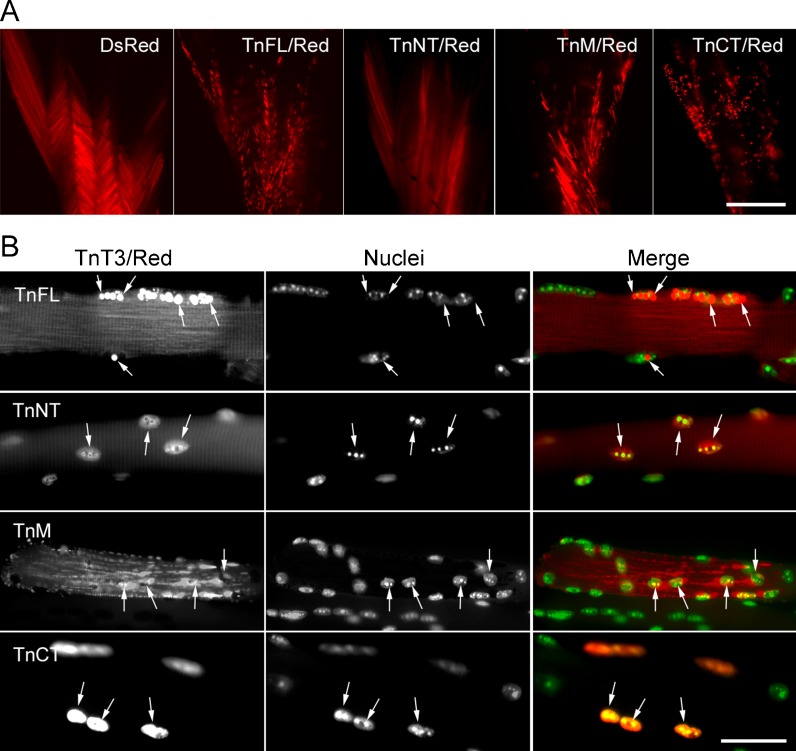
To examine whether TnT3/DsRed fusion constructs show subcellular distribution patterns similar to those observed in the C2C12 cells in skeletal muscle fiber in vivo, we performed in vivo electroporation experiments in mouse FDB muscle. As early as 3 days after electroporation, we observed expression of all constructs. When dissected, whole FDB muscles were observed at low magnification (×4 objective), the TnNT/DsRed and control DsRed showed a similar diffuse distribution throughout the muscle, while the TnFL/DsRed, TnM/DsRed, and TnCT/DsRed showed mainly a punctate distribution in addition to some large patches and diffuse background. The TnFL/DsRed and particularly the TnCT/DsRed construct showed much finer puncta, reflecting possible nuclear localization (Fig. 5a).
Fig. 5.
Subcellular localization of TnT3/DsRed proteins in mouse FDB fibers. TnT3/DsRed constructs and control plasmids were electroporated in vivo into the mouse FDB muscle. a Expression pattern of all constructs in whole isolated muscle. Control DsRed and TnNT/DsRed were expressed evenly throughout the muscle. In contrast, TnFL/DsRed, TnM/DsRed, and TnCT/DsRed showed a punctate distribution. b Higher magnification imaging analysis of individual myofibers showed that both TnFL/DsRed and TnCT/DsRed localized in some myonuclei. Additionally, TnFL/DsRed showed weak striated pattern. Notably, these two constructs localized at different subnuclear domains as revealed by their different co-localization pattern with Hoechst 33342 DNA staining. TnNT/DsRed also expressed in the myonuclei, but the myonuclear DNA staining pattern was not affected. TnM/DsRed mainly localized in the cytoplasm as small puncta, while most of it was highly ordered and reflects binding to myofibrils. Arrows point to nuclei in red fluorescence-positive areas. Scale bars, 800 μm (a), 50 μm (b). Fibers are representative of at least 200 fibers in each group from 2 to 13 experiments (DsRed, n = 3; TnFL/DsRed, n = 5; TnNT/DsRed, n = 6; TnM/DsRed, n = 2; TnCT/DsRed, n = 13)
We then dissected single FDB muscle fibers to observe this subnuclear localization in more detail. Isolated fibers were stained with Hoechst 33342 to label DNA, which allowed us to follow the nuclear localization of various TnT3/DsRed constructs (Fig. 5b). At high magnification (×40 objective), TnFL/DsRed and TnCT/DsRed localized in some myonuclei. Notably, TnFL/DsRed localized mainly between the strongest DNA-stained puncta (heterochromatin), while TnCT/DsRed mainly co-localized with the DNA stain. Also, TnFL/DsRed showed a striated pattern throughout the fiber, where fluorescence was dimmer than in the nucleus. In contrast, TnCT/DsRed localized only in the myonuclei (Figs. 5b and S4).
TnNT/DsRed was also expressed throughout the fiber, including the myonuclei, yet the myonuclear DNA staining pattern was not affected. TnM/DsRed mainly localized in the cytoplasm and showed a punctate striation pattern, reflecting its binding to the myofiber thin filaments. Like TnNT/DsRed, control DsRed expression was distributed throughout the fiber (Fig. S5).
