Abstract
Regulated by pH, membrane-anchored proteins E and M play a series of roles during dengue virus maturation and membrane fusion. Our atomic model of the whole virion from cryo electron microscopy at 3.5Å resolution reveals that in the mature virus at neutral extracellular pH, the N-terminal 20-amino acid segment of M (involving three pH-sensing histidines) latches and thereby prevents spring-loaded E fusion protein from prematurely exposing its fusion peptide. This M latch was fastened at an earlier stage, during maturation at acid pH in the trans-Golgi network. At a later stage, to initiate infection in response to acid pH in the late endosome, M releases the latch and exposes the fusion peptide. Thus, M serves as a multistep chaperone of E to control the conformational changes accompanying maturation and infection. These pH-sensitive interactions could serve as targets for drug discovery.
Keywords: cryo electron microscopy, flavivirus, bio-threat agent, enveloped viruses, chaperone, viral maturation
Introduction
Dengue virus is a prevalent mosquito-borne flavivirus that is endemic across tropical and subtropical regions, causing diseases ranging from self-limiting fever to lethal hemorrhagic fever and shock. Each year, more than 50 million people are infected 1. Dengue virus is also a potential bio-threat agent 2. Currently, there are neither licensed vaccines nor specific antiviral therapies against dengue infection. Indeed, the spread of dengue virus is recognized as a major urban public health concern by the World Health Organization 1.
Viral membrane proteins play critical roles during the life cycle of enveloped viruses like dengue virus, particularly during entry into a host cell. The two dengue virus membrane proteins, E and M, undergo dramatic structural changes from the immature to the mature, fusogenic form of the virion 3 and then again at the time of infection. These two proteins are expressed in a polyprotein that is cleaved to yield the precursor of M (prM) and E 4. Then, upon exposure to the neutral pH of the endoplasmic reticulum, prM binds to the E protein to form the ‘spiky immature’ form of the virus 3, the spikes consisting of trimers of the outward pointing domain II of E. E and prM then undergo further maturation that includes three steps: (i) triggered by low pH in the trans-Golgi network (TGN), formation of pr-stabilized dimers of E lying on the surface, producing the ‘smooth immature’ virus 3; (ii) cleavage of prM by the furin protease in the TGN into the pr and M portions 4; and (iii) triggered by neutral pH in the extracellular space, shedding of pr upon release from the cell to yield the fusion-competent, ‘smooth mature’ virion 3. Later, during infection, dengue virus enters the cell through receptor-mediated endocytosis, and fusion of virus membrane with endosomal membrane is triggered by low pH in the late endosome 5,6. Although cryoEM has provided low-resolution in situ structures of the immature viruses and of the mature virion 7,8, and X-ray crystallography has provided high resolution ex situ structures of some domains of E and M 4,9-11, what is not known is how changes in pH bring about these structural – and transitional – transformations.
We set out to explain the interplay between E and M during viral maturation and infection by solving the cryoEM structure of a native, mature dengue virus. Here we report the 3.5Å structure of the mature dengue virion in its native form as determined by cryoEM single particle reconstruction. We discover a latch-type interaction between E and M, mediated by pH-sensing histidine residues, that holds E in place and prevents premature exposure of its fusion peptide. This structure also provides insight into histidine-based, pH-sensitive, maturational processes that spring-load E for later exposure of its fusion peptide at the right time, move M to the latch site, and engage the M latch on E. Thus, the structure reveals that, in response to shifts in pH, M chaperones E through the dramatic conformational changes required for several stages of dengue virus maturation and infection.
Results
Structural validation
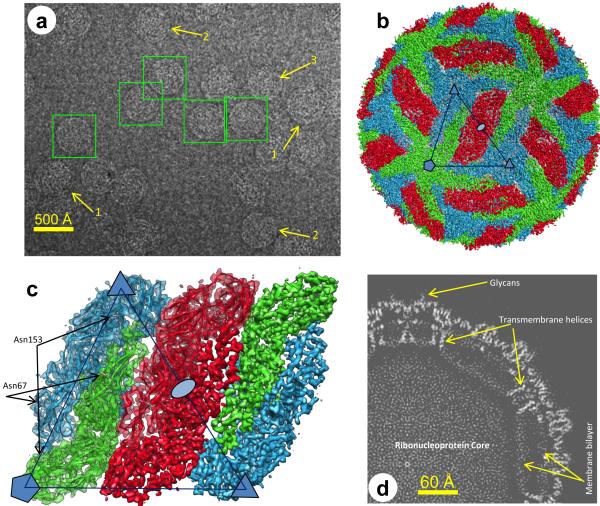
CryoEM micrographs of purified dengue virions recorded in a Titan Krios microscope show spherical (mature) particles (boxed in Fig. 1a) among partially mature, irregular or incomplete ones (arrows #1, #2, #3 in Fig. 1a), as previously observed 12. We eliminated these partially mature virions as much as possible through visual inspection and obtained 32,596 spherical particles from 1103 films. Subsequently, we computationally selected 9288 ‘good’ particles (Supplementary Fig. 1a) that strictly conform to icosahedral symmetry by use of a global orientation-center search method based on a multi-path simulated annealing algorithm 13. The final reconstruction (Fig. 1b-d and Supplementary Movie 1) has an effective resolution of 3.5 Å14 (Supplementary Fig. 1b) and is validated by identification of amino acids in the density map (Supplementary Figs. 2-4).
Figure 1. Overview of the cryoEM structure of the dengue virion.