Endogenous TnT3 and its fragments change expression pattern with aging
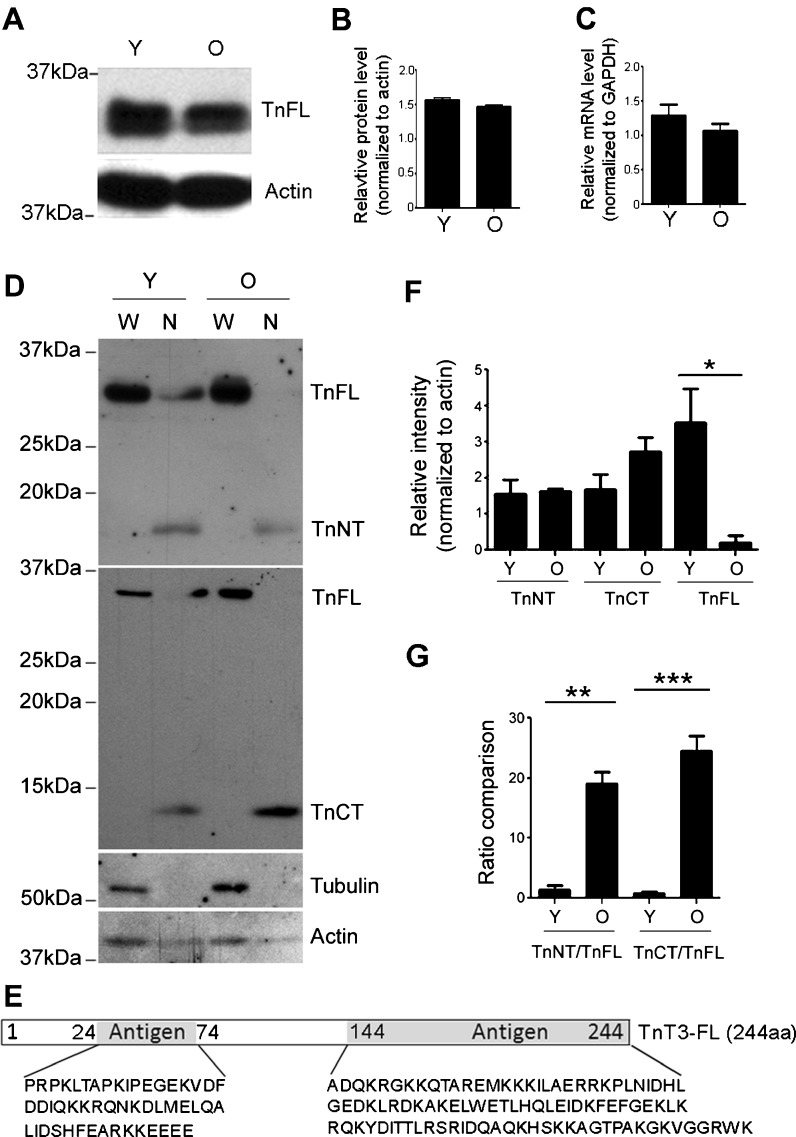
To examine age-related changes in endogenous TnT3 expression and nuclear localization, we collected the whole cell lysis as well as the nuclear fractions from young (3-month) and old (26–28-month) FVB mouse TA muscle for immunoblotting. We found slightly decreased TnFL expression in the old compared to the young group (Fig. 6a, b). Consistently, TnT3 mRNA also decreased slightly with aging, as measured by quantitative real-time PCR (Fig. 6c). Whole cell lysis and nuclear fractions from young and old mice were further analyzed by immunobloting with TnT3, α-tubulin, and pan-actin antibodies. As expected, α-tubulin was found mainly in the whole cell lysis pool, while actin was in both fractions. Interestingly, besides full-length TnT3 (TnFL, 35 kDa), two smaller bands of 18 and 13 kDa were detected in the nuclear fractions. These bands were specific to NH2-terminal and the COOH-terminal epitope-targeted antibodies, respectively, and thus likely represent endogenous NH2-terminal and COOH-terminal fragments of full-length TnT3 (Fig. 6d, e).
Fig. 6.
Western blot analysis of TnT3 and its fragments in young (3-month) and old (26–28-month) FVB mouse skeletal muscle. a TnT3 in TA whole muscle lysate, detected by a C-terminal-targeting antibody, shows decreased TnFL in old (O) compared to young (Y) mice. b TnFL expression was normalized to actin. c qPCR showing a slight decrease in TnT3, normalized to GADPH, mRNA in old (n = 5) compared to young (n = 6) mice. d Whole cell lysis (W) and nuclear protein fractions (N) from young and old mice were analyzed using TnT3 NT and CT antibodies. (e) The specific TnT3 N- and C-terminal regions targeted by the antibodies. f Normalized (against actin) TnNT, TnCT, and TnFL in the nuclear extracts were compared between young and old groups. TnCT tended to increase in the old nuclear extracts, and TnFL decreased dramatically (*P < 0.05, n = 3). TnNT did not change significantly. g The nuclear ratio between TnNT or TnCT and TnFL in both age groups was compared (**P = 0.012; ***P < 0.001, n = 3)
We compared the age groups for the relative amount (normalized to actin) of TnNT, TnCT, and TnFL in the nuclear extracts. While TnNT did not differ, TnCT was 50% greater while TnFL was dramatically decreased in the old group (P < 0.05, n = 3) (Fig. 6d, f). The nuclear ratio between TnNT or TnCT and TnFL increased significantly in the old group (P = 0.012 and P < 0.001, respectively, n = 3) (Fig. 6d, g).
Expression of TnT3/DsRed proteins in mouse skeletal muscle in vivo affects myonuclear morphology and DNA staining pattern
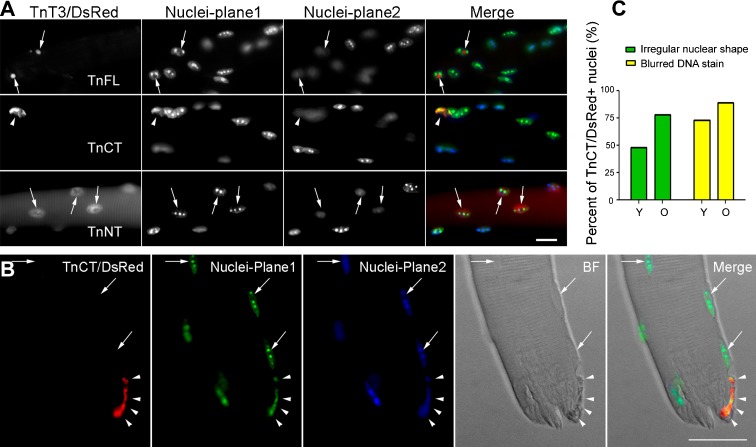
As TnFL/DsRed and TnCT/DsRed affect the shape of C2C12 nuclei and associate with condensed DNA in myonuclei differentially, we compared and quantified their effects on myonuclear shape and DNA staining pattern in muscle fibers electroporated in vivo. TnFL/DsRed-expressing fibers from young mice exhibited irregular nuclei 1 week after electroporation but showed no co-localization with the strongest DNA puncta. In contrast, myonuclei in TnCT/DsRed-transfected fibers were indented and fractured, and the DNA staining more diffuse than in untransfected neighboring myonuclei. As a control, TnNT/DsRed affected neither myonuclear shape nor the DNA staining pattern (Fig. 7a). Based on our finding that the endogenous nuclear fraction of TnFL decreased dramatically with aging, while TnCT increased, we compared the effect of TnCT/DsRed on the myonuclei from young and old mice. TnCT/DsRed-transfected fibers from old mice showed elongated, broken myonuclei, with a general loss of condensed DNA staining (heterochromatin) (Fig. 7b). The effect of TnCT/DsRed on nuclei shape and DNA pattern was greater in old fibers than young fibers (Fig. 7c).
Fig. 7.
Transient expression of TnT3/DsRed proteins in mouse skeletal muscle in vivo affects myonuclear phenotype. a TnFL, TnCT, and TnNT nuclear expression in young FDB muscle fibers. Nuclei staining pattern was examined at two planes. Arrows indicate nuclei DNA staining in a clear, sharp punctate pattern; arrow heads indicate a blurred DNA staining pattern in the TnCT/DsRed-positive nucleus. b TnCT/DsRed expression in FDB muscle fibers from old FVB mice. Under the brightfield (BF) view, the TnCT/DsRed-positive nucleus showed abnormal shape and DNA stain (arrow heads) compared to neighboring normal untransfected nuclei (arrows). c Effects of TnCT/DsRed on nuclear shape and DNA staining pattern in young (Y, 271 nuclei) and old (O, 156 nuclei) muscle fibers from two experiments. Scale bars, 25 μm (a), 100 μm (b)
Overexpressing TnT3/DsRed fusion proteins results in apoptosis
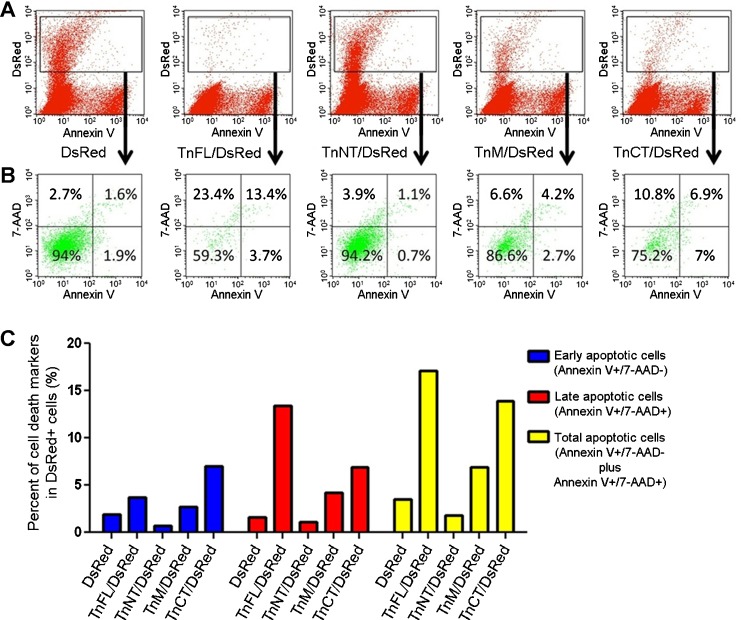
To verify whether TnT3 constructs had toxic effects, we performed Annexin V and 7-AAD staining to detect early and late apoptotic cells (Gil et al. 2011). The flow cytometry data (Fig. 8) demonstrate that TnNT/DsRed did not increase early or late apoptosis in the DsRed-positive cells compared to DsRed-transfected control cells. However, early and late apoptosis increased dramatically in cells expressing TnFL/DsRed or TnCT/DsRed. TnM/DsRed also showed a weaker toxic effect than TnFL/DsRed or TnCT/DsRed cells. The strong cell toxicity of both TnFL/DsRed and TnCT/DsRed was also confirmed using the same flow cytometry assay to test the NIH3T3 fibroblast cultures (Fig. S6).
Fig. 8.
TnCT/DsRed and TnFL/DsRed overexpression induces apoptosis in C2C12 cells. Cells were analyzed with a FACSCalibur flow cytometer 48 h posttransfection. Data were collected on at least 100,000 freshly stained cells. Representative analyses of 7-AAD and Annexin V staining (b) followed pregating on DsRed (a). b Results were plotted as fluorescence intensity of Annexin V as a function of fluorescence intensity of 7-AAD. The numbers in each square represent the percentage of Annexin V−/7-AAD−(viable cells, left bottom corner); Annexin V+/7-AAD−(early apoptotic cells, right bottom corner); Annexin V+/7-AAD+(late apoptotic cells, right top corner); and Annexin V−/7-AAD+(broken cells, left top corner) in the DsRed-positive cell population. Data shown are representative of two independent experiments. c The percent of total apoptotic cells was obtained by adding early (Annexin V+/7-AAD−) and late apoptotic cells (Annexin V+/7-AAD+)
Discussion
TnT is well known to mediate the interaction between the Tn complex and Tm, which is essential for the contraction of calcium-activated striated muscle (Perry 1998). This regulatory function takes place in the myoplasm, where TnT binds Tm. Recent studies support alternative roles for TnI. During Drosophila development, TnI and Tm were found in the nucleus and to regulate chromosomal integrity (Sahota et al. 2009). Additionally, fast skeletal muscle TnI may also localize in the mammalian cell nucleus and work as a co-activator of the nuclear receptor-estrogen receptor-related alpha (Li et al. 2008).
Here, we report the novel finding that TnT—specifically, fast skeletal muscle TnT3—localizes in the nucleus of myofibers as an intact or fragmented protein. The relationship with nucleolar structures and RNA polymerases implies a role for TnT3 in RNA transcription regulation. Their cytotoxic effects, their relative abundance in young and old skeletal muscle myonuclei, and differences between intact TnT3 and the COOH-terminal fragment in subnuclear localization all support the idea that TnT3 plays a surprising and critical role in aging skeletal muscle and justify further studies.
Evidence for TnT3 nuclear localization
TnFL/DsRed showed a striated distribution in the sarcoplasm and/or the nucleus of mouse FDB myofibers. The competition between expressed and endogenous TnT3 may explain the weak striated pattern. In addition, several lines of evidence support nuclear localization of TnT3. First, in the western blot assay of muscle nuclear protein fractions, two specific antibodies, targeting either the TnT3 NH2- or COOH-terminal epitope, detected endogenous TnT3. These antibodies detected two TnT3 fragments with different molecular weights that supposedly contain the domain carrying either the NH2-terminal or COOH-terminal epitope, respectively. Second, overexpressing full-length TnT3 or fragments with COOH-terminally tagged DsRed showed distinct subcellular localization. Not only did the overexpressed TnT3 localize in the nuclei of undifferentiated C2C12 myoblasts or other non-muscle cells (NIH3T3 fibroblasts) but also in myonuclei in vivo. Similar subcellular localization observed with GFP NH2-terminally tagged to TnT3 in C2C12 cells (Fig. S3) deters the possibility that TnT3 nuclear localization is an artifact resulting from the expression of the fusion protein or the tag itself. In addition, truncation analysis indicates that the nuclear localization signal resides in the COOH-terminal region. Third, fibrillarin immunofluorescence staining showed slight differences between the subnuclear localization of full-length TnT3 or its COOH-terminal fragment and that of the nucleolus, consistent with reports that the NH2-terminal region may affect the COOH-terminal region’s function (Wei and Jin 2011) or protein conformation. The fusion TnT3 protein had much higher level in the nucleus than that incorporated into the myofibrils; therefore, the structural manipulation might have altered the functional behavior of TnT3. This observation may implicate that misfolded or damaged TnT3 would have increased tendency to translocate to the nucleus. Although we have not tested the hypothesis that the COOH-terminal region meditates TnT3 nuclear translocation through interaction with TnI and Tm, they have been reported in the nucleus (Li et al. 2008; Sahota et al. 2009). Conversely, TnT may regulate TnI and Tm nuclear translocation. Considering the high degree of conservation in this region across species (Jin et al. 2008), future work should determine the nuclear localization signal for other TnT isoforms.