(a) CryoEM images. Particles in boxes were chosen for further processing after excluding partially mature (1), irregularly shaped (2) or incomplete (3) ones. (b) Surface rendering of the cryoEM density map. E:M heterodimers of the same color are equivalent by icosahedral symmetry. Heterodimers of different colors are quasi-equivalent, with green E:M dimers falling on the icosahedral 5-fold axes, blue on the 3-fold, and red on the 2-fold. (c) Close-up view of a rhombus-shaped group of six E:M dimers with all of the parts of two asymmetric units. One asymmetric unit, containing all of the parts of three E:M dimers, is outlined by the large triangle. Densities in the right half are shown as surface representation, whereas densities in the left half are shown as semi-transparent surfaces with ribbon diagrams of their atomic models superimposed. The glycosylated Asn67 and Asn153 of E are indicated. (d) The central slab (7.7 Å-thick) of the density map perpendicular to a 3-fold symmetry axis. The membrane bilayer appears more polygonal than circular, with transmembrane helices at its corners. See also Supplementary Figure 1 and Supplementary Movie 1.
Overall structure
Inside the envelope, the capsid of the virus is disordered, as described previously 7,8. On the surface of the virion, Asn67 and Asn153 of every E subunit are glycosylated (Fig. 1c), as previously observed 9,11. We built an atomic model for the asymmetric unit (Fig. 2a,b and Supplementary Fig. 2,5), which contains three E subunits and three M subunits (Fig. 1c). The three quasi-equivalent copies of E are very similar to each other, as are those of M, as illustrated by their superposition in Supplementary Figure 5a. The averaged root-mean squared deviation of the locations of the Cα atoms among the three quasi-equivalent copies of E and M is only 1.2 Å, the largest difference being in the loop that connects domains II and III. Therefore, we averaged these three copies (Supplementary Movie 1).
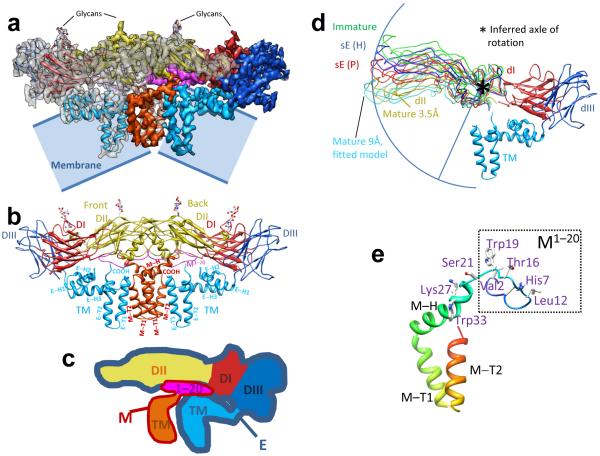
Figure 2. Atomic model of the E:M:M:E heterotetramer.
(a) Side view of the averaged heterotetramer. (b) Side view of the atomic model of the tetramer shown in ribbon with glycans at Asn63 and Asn157 of E shown as sticks. The M1-20 loop binds to a groove in E (see also Fig. 4a,b). (c) The color scheme of the domains of E:M follows previous work 8-11: for E, red for domain I (dI), yellow for domain II (dII) and blue for domain III (dIII), comprising the three parts of the ectodomain; cyan for the transmembrane domain (TM); for M, magenta for the first 20 amino acids (M1-20) (ectodomain) and orange for the transmembrane domain (TM). (d) Hinge in dII of E. The blue arc, centered on a β-hairpin of dII (asterisk), connects the tips of dII in the various conformations: sE(H): solubilized E Harvard crystal structure 9; sE(P): solubilized E Purdue crystal structure 11; Immature: E ectodomain crystal structure 11; Mature 3.5Å: our in situ atomic structure of full-length E in the cryoEM structure; Mature 9Å: pseudo-atomic model obtained by fitting to a 9-Å mature virion cryoEM structure 11. TM (transmembrane domain), dI and dIII from our cryoEM structure. (e) Ribbon model of M color-coded from blue at the N-terminus through red at the C-terminus, with key residues mentioned in the text shown as sticks. See also Supplementary Figures 2-5 and Supplementary Movie 1.
Mature dengue virion has a smooth icosahedral outer surface covered by E protein, visible in Figure 1b and in Supplementary Movie 1, with M protein underneath. Five E monomers surround each 5-fold axis (marked by a pentagon, Fig. 1b) in the shape of a starfish, three surround each 3-fold axis (marked by small triangles), and two surround each 2-fold axis (marked by an oval) (Supplementary Movie 1), as established in previous studies at resolutions of 9 Å (ref. 8) and 24 Å (ref. 7). Each of the large triangles in Figure 1b,c outlines an asymmetric unit. Two adjacent triangles contain three E:M:M:E heterotetramers of membrane proteins E and M, making one ‘rhombic raft’ (Fig. 1c and Supplementary Movie 1). E:M:M:E heterotetramers bind neighboring E:M:M:E heterotetramers (Fig. 1b) through E to E interactions, mainly hydrophilic (Supplementary Table 1) at interfaces along the lateral edges of E. These E and M proteins anchor to an underlying lipid bilayer envelope through their transmembrane helices E-T1, E-T2, M-T1 and M-T2 (Figs. 1d, 2b and Supplementary Figs. 3,4, 5b, 6a). Apart from the last three residues of M at its C-terminal, all residues of E and M are ordered in the structure, thus permitting atomic modeling for both full-length proteins – leaving out the last three amino acids of M (Fig. 2a,b, Supplementary Fig. 2,3 and Supplementary Movie 1).
In situ structure of E
The in situ structure of the full-length dengue E protein contains four domains, the transmembrane (TM) domain (cyan in Supplementary Fig. 2) and the domains I, II and III (red, yellow and blue in Supplementary Fig. 2) that comprise the ectodomain (Fig. 2a,b). Our atomic model of the TM domain of E, which anchors domain III to the membrane, consists of three perimembrane helices E-H1, E-H2 and E-H3 at the N-terminal and two transmembrane helices E-T1 and E-T2, all interlinked by loops (Figs. 2b, 3a and Supplementary Figs. 3, 5a,d,e).
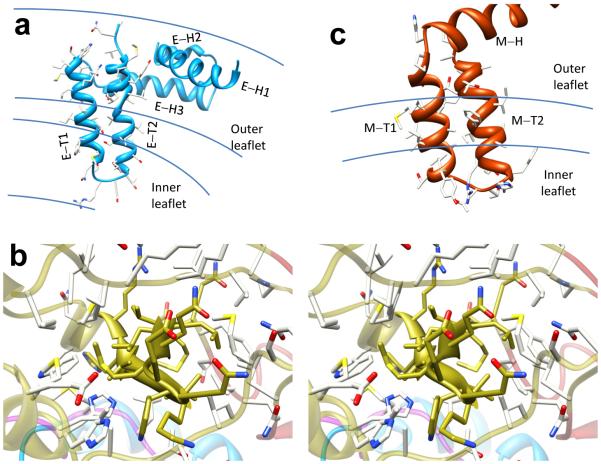
Figure 3. Hydrophobic interactions.
(a) Close-up view of the transmembrane helices (E-T1 and E-T2) and perimembrane helices (E-H1 and E-H2) of E protein (cyan ribbons). The boundaries of the outer and inner leaflets of the phospholipid bilayer are marked. (b) Stereo view of the hydrophobic environment of the β-hairpin (solid golden ribbon with sticks) in dII of E. This domain (atoms colored golden for C, red for O, blue for N, yellow for S) is shown together with surrounding environment (semi-transparent ribbons with sticks, atoms colored white for C, red for O, blue for N, yellow for S) to illustrate hydrophobic interactions. Except for residues at the top and bottom surfaces of the protein, almost all the residues of the hairpin and its surrounding residues are hydrophobic, as indicated by the atom types. (c) Close-up view of the transmembrane helices (M-T1 and M-T2) and the perimembrane helix (M-H) of M protein (orange ribbons). The sticks represent atomic models of selected side chains. See also Supplementary Figure 6a.
We can also identify structural elements crucial to the movement of E’s domains during viral maturation and membrane fusion by comparing our in situ structure of E at atomic resolution to other available structures (including pseudo-atomic models) of the ectodomain of E (ref. 9,11). Superposition of all of these structures by matching Cα atoms of their domain I reveals a relative rotation of domain II (Fig. 2d), with domain I held in place by interaction with the domain III, anchored to the TM domain, the latter anchored to the membrane 9,10. We propose that the β hairpin (Val197 to Val208, marked by an asterisk in Fig. 2d; β strands f and g in Supplementary Fig. 5c) is the axle of this rotation, as illustrated by the arc that connects the tips of all but one of the domain II structures in Figure 2d. (The exception is the 9-Å pseudo-atomic model 11.) The location of this hairpin (as measured from Val197) is ~12 Å away from the previously suggested ‘hinge’ of Gly190 11.
We propose that rotation about this axle would enable the conformational change of E required for the virus to begin fusion with endosomal membrane and release its core into the cytoplasm. The richness of hydrophobic residues on and around this hairpin (golden ribbon in Fig. 3b) is conducive of such a rotation. Indeed, small molecules that bind to the cavity next to this hairpin have been shown to block viral entry 15,16, presumably by hindering the relative rotation about the β hairpin and preventing conformational change required for fusion.
In situ structure of M protein
In our cryoEM structure, 72 of the 75 residues of the full-length M protein are resolved (purple and orange in Fig. 2a-c and Supplementary Fig. 2). M has a single domain with three portions (Fig. 2e and Supplementary Fig. 3): an extended N-terminal loop (a.a. 1-20, named M1-20), an amphipathic perimembrane helix (a.a. 21-40, named M-H), and a pair of transmembrane helices (a.a. 41-75, named M-T1 and M-T2) (Fig. 3c and Supplementary Fig. 6a). The perimembrane and transmembrane helices serve to anchor the M protein to the membrane.
Val2 of M1-20 (Fig. 2e) is bound in a pocket of E (pocket 1 in Fig. 4a,b). M1-20 also contains Thr16 (Fig. 2e), where the loop bends and turns to the other side of helix αB of E (Supplementary Fig. 5b,c). This bend is very close to the ‘hole’ formed between two E subunits (Supplementary Fig. 7), a feature described previously 8 and discussed further below.
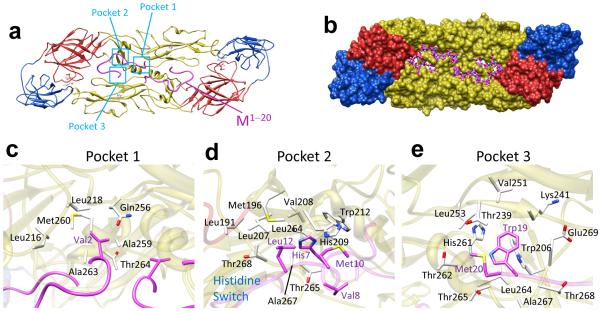
Figure 4. Key interactions between E and M.
(a,b) E:M:M:E heterotetramer viewed from inside the virus. The ribbon model (a) shows three pockets (cyan boxes) on E where M binds. (Transmembrane domains omitted.) The space-filling model (b) of E shows the groove where M (stick model: atoms C-magenta, N-blue, O-red, S-yellow, H-white) binds. See Supplementary Movie 2. (c-e) Enlargement of pockets 1-3 viewed along directions that best depict interactions. (c) Val2 and the first few residues of M sit in a big cavity in the inner surface of E. (d) His7, Met10 and Leu12 of M form a hydrophobic core with neighboring residues in E. The two opposing histidine residues (H7 of M and H209 of E), when protonated at low pH, repel each other. (e) A conserved Trp19 (W19) from M inserts into a deep recess along the E:E dimer interface that includes the partially conserved H261 of E (Supplementary Fig. 9). In stick models, atom types are colored as in b. See also Supplementary Movie 2 and Supplementary Figure 8.