TnT3 subnuclear localization and myonuclear apoptosis and/or dysfunction
Effects on cell apoptosis vary among the TnT3 regions when expressed in C2C12 myoblasts or NIH3T3 fibroblasts, consistent with a recent finding from the Jin group on cytotoxicity of the non-myofilament-associated TnT fragments, including the NH2-terminal variable region from human cardiac TnT and the highly conserved middle and COOH-terminal regions from the mouse slow TnT (Jeong et al. 2009). We generated a TnT3 fragment with a molecular weight similar to that detected by the TnT3 COOH-terminal epitope antibody by subcloning the last 85 amino acids of the COOH-terminal region. Although our COOH-terminal region is 25 amino acids shorter and the middle region 13 amino acids shorter than that studied by the Jin group, sequence comparison indicated that the middle and COOH-terminal regions we chose are highly conserved among TnT isoforms, possibly explaining the similar toxicity of these TnT constructs. Our findings confirm that middle and COOH-terminal regions are not only conserved in the peptide sequence but also in their function (apoptosis). Note that the DsRed conjugation enabled us to analyze TnT3 subnuclear localization and to elucidate the mechanism involved in cytotoxicity, which is probably mediated by nuclear translocation and nucleoli targeting.
The nucleolus is a large nuclear domain with a contrasted structure directly involved in ribosome biosynthesis (Sirri et al. 2008). Its function is tightly linked to cell growth, proliferation, and other important cell events, including cell cycle regulation, senescence, and stress responses (Visintin and Amon 2000; Guarente 1997; Kennedy et al. 1997; Sherr and Weber 2000; Olson 2004). Changes in nucleolar organization are widely thought to represent diagnostic markers for different pathologies (Zimber et al. 2004). A recent proteomic study identified hundreds of human nucleolar proteins (Andersen et al. 2005), among them, fibrillarin, which is known to play an important role in pre-rRNA processing during ribosomal biogenesis. Knocking down fibrillarin in mammalian cells has been reported to lead to abnormal nuclear morphology and inhibition of cell proliferation (Amin et al. 2007). TnT3/DsRed or TnCT/DsRed transfection alters fibrillarin location, which may lead to abnormal nuclear shape and, consequently, cell toxicity. Nuclear morphology has been shown to influence fundamental aspects of nuclear function, including chromatin remodeling and gene transcription (Dalby et al. 2007; Itano et al. 2003; Lammerding et al. 2004; Thomas et al. 2002). Changes in myonuclear morphology, including shrinkage, fracture, and fragmentation, indicate cell death. Our flow cytometry results in C2C12 myoblasts, and NIH3T3 fibroblasts further support this link between apoptosis and altered nuclear morphology. Further investigation of TnT3’s downstream partners will clarify the mechanism that activates the cell death pathway.
TnT3 nuclear imbalance and muscle aging
Myonuclear apoptosis and structural changes related to impaired RNA processing occur during muscle aging as reported for sarcopenia (Malatesta et al. 2009, 2010; Alway and Siu 2008; Dirks and Leeuwenburgh 2002). Although apoptotic nuclei have been documented in skeletal myofibers, the precise signaling mechanisms leading to nuclear loss are not fully understood. Sarcopenia-related apoptosis has been shown to be regulated differently in fast- and slow-twitch muscles; type II fast muscle is more affected and exhibits fewer satellite cells (Rice and Blough 2006; Verdijk et al. 2007). Our findings on TnT3 nuclear localization and cytotoxicity provide new insight into this preferential type II fast fiber atrophy/loss with aging. TnT3 is a muscle-specific fast isoform, and its expression has been related to the fast myosin isoforms at the single fiber level (Brotto et al. 2006). Our findings that nuclear TnT3 COOH-terminal fragment expression increases with aging and that this fragment has several negative effects to the cell offer a plausible link to sarcopenia.
We have found that the nuclear full-length TnT3 decreases while its fragments increase with aging, thus increasing the ratio between TnT3 fragments, in particular the COOH-terminal fragments. We hypothesize that this imbalance plays a physiological role in muscle nuclei. Muscle electroporation was relatively inefficient in inducing myonuclear expression, so calculating the number of lost nuclei and whether muscle force was affected was difficult. The differential consequences of transfecting full-length TnT3 or the COOH-terminal region construct may be explained by their relationship with the nucleolus and different nucleolar proteins, including fibrillarin and RNA polymerases I and II. RNA Pol I subunits are enriched in the nucleolar FC region, and RNA Pol II activity is largely absent from this area and localizes in the nucleoplasm (Hernandez-Verdun 2006; Boisvert et al. 2007; Sirri et al. 2008). Our study found TnT3/DsRed mainly surrounded by the endogenous fibrillarin (similar to Pol I), which formed the “nucleolar cap” structure (Fox and Lamond 2010; Shav-Tal et al. 2005) and recruited endogenous Pol II to the nucleolar area. In contrast, the TnCT/DsRed mainly co-localized with fibrillarin and Pol I, but not Pol II, in the nucleolar area, with some remaining in the nucleoplasm (as Pol I does), where the Pol II is mainly distributed. Transcription inhibition with ActD or DRB demonstrated that TnT3 and the COOH-terminal fragments are associated with RNA transcription activities (van Koningsbruggen et al. 2004) and interact with Pol I and Pol II differentially. These differences in subnuclear localization are also reflected by the DNA staining pattern: TnT3/DsRed clearly localizes between the condensed heterochromatin DNA stain, while TnCT/DsRed showed less sharp staining (Figs. 5b and 7a). Theoretically, therefore, the fragments function differently, considering that most subnuclear bodies, including Cajal and PML bodies and nuclear speckles, reside in the interchromatin space (Lamond and Spector 2003). With the reported modifying effects of the TnT NH2-terminal region on middle and COOH-terminal region functions (Wei and Jin 2011), further identification of nuclear partners that may bind to full-length TnT3 or the COOH-terminal region will elucidate their functional differences.