The perimembrane helix (M-H) starts at Ser21 and strikes the outer leaflet at an angle, with a kink at Lys27 (Fig. 2e and Supplementary Fig. 3a). The first part (before the kink), mainly hydrophilic but with one tryptophan (Trp26) inserted into the membrane, is slightly outside the head-group region (Supplementary Fig. 6a). The second part (after the kink), buried in this head-group region, is amphipathic, like the perimembrane helices of the E protein.
The two transmembrane helices M-T1 and M-T2 are shorter than the transmembrane helices of E (Fig. 2b and Supplementary Fig. 5a,b). Unlike those in E, these two transmembrane helices in M contain mostly hydrophobic residues, with the exception of several hydrophilic residues in the head group region of the lipid (Fig. 3c and Supplementary Fig. 6a). The last resolved residue of M, Pro72, is located at the edge of the head group region of the outer leaflet.
Membrane-protein interactions
The discernible features in the membrane density of the reconstruction suggest the existence of some lateral order among the phospholipids of the lipid bilayer (Supplementary Fig. 6b,c), as is the case for bacteriophage PRD1 17. The membrane is bent to an angular shape at the distal ends of the transmembrane helices of E and M, where the membrane thickness is reduced from 42 Å to 30 Å (Fig. 1d, Supplementary Fig. 6d,e) because of the short lengths of these transmembrane helices. Indeed, for each transmembrane helix of M, there are just four helical turns, and for each transmembrane helix of E, there are just five. This angular membrane shape is in sharp contrast to the spherical membrane shape observed in alphaviruses 18, where the transmembrane helices are noticeably longer (seven helical turns), cross a fully relaxed envelope and reach underlying capsid proteins.
The two transmembrane helices E-T1 and E-T2 are oriented vertically and span the hydrophobic region of the membrane, with their interconnecting loops buried within the head-group region of the inner leaflet (Fig. 2b, 3a and Supplementary Fig. 6a). They form a coiled-coil (Fig. 3a), with hydrophobic surfaces facing outward and multiple hydrophilic residues (Thr and Ser) facing inward, reminiscent of the configuration of a leucine zipper – although with hydrophilic residues (Thr and Ser) for interaction between the coiled α-helices instead of hydrophobic residues (Leu). These interactions explain why a functional mosaic cannot be constructed from E-T1 and E-T2 of two different viruses 19.
The three perimembrane helices of E (E-H1, E-H2 and E-H3) and one perimembrane helix of M (M-H) lie horizontally among the head groups of the outer leaflet of the envelope (Fig. 3a,c and Supplementary Fig. 6a). Indeed, our structure reveals that helices E-H1 and E-H3 are amphipathic, with top halves containing mainly hydrophilic residues facing outside and bottom halves containing mainly hydrophobic residues and interacting with the alkyl groups of the membrane. The smallest of the perimembrane helices, E-H2, is a two-turn 310 helix that was not resolved in the 9Å structure 8. It extends slightly outside the head-group region and contains mainly hydrophilic residues.
Interactions between E and M
M and E proteins interact primarily through hydrophobic contacts. M1-20 lines a groove in the side of E that faces the membrane on the viral envelope (Fig. 4a,b, Supplementary Fig. 8a and Supplementary Movie 2). The area of the buried surface between M and the ectodomain of E is 1138 Å2 (Fig. 4b). Surprisingly, despite the extensive interactions, we find no hydrogen bonds between M and the ectodomain of E. Their binding affinity comes from three hydrophobic interactions: Firstly, Val2 of M (Fig. 2e and Supplementary Fig. 3) inserts into a pocket in E (pocket 1 in Fig. 4a,c, Supplementary Fig. 8b,c, and Supplementary Movie 2) formed by its Leu216, Leu218 and Met260 on one side of helix αB of E. Secondly, His7, Met10 and Leu12 of M (Fig. 2e and Supplementary Fig. 3) form another hydrophobic core with residues of E that include His209 and Trp212 (pocket 2 in Fig. 4a,d, Supplementary Fig. 8d,e and Supplementary Movie 2). Thirdly, Trp19 of M (Fig. 2e and Supplementary Fig. 3) is encompassed by a deep recess (pocket 3 in Fig. 4a,e, Supplementary Fig. 8f,g and Supplementary Movie 2) that includes Trp206 and His261 (on the other side of helix αB of the same E monomer noted in pocket 1). These hydrophobic interactions provide the stability of mature dengue virus.
Indeed, many of the aforementioned residues are highly conserved among all flaviviruses or among just all dengue viruses (Supplementary Fig. 9). Of M, His7, Leu12 and Trp19 are strictly conserved among all flaviviruses (with a single exception of a Met in Powassan tick-borne encephalitis in the position corresponding to Leu12), and Val2 can be replaced only with residues of branched hydrophobic side chains (i.e., Leu and Ile). For pocket 1, Leu216 and Leu218 of E are strictly conserved among flaviviruses, and Met260 is conserved among different strains of dengue virus. For pocket 2, Trp212 of E is strictly conserved, and His209 is strictly conserved except for yellow fever and St. Louis encephalitis. For pocket 3, Trp206 of E can only be replaced by an aromatic phenylalanine, and His261 is conserved among dengue and Japanese encephalitis subtypes. The conservation of these residues is consistent with critical roles played by these interactions.