In conclusion, fast skeletal muscle TnT3 is localized in the nucleus and is found predominantly at full length in the young, while its fragments predominate in the old, which may lead to aging-related myonuclear death or dysfunction. In addition to its potential clinical use as a diagnostic marker of muscle damage, TnT may be a therapeutic target to prevent muscle aging and damage. In either case, blocking TnT fragmentation and/or nuclear translocation will be considered in future work focusing on the regulatory mechanism of TnT3 fragmentation and the role of the proteases—calpains and caspases, both reported to cleave different TnTs (Dargelos et al. 2007, 2008; Wei and Jin 2011). Since the COOH-terminal region is highly conserved among isoforms and across species, further work should also compare the cardiac and slow muscle isoforms.
Electronic supplementary material
Construction of TnT3/DsRed cDNAs by PCR and subcloning. Primers targeting different regions (arrows) of the mouse TnT3 cDNA were designed with HindIII or SacII restriction enzyme cutting sequences overhanging at the 5′ end of each primer. PCR products were subcloned into pDsRed2-N1 multiple cloning sites (MCS) after HindIII and SacII digestion to finally construct TnFL/DsRed, TnNT/DsRed, TnM/DsRed, and TnCT/DsRed, respectively (TIFF 5576 kb)
Flowchart representing sample preparation for whole cell lysis, cytosolic, myofibrillar, and nuclear fractions from mouse TA skeletal muscle. Fractions were subsequently analyzed by western blotting (Fig. 1a) to confirm the successful separation of nuclear fraction from myofibrillar fraction and the presence of endogenous nuclear TnT3. Trichloroacetic acid (TCA) was used to precipitate and concentrate the total protein from W, C, and N fractions (TIFF 1245 kb)
Subcellular localization of N-terminally GFP-tagged TnT3 in C2C12 cells. When transiently overexpressed in the C2C12 myoblasts, GFP/TnFL shows similar nuclear localization to that of TnFL/DsRed. Scale bar, 20 μm (TIFF 1503 kb)
TnFL/DsRed, but not TnCT/DsRed, showed a striated distribution pattern in addition to its myonuclear localization when transiently overexpressed in myofibers. FVB fibers transiently transfected with TnFL/DsRed or TnCT/DsRed were isolated and imaged under fluorescence microscope. By overexposure, with the nuclear DsRed saturated, only the TnFL/DsRed transfected fibers showed a striated fluorescent pattern. Scale bar, 50 μm (TIFF 6511 kb)
DsRed overexpression in mouse FDB muscle fiber. Control DsRed protein diffuses throughout the muscle fiber. Scale bar, 50 μm (TIFF 4968 kb)
TnCT/DsRed and TnFL/DsRed overexpression induces apoptosis in NIH3T3 fibroblasts. As with C2C12, NIH3T3 cells were analyzed with a FACSCalibur flow cytometer 48 h posttransfection. Data were collected on at least 100,000 freshly stained cells. Representative analyses of 7-AAD and Annexin V staining (B) followed pregating on DsRed (A). B Results were plotted as fluorescence intensity of Annexin V as a function of fluorescence intensity of 7-AAD. The numbers in each square are explained for Fig. 8. C The percent of total apoptotic cells was obtained by adding early and late apoptotic cells (TIFF 6362 kb)
Acknowledgments
This study was supported by grants from the National Institutes of Health/National Institute on Aging (AG13934, AG033385, AG15820, and FIRCA-BB TW008091) and the Muscular Dystrophy Association (MDA #33149) to Osvaldo Delbono and the Wake Forest Claude D. Pepper Older Americans Independence Center (P30-AG21332).
Abbreviations
- TnT3
Troponin T3
- Tm
Tropomyosin
- ActD
Actinomycin D
- Pol
Polymerase
- PML
Promyelocytic leukemia
References
- Alway SE, Siu PM. Nuclear apoptosis contributes to sarcopenia. Exerc Sport Sci Rev. 2008;36(2):51–57. doi: 10.1097/JES.0b013e318168e9dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MA, Matsunaga S, Ma N, Takata H, Yokoyama M, Uchiyama S, Fukui K. Fibrillarin, a nucleolar protein, is required for normal nuclear morphology and cellular growth in HeLa cells. Biochem Biophys Res Commun. 2007;360(2):320–326. doi: 10.1016/j.bbrc.2007.06.092. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433(7021):77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Barbieri M, Ferrucci L, Ragno E, Corsi A, Bandinelli S, Bonafe M, Olivieri F, Giovagnetti S, Franceschi C, Guralnik JM, Paolisso G. Chronic inflammation and the effect of IGF-I on muscle strength and power in older persons. Am J Physiol Endocrinol Metab. 2003;284(3):E481–E487. doi: 10.1152/ajpendo.00319.2002. [DOI] [PubMed] [Google Scholar]
- Biesiadecki BJ, Jin JP. Exon skipping in cardiac troponin T of turkeys with inherited dilated cardiomyopathy. J Biol Chem. 2002;277(21):18459–18468. doi: 10.1074/jbc.M200788200. [DOI] [PubMed] [Google Scholar]
- Biesiadecki BJ, Chong SM, Nosek TM, Jin JP. Troponin T core structure and the regulatory NH2-terminal variable region. Biochemistry. 2007;46(5):1368–1379. doi: 10.1021/bi061949m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbrair A, Wang ZM, Messi ML, Enikolopov GN, Delbono O. Nestin-GFP transgene reveals neural precursor cells in adult skeletal muscle. PLoS One. 2011;6(2):e16816. doi: 10.1371/journal.pone.0016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8(7):574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Brooks SV, Faulkner JA. Maximum and sustained power of extensor digitorum longus muscles from young, adult, and old mice. J Gerontol. 1991;46(1):B28–B33. doi: 10.1093/geronj/46.1.B28. [DOI] [PubMed] [Google Scholar]
- Brotto MA, Biesiadecki BJ, Brotto LS, Nosek TM, Jin JP. Coupled expression of troponin T and troponin I isoforms in single skeletal muscle fibers correlates with contractility. Am J Physiol Cell Physiol. 2006;290(2):C567–C576. doi: 10.1152/ajpcell.00422.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buford TW, Anton SD, Judge AR, Marzetti E, Wohlgemuth SE, Carter CS, Leeuwenburgh C, Pahor M, Manini TM. Models of accelerated sarcopenia: critical pieces for solving the puzzle of age-related muscle atrophy. Ageing Res Rev. 2010;9(4):369–383. doi: 10.1016/j.arr.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson ME, Conboy IM. Loss of stem cell regenerative capacity within aged niches. Aging Cell. 2007;6(3):371–382. doi: 10.1111/j.1474-9726.2007.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra M, Montgomery DE, Kim JJ, Solaro RJ. The N-terminal region of troponin T is essential for the maximal activation of rat cardiac myofilaments. J Mol Cell Cardiol. 1999;31(4):867–880. doi: 10.1006/jmcc.1999.0928. [DOI] [PubMed] [Google Scholar]