Remarkably, there are three histidine residues, His7 of M in pocket 2, His209 of E in pocket 2 and His261 of E in pocket 3, involved in the E:M interactions discussed above. Because the theoretical pKa of the side chain of histidine is 6, these interactions would be abolished or weakened at pH below 6, when histidine residues are protonated and become positively charged. Specifically, the two closely placed, strictly conserved histidine residues His7 of M and His209 of E, both in pocket 2, would repel each other when both are positively charged (Fig. 4d and Supplementary Fig. 8d,h), impelling dissociation of E from M. This pH-dependent character of histidines allows them to act as pH sensors. We propose that at physiological pH in the bloodstream during viral transmission (either in its mosquito vector or within a human body), M binds to and holds E in the dimer form pointing along the surface, providing stability for the virus as revealed in our structure; however, at the acidic pH in the late endosome, M frees E and allows it to transition into the outward-pointing, trimeric, fusogenic form, facilitating viral entry.
Although structure-based mutagenesis studies of E have demonstrated the critical importance of histidines to its conformational change triggered by low pH 20,21, the three histidines identified above have been largely overlooked in dengue virus. However, in confirmation of this mechanism, mutation of the counterparts of dengue virus His209 and His261 – namely His214 and His263 in West Nile virus (WNV) – individually to glutamines, reduces cell entry by engineered single round infectious particles 22.
DISCUSSION
A model for maturation and infection
Our atomic structure of the mature virion, together with previous models of the immature particle and the post-fusion form of E (ref. 3,4,9-11,23,24), can fill out the picture of dengue virus maturation, through multiple pH-sensitive stages (Fig. 5).
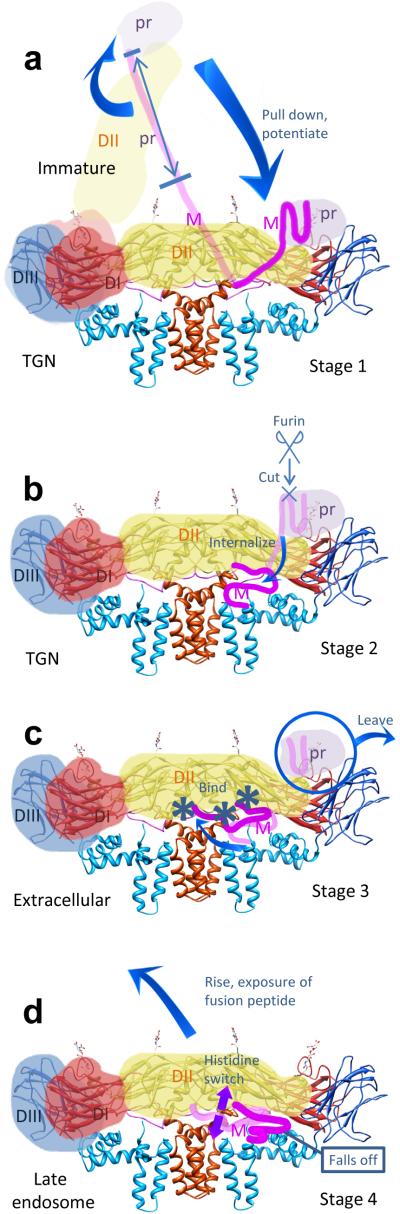
Figure 5. Proposed mechanisms for maturation (Stages 1-3) and exposure of the fusion peptide of E required for infection (Stage 4).
See text for details. (a) Spiky immature virus. (b) Smooth immature virus. (c) Smooth mature virus. (d) Exposure of fusion peptide.
At neutral pH in the endoplasmic reticulum, in the immature spiky virus (Stage 1 in Fig. 5a) the three domains II of each E trimer point outward 3,4,23. Low pH in the TGN triggers rotation of each of the three domains II to point along the surface, where pairs of domains II join to yield E dimers, producing the smooth character of the smooth immature form of the virus (Stage 2 in Fig. 5b) 3,4,23. We suggest that this rotation is effected by prM in the following manner: The unfolded portion of pr (the blue double-headed arrow in Stage 1) acts like a ‘drawstring’ that is attached to the folded head of pr, represented by the light grey in Stage 1. As the drawstring tightens, induced by the low pH in the TGN, the folded head pulls domain II of E down and spring-loads it (Stage 2). This repositioning also presents the furin cleavage site on the prM to the furin protease 3,4,23 (Stage 2).
After prM lowers E, its pr portion binds both E molecules within the newly formed dimer and latches them in this spring-loaded, lowered position 3,4,23. Subsequent furin cleavage of prM separates pr from M and allows M1-20 to internalize below the E dimer (to the same side as the rest of M (a.a. 21-75), which includes M’s transmembrane domain anchored to the membrane), presumably by passing through the ‘hole’ 8 between the two E monomers in their dimer (Supplementary Fig. 7a) (Stage 2 to Stage 3 in Fig. 5c). [The hole had been guarded by two histidine residues (His27 and His244), one from each E in the dimer, that line the walls of the hole, acting as a double door. The double door had opened under low pH conditions in the TGN due to electrostatic repulsion (Supplementary Fig. 7b). After the M1-20 part of M had passed through, the double door closed at physiological pH in the extracellular environment due to hydrophobic interaction among these two histidines and a neighboring phenylalanine (Phe279) (Supplementary Fig. 7a), as shown by the atomic structure of the mature virion.]
After the virion leaves the cell, it encounters a higher, physiological pH in the extracellular environment and becomes mature (Stage 3 in Fig. 5c). In this step, due to the physiological pH that is higher than the pKa of histidines, the charges on the three histidines along the E:M interface, H7 of M, H209 of E and H261 of E, are removed. The deprotonation of these histidine residues allows hydrophobic interactions that bind M and E (Stage 3). Consequently, M will take the place of pr in latching E along the envelope, with E in the spring-loaded, mature form. A similar deprotonation also disrupts the binding between His244 (the same His lining the double door) of E and Asp63 of pr, leading to the dissociation between E and pr (ref. 4). The virus thus sheds pr to become a mature virion (Stage 3).
Therefore, we propose that M acts not only as a stabilizer but also as a latch, anchored by its transmembrane domain, that restrains the E protein from rising. Premature rising would be catastrophic for the virus by exposing the fusogenic peptide early, rendering the virus unable to fuse with endosomal membrane and release its core into the cytoplasm for replication. At the right time, when the virus encounters the low pH of the late endosome, the now protonated histidine residues allow dissociation between E and M, leading to timely ‘unlatching’ of the E subunits that rise by a rotation by the ectodomain of E about its anchor to its transmembrane domain. This rise is accompanied by a rotation of domain II with respect to domain I – about the axle described in Figs. 2d and 3c – that permits the formation of fusogenic E trimers (Stage 4 in Fig. 5d). This latch had also stored free energy in E to be used for the rising of E. Many of the details of this model, employing the four stages and the mechanisms that transform one stage into another, await experimental testing by structure-based mutations, particularly of the different His residues, along with structure determination at the different stages.
The controlled release of E and the precise timing of the formation of the fusogenic trimer may be key features of flaviviruses that are critical to their biological function, resembling the E1-E2 cooperation in alphaviruses. Interestingly, yellow fever virus bears a negatively charged residue (Asp209) in pocket 2 (Fig. 4d, H209 in Supplementary Fig. 8h), which attracts rather than repulses M at low pH. Therefore, yellow fever virus might use a different mechanism for maturation and triggering by low pH.
Fusion strategies and countermeasures
Viral membrane fusion proteins face two challenges to achieve productive fusion with host cells: First, every fusion protein has to arrest in a high energy, pre-fusion form during folding, prior to reaching its low energy, post-fusion form. Second, those fusion proteins that are pH-sensitive must distinguish the low pH in the TGN during egress from the low pH in the late endosome during infection. Fusion should occur in the latter stage but not the former. The three classes of viral fusion protein overcome these challenges with different strategies.
Class I fusion proteins, as found in influenza viruses and HIV, employ a hidden knife strategy. They overcome the first challenge by expressing their fusogenic peptides in the middle of an inactive precursor whose cleavage, either in the TGN (HIV) or later (influenza) leaves this peptide at a new N-terminus to prime fusion 6. Those that are pH sensitive, like influenza haemagglutinin, overcome the second challenge by passing through the low pH in the TGN in an uncleaved form. Later, the low pH in the late endosome during infection exposes the fusion peptide from a primed (cleaved) virus. Exposure of the fusion peptide by pH-insensitive fusion proteins, as found in HIV, is triggered during infection by binding to membrane receptors.
Class III fusion proteins, as found in rhabdoviruses, baculoviruses and some herpesviruses, employ a reversible form strategy, specifically a reversible pH-driven conformational change. The fusion protein takes its fusion incompetent ‘post-fusion’ form through the TGN but adopts its fusion competent ‘pre-fusion’ form at higher pH outside the cell, as reviewed in ref. 25.
Our structure reveals how the class II viral fusion protein in dengue virus employs a multistep chaperone strategy 5, involving a second membrane protein (M) that is used to aid in the folding, trafficking and function of the fusion protein (E). One review suggested that the action of a protein like M in alphavirus might be conceptualized as a chaperone 5. Because M in dengue virus performs many roles in a complicated, multistep process widely dispersed over space and time, because it cannot be recycled as a result of cleavage and irreversible conformational changes, because its anchoring in the membrane is critical to its function, and because it responds to conditions – pH in the TGN, the extracellular environment, and the endosome – we describe M as a membrane-anchored, pH-sensing, multistep chaperone.
For these fusion strategies, countermeasures can be devised against viral infection. The special dependency of Class II fusion proteins on chaperone proteins may be their Achilles heel. Indeed, in addition to providing insight into the mechanisms of viral maturation and fusion, our structure identifies specific interactions between the dengue virus fusion protein and its chaperone protein in atomic detail that are critical for maturation and infection. These specific interactions are potential targets for future therapeutic intervention. Indeed, the small peptide enfuvirtide blocks pre-fusion folding of the gp41 fusion protein in HIV with itself by competing for a binding site on gp41 27. By contrast, small molecule analogs of dengue M protein that block its access to any or all of the three pockets in E might disrupt the function of the chaperone protein M and thereby abolish dengue virus maturation and/or trigger premature exposure of the fusogenic peptide of E from the mature virus. Such small molecules could serve as novel leads for drug discovery.
Online Methods
Isolation of dengue virus
C6/36 cells were cultured at 33°C in the presence of 5% CO2. During cell passaging, we detached cells from flasks by vigorously shaking each flask a few times, avoiding exposure of cells to trypsin. Twenty-seven Corning T125 flasks, each containing C6/36 cells in 50ml of medium, were infected by dengue virus type 2 New Guinean strain. At 5 days post infection, cell culture medium was collected and centrifuged with a Beckmann centrifuge (11,000g) for 50 minutes to pellet and discard large debris. PEG-8000 was added to the supernatant to a final concentration of 7% (w/w). The sample was left at 4°C for 8 hours and subsequently centrifuged with a Beckman centrifuge (11,000g) for 45 minutes to collect the virus-containing pellet. The virus was resuspended in TNE buffer (50mM Tris, 140mM NaCl, 5mM EDTA, pH 7.4) by soaking the pellet in the buffer for 20 mins. The resuspended sample was then loaded at the top of a glycerol-potassium tartrate gradient (10% – 40% potassium tartrate, 30% – 7.5% glycerol, from top to bottom) and centrifuged for 12 hours at 120,000g (Beckman Coulter SW41) at 4°C. A band was located at about 3/4 distance from the top of the gradient. The gradient material above the band was removed with a pipette; then, the virus-containing band was carefully collected with another pipette. The collected viral sample (1 ml) was diluted to ~12 ml by TNE buffer and pelleted for 2 hours at 120,000g (Beckman Coulter SW41) at 4°C to remove gradient material and to concentrate the sample. The pelleted viral sample was resuspended in 100 μl of TNE buffer for cryoEM.