- Dalby MJ, Gadegaard N, Herzyk P, Sutherland D, Agheli H, Wilkinson CD, Curtis AS. Nanomechanotransduction and interphase nuclear organization influence on genomic control. J Cell Biochem. 2007;102(5):1234–1244. doi: 10.1002/jcb.21354. [DOI] [PubMed] [Google Scholar]
- Dargelos E, Brule C, Combaret L, Hadj-Sassi A, Dulong S, Poussard S, Cottin P. Involvement of the calcium-dependent proteolytic system in skeletal muscle aging. Exp Gerontol. 2007;42(11):1088–1098. doi: 10.1016/j.exger.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Dargelos E, Poussard S, Brule C, Daury L, Cottin P. Calcium-dependent proteolytic system and muscle dysfunctions: a possible role of calpains in sarcopenia. Biochimie. 2008;90(2):359–368. doi: 10.1016/j.biochi.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Day K, Shefer G, Shearer A, Yablonka-Reuveni Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev Biol. 2010;340(2):330–343. doi: 10.1016/j.ydbio.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degens H, Alway SE. Skeletal muscle function and hypertrophy are diminished in old age. Muscle Nerve. 2003;27(3):339–347. doi: 10.1002/mus.10314. [DOI] [PubMed] [Google Scholar]
- Degens H, Alway SE. Control of muscle size during disuse, disease, and aging. Int J Sports Med. 2006;27(2):94–99. doi: 10.1055/s-2005-837571. [DOI] [PubMed] [Google Scholar]
- Delbono O (2011) Expression and regulation of excitation–contraction coupling proteins in aging skeletal muscle. Curr Aging Sci (in press) [DOI] [PMC free article] [PubMed]
- DiFranco M, Neco P, Capote J, Meera P, Vergara JL. Quantitative evaluation of mammalian skeletal muscle as a heterologous protein expression system. Protein Expr Purif. 2006;47(1):281–288. doi: 10.1016/j.pep.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Dirks A, Leeuwenburgh C. Apoptosis in skeletal muscle with aging. Am J Physiol Regul Integr Comp Physiol. 2002;282(2):R519–R527. doi: 10.1152/ajpregu.00458.2001. [DOI] [PubMed] [Google Scholar]
- Dousset T, Wang C, Verheggen C, Chen D, Hernandez-Verdun D, Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell. 2000;11(8):2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng HZ, Biesiadecki BJ, Yu ZB, Hossain MM, Jin JP. Restricted N-terminal truncation of cardiac troponin T: a novel mechanism for functional adaptation to energetic crisis. J Physiol. 2008;586(14):3537–3550. doi: 10.1113/jphysiol.2008.153577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2(7):a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC. Structural mechanism of muscle contraction. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- Gil C, Falques A, Sarro E, Cubi R, Blasi J, Aguilera J, Itarte E. Protein kinase CK2 associates to lipid rafts and its pharmacological inhibition enhances neurotransmitter release. FEBS Lett. 2011;585(2):414–420. doi: 10.1016/j.febslet.2010.12.029. [DOI] [PubMed] [Google Scholar]
- Gomes AV, Guzman G, Zhao J, Potter JD. Cardiac troponin T isoforms affect the Ca2+ sensitivity and inhibition of force development. Insights into the role of troponin T isoforms in the heart. J Biol Chem. 2002;277(38):35341–35349. doi: 10.1074/jbc.M204118200. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Delbono O. Age-dependent fatigue in single intact fast- and slow fibers from mouse EDL and soleus skeletal muscles. Mech Ageing Dev. 2001;122(10):1019–1032. doi: 10.1016/S0047-6374(01)00229-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Messi ML, Delbono O. The specific force of single intact extensor digitorum longus and soleus mouse muscle fibers declines with aging. J Membr Biol. 2000;178(3):175–183. doi: 10.1007/s002320010025. [DOI] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80(2):853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Guarente L. Link between aging and the nucleolus. Genes Dev. 1997;11(19):2449–2455. doi: 10.1101/gad.11.19.2449. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem Cell Biol. 2006;125(1–2):127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- Itano N, Okamoto S, Zhang D, Lipton SA, Ruoslahti E. Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus. Proc Natl Acad Sci USA. 2003;100(9):5181–5186. doi: 10.1073/pnas.0531397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong EM, Wang X, Xu K, Hossain MM, Jin JP. Nonmyofilament-associated troponin T fragments induce apoptosis. Am J Physiol Heart Circ Physiol. 2009;297(1):H283–H292. doi: 10.1152/ajpheart.01200.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Moreno R, Wang ZM, Gerring RC, Delbono O. Sarcoplasmic reticulum Ca2+ release declines in muscle fibers from aging mice. Biophys J. 2008;94(8):3178–3188. doi: 10.1529/biophysj.107.118786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Moreno R, Wang ZM, Messi ML, Delbono O. Sarcoplasmic reticulum Ca2+ depletion in adult skeletal muscle fibres measured with the biosensor D1ER. Pflugers Arch. 2010;459(5):725–735. doi: 10.1007/s00424-009-0778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JP, Root DD. Modulation of troponin T molecular conformation and flexibility by metal ion binding to the NH2-terminal variable region. Biochemistry. 2000;39(38):11702–11713. doi: 10.1021/bi9927437. [DOI] [PubMed] [Google Scholar]