CryoEM imaging and initial structure determination
Each aliquot (~2.5 μl) of freshly prepared dengue virus sample was placed onto a Quatifoil 2/1 grid (Quatifoil GmbH, Germany), blotted with filter paper, and plunged into liquid nitrogen-cooled liquid ethane to make cryoEM grids. CryoEM images were first recorded as focal pairs (targeted defocus values of −1μm for close-to-focus images and −2.5μm for far-from-focus images) on a 16 megapixel CCD camera (TVIPS) in a Polara G2 cryoEM instrument (FEI Company) operated at 300kV. These images have a magnification of 93,000x and a pixel size of 0.97 Å/pixel. The measured defocus values of these images range from −0.3μm to −2.2μm, as determined manually using ctfit of EMAN 28. The imaging electron dosage was 17 e-/Å2. Approximately 40,000 particles were selected from 5000 focal pairs using winboxer of IMIRS package 29,30. This dataset was used to obtain a 7-Å resolution reconstruction with IMIRS package 29,30 from a final dataset of ~3300 particles.
To improve the resolution of our reconstruction, we subsequently took images of the frozen grids from the same batch of samples in an FEI Titan Krios cryo electron microscope operated at 300 kV. These images were recorded at a calibrated magnification of 57,500x (59,000x nominal magnification) on Kodak SO-163 Electron Image Films, with a dosage of 25 e−/Å2. The defocus values of these cryoEM images range from −0.7μm to −2.5μm, as determined automatically with CTFFIND331.
Micrographs were digitized in Nikon CoolScan scanners at a pixel size of 6.35 μm on the film. Considering the calibrated magnification of 57,500x, the pixel size of the digitized images is 1.104 Å/pixel. Approximately 32,569 particles were selected from the 1103 films by the boxer program in EMAN package 28 with the assistance of ETHAN 32 automatic boxing.
CryoEM structure refinement
To further improve the resolution of the 3D reconstruction, we processed the Titan Krios particle images with a recently developed procedure, global orientation-center search by multi-path simulated annealing (MPSA) 13, as implemented in EMAN 28. In this procedure, we subjected all of the selected particle images through an iterative process. The 7-Å resolution structure from the Polara dataset was used as the starting model. In each iteration, the orientation and center of each particle were determined by MPSA global search through seven trials, each trial starting with a different initial guess. If for four of the seven trials the determined orientation and center converged, we considered this particle ‘good’. The tolerances for orientation and center convergence were 3° and 3 pixels, respectively. In this way, we determined the orientation and center parameters for each particle and ruled out ‘bad’ particles. ‘Good’ particles were used for 3D reconstruction with the make3d module of EMAN without class-averaging. The reconstruction was subjected to Fourier amplitude scaling similar to ref. 33, temperature (B) factor sharpening (we used a B factor of 40 Å2), low-pass filtering (to the current best resolution) and solvent flattening, and then the resulting density map was used as the reference of the next iteration of refinement. The overall result of these manipulations is comparable to a simple B factor sharpening with a B factor of 240 Å2; using Fourier amplitude scaling allows us to generate a map with more natural structure factors (Fourier amplitudes). To avoid possible model bias, no atomic model was referred to or used throughout this iterative refinement process of the density map and only the cryoEM reconstruction obtained from the experimental cryoEM images was used as the model for refinement. Upon reaching convergence of the refinement and no further improvement in the resolution of the 3D reconstruction based on Fourier shell correlation and on evaluation of side chain densities map, a final map was reconstructed from 9288 ‘good’ particle images from the Titan Krios dataset. A total of 16 iterations of MPSA refinement were performed to reach convergence. 12 iterations were done with data from the first imaging session to refine the structure to 4.2-Å resolution. 4 more iterations were done with all data to refine the structure to the final resolution. Astigmatic CTF correction was performed.
The final map was used to derive an intermediate atomic model (see next section). This intermediate model was Fourier transformed and the resultant Fourier amplitudes were radially averaged to produce a one-dimensional ‘structure factor’ profile as a function of spatial frequency. Fourier amplitude scaling was done by using this ‘structure factor’ profile to suppress noise. After the amplitude scaling, a temperature factor of 40 Å2 was deconvoluted from the map to sharpen high resolution features, and the resulting map was low-passed to 3.3 Å with cosine apodization at the edge of Fourier truncation.
Building the atomic model for the virion and the averaged heterotetramer
We first averaged the density of the three heterotetramers with each rhombic region (see Fig. 1c). We used Coot 34 and REMO 35 to build the atomic models for E and M proteins based on this averaged density map. The protein backbone was first traced with the ‘baton’ tool in Coot. The resulting Cα model was converted into a full-atom model with REMO. We used the CNS package 36 to refine the E:M:M:E heterotetramer structure by pseudo-crystallographic methods as previously described 37 with its two-fold symmetry as an non-crystallographic symmetry (NCS) restraint. One half of the tetramer, containing one copy of E and one of M, was fitted into the density of each of the three copies of E and M in an asymmetric unit. The resulting atomic model was further refined by the CNS package 36 against the map of the entire virion, with icosahedral symmetry as an NCS constraint.