- Jin JP, Zhang Z, Bautista JA. Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit Rev Eukaryot Gene Expr. 2008;18(2):93–124. doi: 10.1615/CritRevEukarGeneExpr.v18.i2.10. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89(3):381–391. doi: 10.1016/S0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- Klitgaard H, Mantoni M, Schiaffino S, Ausoni S, Gorza L, Laurent-Winter C, Schnohr P, Saltin B. Function, morphology and protein expression of ageing skeletal muscle: a cross-sectional study of elderly men with different training backgrounds. Acta Physiol Scand. 1990;140(1):41–54. doi: 10.1111/j.1748-1716.1990.tb08974.x. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113(3):370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4(8):605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lannergren J, Westerblad H. The temperature dependence of isometric contractions of single, intact fibres dissected from a mouse foot muscle. J Physiol. 1987;390:285–293. doi: 10.1113/jphysiol.1987.sp016700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson L. Morphological and functional characteristics of the ageing skeletal muscle in man. A cross-sectional study. Acta Physiol Scand Suppl. 1978;457:1–36. doi: 10.1111/j.1748-1716.1978.tb06041.x. [DOI] [PubMed] [Google Scholar]
- Larsson L, Ansved T. Effects of ageing on the motor unit. Prog Neurobiol. 1995;45(5):397–458. doi: 10.1016/0301-0082(95)98601-Z. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen B, Chen J, Lou G, Chen S, Zhou D. Fast skeletal muscle troponin I is a co-activator of estrogen receptor-related receptor alpha. Biochem Biophys Res Commun. 2008;369(4):1034–1040. doi: 10.1016/j.bbrc.2008.02.147. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Perdoni F, Muller S, Zancanaro C, Pellicciari C. Nuclei of aged myofibres undergo structural and functional changes suggesting impairment in RNA processing. Eur J Histochem. 2009;53(2):97–106. doi: 10.4081/ejh.2009.e12. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Perdoni F, Muller S, Pellicciari C, Zancanaro C. Pre-mRNA processing is partially impaired in satellite cell nuclei from aged muscles. J Biomed Biotechnol. 2010;2010:410405. doi: 10.1155/2010/410405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E, Privitera G, Simili V, Wohlgemuth SE, Aulisa L, Pahor M, Leeuwenburgh C. Multiple pathways to the same end: mechanisms of myonuclear apoptosis in sarcopenia of aging. ScientificWorldJournal. 2010;10:340–349. doi: 10.1100/tsw.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse CI, Thom JM, Davis MG, Fox KR, Birch KM, Narici MV. Reduced plantarflexor specific torque in the elderly is associated with a lower activation capacity. Eur J Appl Physiol. 2004;92(1–2):219–226. doi: 10.1007/s00421-004-1056-y. [DOI] [PubMed] [Google Scholar]
- Morse CI, Thom JM, Reeves ND, Birch KM, Narici MV. In vivo physiological cross-sectional area and specific force are reduced in the gastrocnemius of elderly men. J Appl Physiol. 2005;99(3):1050–1055. doi: 10.1152/japplphysiol.01186.2004. [DOI] [PubMed] [Google Scholar]
- Narici MV, Maganaris CN. Adaptability of elderly human muscles and tendons to increased loading. J Anat. 2006;208(4):433–443. doi: 10.1111/j.1469-7580.2006.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogut O, Jin JP. Expression, zinc-affinity purification, and characterization of a novel metal-binding cluster in troponin T: metal-stabilized alpha-helical structure and effects of the NH2-terminal variable region on the conformation of intact troponin T and its association with tropomyosin. Biochemistry. 1996;35(51):16581–16590. doi: 10.1021/bi961712y. [DOI] [PubMed] [Google Scholar]
- Olson MO (2004) Sensing cellular stress: another new function for the nucleolus? Sci STKE 2004 (224):pe10 [DOI] [PubMed]
- Onambele GL, Narici MV, Maganaris CN. Calf muscle-tendon properties and postural balance in old age. J Appl Physiol. 2006;100(6):2048–2056. doi: 10.1152/japplphysiol.01442.2005. [DOI] [PubMed] [Google Scholar]
- Pan BS, Gordon AM, Potter JD. Deletion of the first 45 NH2-terminal residues of rabbit skeletal troponin T strengthens binding of troponin to immobilized tropomyosin. J Biol Chem. 1991;266(19):12432–12438. [PubMed] [Google Scholar]
- Payne AM, Zheng Z, Gonzalez E, Wang ZM, Messi ML, Delbono O. External Ca(2+)-dependent excitation–contraction coupling in a population of ageing mouse skeletal muscle fibres. J Physiol. 2004;560(Pt 1):137–155. doi: 10.1113/jphysiol.2004.067322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SV. Troponin T: genetics, properties and function. J Muscle Res Cell Motil. 1998;19(6):575–602. doi: 10.1023/A:1005397501968. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355(6362):730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Reiser PJ, Greaser ML, Moss RL. Developmental changes in troponin T isoform expression and tension production in chicken single skeletal muscle fibres. J Physiol. 1992;449:573–588. doi: 10.1113/jphysiol.1992.sp019102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renganathan M, Messi ML, Delbono O. Overexpression of IGF-1 exclusively in skeletal muscle prevents age-related decline in the number of dihydropyridine receptors. J Biol Chem. 1998;273(44):28845–28851. doi: 10.1074/jbc.273.44.28845. [DOI] [PubMed] [Google Scholar]
- Rice KM, Blough ER. Sarcopenia-related apoptosis is regulated differently in fast- and slow-twitch muscles of the aging F344/N×BN rat model. Mech Ageing Dev. 2006;127(8):670–679. doi: 10.1016/j.mad.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Runge M, Rittweger J, Russo CR, Schiessl H, Felsenberg D. Is muscle power output a key factor in the age-related decline in physical performance? A comparison of muscle cross section, chair-rising test and jumping power. Clin Physiol Funct Imaging. 2004;24(6):335–340. doi: 10.1111/j.1475-097X.2004.00567.x. [DOI] [PubMed] [Google Scholar]
- Sahota VK, Grau BF, Mansilla A, Ferrus A. Troponin I and tropomyosin regulate chromosomal stability and cell polarity. J Cell Sci. 2009;122(Pt 15):2623–2631. doi: 10.1242/jcs.050880. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16(5):2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Rauner G, Yablonka-Reuveni Z, Benayahu D. Reduced satellite cell numbers and myogenic capacity in aging can be alleviated by endurance exercise. PLoS One. 2010;5(10):e13307. doi: 10.1371/journal.pone.0013307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10(1):94–99. doi: 10.1016/S0959-437X(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129(1):13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu PM, Pistilli EE, Murlasits Z, Alway SE. Hindlimb unloading increases muscle content of cytosolic but not nuclear Id2 and p53 proteins in young adult and aged rats. J Appl Physiol. 2006;100(3):907–916. doi: 10.1152/japplphysiol.01012.2005. [DOI] [PubMed] [Google Scholar]