We then built atomic models for the glycans at Asn67 and Asn153. Atomic models for a single sugar of N-acetyl-glucosamine (NAG) and a disaccharide with two NAGs were built for Asn67 and Asn153, respectively. Densities for additional sugars on these two glycosylation sites exist but are poorly ordered and were therefore not modeled. These additional sugars are more apparent in lower resolution density maps, suggesting their flexibility.
The full model was refined again in CNS as described above (R-factor: 29.3%, see R-factors of individual resolution bins in Supplementary Table 2). We also added sugars to the atomic model for the averaged tetramer and refined that model. The final R-factor for the averaged tetramer is 28.8% at 3.5 Å.
Supplementary Material
Acknowledgments
We thank V. Vordam of Centers for Disease Control Dengue Branch (San Juan, Puerto Rico) for providing the viral stock and advising about cell culture, I. Atanasov and W. H. Hui for participation in data acquisition, J. Jiang for suggestions in data processing, and UCLA undergraduate students K. M. Lau, J. Chen and K. Chen and B. K. Zhou of Beverly Vista School for scanning photographic films and boxing particles. We are also grateful to A. Paredes of the US Food and Drug Administration and J.-Q. Zhang of Sun Yat-Sen University for preliminary efforts in viral preparation. This work is supported in part by grants from the US National Institutes of Health grant GM071940 (to ZHZ), National Natural Science Foundation of China (NSFC) grant 30928003 and 30725017 (to GB), NSFC 30470085 and 30870480 (to QZ).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Author Contributions Z.H.Z., X.Z., P.G. and X.Y. designed experiments. J.M.B., X.Z. and X.Y. cultured cells and purified virus samples. X.Z., X.Y., P.G., J.M.B. and Z.H.Z. obtained cryoEM images. Z.H.Z., J.M.B. and X.Z. participated in the image processing and 3D reconstruction from the Polara data. X.Z. obtained a 7-Å structure from the Titan Krios data. P.G. refined the structure to 3.5-Å resolution and built the atomic models. P.G., X.Z. and Z.H.Z. interpreted the structure and drafted the manuscript. P.G., X.Z., Z.H.Z. and S.S. finalized the manuscript. G.B. and Q.Z. participated in discussion and interpretation of the results. All authors reviewed the final manuscript.
Accession Numbers
The atomic coordinates of the mature dengue virion have been deposited with the Protein Data Bank (accession code 3J27). The cryoEM density maps of the virion and the averaged tetramer have been deposited with the Electron Microscopy Data Bank (EMDB) (accession code 3J27 and EMD-5499, respectively).
References
- 1.WHO . Dengue: guidelines for diagnosis, treatment, prevention and control. New edition. WHO; Geneva, Switzerland: 2009. [PubMed] [Google Scholar]
- 2.Borio L, et al. Hemorrhagic fever viruses as biological weapons: medical and public health management. Jama. 2002;287:2391–405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- 3.Yu IM, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–7. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 4.Li L, et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–4. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 5.Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Micro. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn RJ, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–12. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100:6986–91. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–9. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–18. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plevka P, et al. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 2011;12:602–6. doi: 10.1038/embor.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Jiang W, Jakana J, Chiu W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J Struct Biol. 2007;160:11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Kaptein SJ, et al. A derivate of the antibiotic doxorubicin is a selective inhibitor of dengue and yellow fever virus replication in vitro. Antimicrob Agents Chemother. 2010;54:5269–80. doi: 10.1128/AAC.00686-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang QY, et al. A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother. 2009;53:1823–31. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cockburn JJ, et al. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature. 2004;432:122–5. doi: 10.1038/nature03053. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, et al. 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. Embo J. 2011;30:3854–63. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz R, et al. The unique transmembrane hairpin of flavivirus fusion protein E is essential for membrane fusion. J Virol. 2011;85:4377–85. doi: 10.1128/JVI.02458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritz R, Stiasny K, Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol. 2008;183:353–61. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroschewski H, Sagripanti JL, Davidson AD. Identification of amino acids in the dengue virus type 2 envelope glycoprotein critical to virus infectivity. J Gen Virol. 2009;90:2457–61. doi: 10.1099/vir.0.011486-0. [DOI] [PubMed] [Google Scholar]
- 22.Nelson S, Poddar S, Lin T-Y, Pierson TC. Protonation of Individual Histidine Residues Is Not Required for the pH-Dependent Entry of West Nile Virus: Evaluation of the “Histidine Switch” Hypothesis. J Virol. 2009;83:12631–12635. doi: 10.1128/JVI.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu IM, et al. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J Virol. 2009;83:12101–7. doi: 10.1128/JVI.01637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Structures of immature flavivirus particles. Embo J. 2003;22:2604–13. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaudin Y. Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Subcell Biochem. 2000;34:379–408. doi: 10.1007/0-306-46824-7_10. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz IC, Allison SL, Heinz FX, Helenius A. Folding and Dimerization of Tick-Borne Encephalitis Virus Envelope Proteins prM and E in the Endoplasmic Reticulum. J. Virol. 2002;76:5480–5491. doi: 10.1128/JVI.76.11.5480-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalezari JP, et al. A phase II clinical study of the long-term safety and antiviral activity of enfuvirtide-based antiretroviral therapy. AIDS. 2003;17:691–698. doi: 10.1097/00002030-200303280-00007. [DOI] [PubMed] [Google Scholar]
- 28.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 29.Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, et al. Atomic Structure of Human Adenovirus by Cryo-EM Reveals Interactions Among Protein Networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 32.Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. Local Average Intensity-Based Method for Identifying Spherical Particles in Electron Micrographs. J Struct Biol. 2000;131:126–134. doi: 10.1006/jsbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463:379–83. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Zhang Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins: Structure, Function, and Bioinformatics. 2009;76:665–676. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 37.Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci U S A. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.