- Snijders T, Verdijk LB, van Loon LJ. The impact of sarcopenia and exercise training on skeletal muscle satellite cells. Ageing Res Rev. 2009;8(4):328–338. doi: 10.1016/j.arr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Szczesna D, Potter JD. The role of troponin in the Ca(2+)-regulation of skeletal muscle contraction. Results Probl Cell Differ. 2002;36:171–190. doi: 10.1007/978-3-540-46558-4_13. [DOI] [PubMed] [Google Scholar]
- Taylor JR, Zheng Z, Wang ZM, Payne AM, Messi ML, Delbono O. Increased CaVbeta1A expression with aging contributes to skeletal muscle weakness. Aging Cell. 2009;8(5):584–594. doi: 10.1111/j.1474-9726.2009.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2002;99(4):1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DR. Sarcopenia. Clin Geriatr Med. 2010;26(2):331–346. doi: 10.1016/j.cger.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- Tomlinson BE, Irving D, Rebeiz JJ. Total numbers of limb motor neurones in the human lumbosacral cord and an analysis of the accuracy of various sampling procedures. J Neurol Sci. 1973;20(3):313–327. doi: 10.1016/0022-510X(73)90193-7. [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S, Dirks RW, Mommaas AM, Onderwater JJ, Deidda G, Padberg GW, Frants RR, van der Maarel SM. FRG1P is localised in the nucleolus, Cajal bodies, and speckles. J Med Genet. 2004;41(4):e46. doi: 10.1136/jmg.2003.012781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdijk LB, Koopman R, Schaart G, Meijer K, Savelberg HH, van Loon LJ. Satellite cell content is specifically reduced in type II skeletal muscle fibers in the elderly. Am J Physiol Endocrinol Metab. 2007;292(1):E151–E157. doi: 10.1152/ajpendo.00278.2006. [DOI] [PubMed] [Google Scholar]
- Visintin R, Amon A. The nucleolus: the magician’s hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12(6):752. doi: 10.1016/S0955-0674(00)00165-4. [DOI] [PubMed] [Google Scholar]
- Wang J, Jin JP. Primary structure and developmental acidic to basic transition of 13 alternatively spliced mouse fast skeletal muscle troponin T isoforms. Gene. 1997;193(1):105–114. doi: 10.1016/S0378-1119(97)00100-5. [DOI] [PubMed] [Google Scholar]
- Wang J, Jin JP. Conformational modulation of troponin T by configuration of the NH2-terminal variable region and functional effects. Biochemistry. 1998;37(41):14519–14528. doi: 10.1021/bi9812322. [DOI] [PubMed] [Google Scholar]
- Wei B, Jin JP. Troponin T isoforms and posttranscriptional modifications: evolution, regulation and function. Arch Biochem Biophys. 2011;505(2):144–154. doi: 10.1016/j.abb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zaal KJ, Sheridan J, Mehta A, Gundersen GG, Ralston E. Microtubule plus-end binding protein EB1 is necessary for muscle cell differentiation, elongation and fusion. J Cell Sci. 2009;122(Pt 9):1401–1409. doi: 10.1242/jcs.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jin JP, Root DD. Binding of calcium ions to an avian flight muscle troponin T. Biochemistry. 2004;43(9):2645–2655. doi: 10.1021/bi035067o. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Biesiadecki BJ, Jin JP. Selective deletion of the NH2-terminal variable region of cardiac troponin T in ischemia reperfusion by myofibril-associated mu-calpain cleavage. Biochemistry. 2006;45(38):11681–11694. doi: 10.1021/bi060273s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal. 2004;16(10):1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Zinna EM, Yarasheski KE. Exercise treatment to counteract protein wasting of chronic diseases. Curr Opin Clin Nutr Metab Care. 2003;6(1):87–93. doi: 10.1097/00075197-200301000-00013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction of TnT3/DsRed cDNAs by PCR and subcloning. Primers targeting different regions (arrows) of the mouse TnT3 cDNA were designed with HindIII or SacII restriction enzyme cutting sequences overhanging at the 5′ end of each primer. PCR products were subcloned into pDsRed2-N1 multiple cloning sites (MCS) after HindIII and SacII digestion to finally construct TnFL/DsRed, TnNT/DsRed, TnM/DsRed, and TnCT/DsRed, respectively (TIFF 5576 kb)
Flowchart representing sample preparation for whole cell lysis, cytosolic, myofibrillar, and nuclear fractions from mouse TA skeletal muscle. Fractions were subsequently analyzed by western blotting (Fig. 1a) to confirm the successful separation of nuclear fraction from myofibrillar fraction and the presence of endogenous nuclear TnT3. Trichloroacetic acid (TCA) was used to precipitate and concentrate the total protein from W, C, and N fractions (TIFF 1245 kb)
Subcellular localization of N-terminally GFP-tagged TnT3 in C2C12 cells. When transiently overexpressed in the C2C12 myoblasts, GFP/TnFL shows similar nuclear localization to that of TnFL/DsRed. Scale bar, 20 μm (TIFF 1503 kb)
TnFL/DsRed, but not TnCT/DsRed, showed a striated distribution pattern in addition to its myonuclear localization when transiently overexpressed in myofibers. FVB fibers transiently transfected with TnFL/DsRed or TnCT/DsRed were isolated and imaged under fluorescence microscope. By overexposure, with the nuclear DsRed saturated, only the TnFL/DsRed transfected fibers showed a striated fluorescent pattern. Scale bar, 50 μm (TIFF 6511 kb)
DsRed overexpression in mouse FDB muscle fiber. Control DsRed protein diffuses throughout the muscle fiber. Scale bar, 50 μm (TIFF 4968 kb)
TnCT/DsRed and TnFL/DsRed overexpression induces apoptosis in NIH3T3 fibroblasts. As with C2C12, NIH3T3 cells were analyzed with a FACSCalibur flow cytometer 48 h posttransfection. Data were collected on at least 100,000 freshly stained cells. Representative analyses of 7-AAD and Annexin V staining (B) followed pregating on DsRed (A). B Results were plotted as fluorescence intensity of Annexin V as a function of fluorescence intensity of 7-AAD. The numbers in each square are explained for Fig. 8. C The percent of total apoptotic cells was obtained by adding early and late apoptotic cells (TIFF 6362 kb)