Abstract
In bony fishes, Bfsp2 orthologues are predicted to possess a C-terminal tail domain, which is absent from avian, amphibian and mammalian Bfsp2 sequences. These sequences, are however, not conserved between fish species and therefore questions whether they have a functional role. For other intermediate filament proteins, the C-terminal tail domain is important for both filament assembly and regulating interactions between filaments. We confirm that zebrafish has a single Bfsp2 gene by radiation mapping. Two transcripts (bfsp2α and bfsp2β) are produced by alternative splicing of the last exon. Using a polyclonal antibody specific to a tridecameric peptide in the C-terminal tail domain common to both zebrafish Bfsp2 splice variants, we have confirmed its expression in zebrafish lens fibre cells. We have also determined the in vitro assembly properties of zebrafish Bfsp2α and conclude that the C-terminal sequences are required to regulate not only the diameter and uniformity of the in vitro assembly filaments, but also their filament–filament associations in vitro. Therefore we conclude zebrafish Bfsp2α is a functional orthologue conforming more closely to the conventional domain structure of intermediate filament proteins. Data mining of the genome databases suggest that the loss of this tail domain could occur in several stages leading eventually to completely tailless orthologues, such as human BFSP2.
Keywords: lens, cytoskeleton, beaded filaments, BFSP1, intermediate filament, evolution
1. Introduction
The consensus secondary sequence characteristics for the family of intermediate filament (IF) proteins (PFAM: 0038) is a central α-helical rod domain of between 310 and 350 residues comprising two substantial α-helical domains punctuated by a linker, linker L12 (Herrmann and Aebi, 2004). This rod domain is positioned between a non-α-helical head domain and a non-α-helical tail domain. Between the different classes of intermediate filament proteins, the head and tail domains show the most sequence diversity, but within each class these sequences are usually well conserved (Nielsen and Jorgensen, 2003; Parry, 2005).
The beaded filament structural protein 2 (Bfsp2) remains an orphan within the intermediate filament family (Kim and Coulombe, 2007; Omary et al., 2006) not least because in mammals and birds it is only one of two IF proteins to lack a C-terminal tail domain (Merdes et al., 1993; Perng and Quinlan, 2005; Perng et al., 2007 and references therein). The other protein is human keratin 19 (Stasiak and Lane, 1987), but its sequence, assembly and expression profile characteristics confirm this to be a member of the type II class of intermediate filament protein. Bfsp2 expression in the lens coincides with the differentiation of epithelial cells into lens fibre cells (Ireland et al., 2000 and references therein) and although the lens is epithelial in origin (eg Alexander, 1937), there is insufficient supporting evidence to include Bfsp2 in any one of the existing IF protein classes (Kim and Coulombe, 2007; Omary et al., 2006). With the publication of the complete cDNA sequence for trout bfsp2 (Binkley et al., 2002), Bfsp2 complies better with the consensus domain structure for intermediate filament family members by possessing a C-terminal non-α-helical tail domain. This however, then poses a number of questions. Is the C-terminal tail domain present in all fish Bfsp2? Is it conserved in sequence amongst fishes? What impact might this have on its assembly characteristics? Depending on the answers to such questions it is conceivable that these C-terminal non-α-helical tail domain sequences could even be redundant in readiness for their complete loss later in tetrapod evolution. The consensus view is that the C-terminal non-α-helical tail domain is not necessary for filament assembly, but regulates the width of the filament (Herrmann et al., 1996) as well as being involved in filament–filament interactions (Bousquet et al., 2001; Leterrier et al., 1996; Lin et al., 2010) and the cytoplasmic distribution of intermediate filaments (Lowrie et al., 2000).
In the mammalian lens, the beaded filaments are believed to be important for the optical properties of the lens (reviewed in Song et al., 2009). This is because the targetted deletion of mouse bfsp2 results in the loss of lenticular optical properties, as demonstrated by both the increase in the back focal length and increased variability for this value for different planes of the knockout lenses (Sandilands et al., 2003). This was caused by the disorganisation of the lens fibre cells (Sandilands et al., 2003). Moreover, the removal of BFSP2 by gene targetting induced a dramatic change in the morphology of the IF cytoskeleton in lens fibre cells (Sandilands et al., 2003, 2004). These data imply that changes in the lens IF cytoskeleton can have dramatic effects upon lens function. This is borne out by the various missense mutations in both BFSP1 and BFSP2 that have been linked to inherited human cataract (reviewed in (Song et al., 2009)). Therefore it is important to investigate how the additional C-terminal non-α-helical tail domain sequences present in the fish orthologues might alter the assembly properties of Bfsp2.
Here we have used database mining to identify the zebrafish bfsp2. We confirm there to be a single gene in zebrafish that is alternatively spliced to produce two protein products (Bfsp2α and Bfsp2β) that differ only in the C-terminal ten residues. We show bfsp2 is expressed in the zebrafish lens using a polyclonal antibody generated to residues 407–419 common to both splice variants. Recombinant Bfsp2α was produced in Escherichia coli and we present data to show that this extra domain is important to the in vitro assembly of Bfsp2 and in common with for example vimentin, another intermediate filament protein expressed in the lens, regulates the width of the intermediate filaments.
2. Materials and methods
2.1. Radiation hybrid (RH) mapping
The zebrafish bfsp2 and bfsp1 genes were radiation hybrid (RH) mapped on the Goodfellow T51 RH panel as described (Dahm et al., 2005; Geisler, 2002) using two and three independent primer pairs, respectively (Table 1). PCRs for RH mapping were done independently in triplicate.
Table 1.
Radiation hybrid mapping of the zebrafish bfsp1 and bfsp2 genes.
| Radiation hybrid (RH) mapping results | primers used |
|---|---|
| zebrafish bsfp2 | |
| 0100000000000000000000100000010000000000000000000000000100000000000000001000001000010000000000 | bfsp2_74s |
| GCAGTGTAGGACGCATGAGC | |
| bfsp2_390as | |
| GACCGCTTTACGATCCAAGC | |
| 0100000000000000000000100000010000000000001000000000000100000100000000000000001000010000000000 | bfsp2_490s |
| GAGAATGTTCAGGCCAATGC | |
| bfsp2_792as | |
| CCAGTGAGAGCGAATGAAGG | |
| zebrafish bsfp1 | |
| 0000000010001000000001000000000000000000000000000001110000000100100001000000001000000000000001 | bfsp1_207s |
| TGAGACAGCAGAGGGTGAGC | |
| bfsp1_592as | |
| GATCCTCTGCCTGTTGATGC | |
| 0000000010001000000001000000010000100000000001000001110000000100100001000000001000000000000001 | bfsp1_1130s |
| CAGACCATCAAGACCACACAGG | |
| bfsp1_1329as | |
| TCCTCGTTCTCGATGATCC | |
| 0000000010001000000001000000010000100000000001000001110000000100100001000000001000000000000001 | bfsp1_1447s |
| GGACATCACCAACGTGAAGC | |
| bfsp1_1806as | |
| GTGGTGGAGACAAAGCATGG |
2.2. Zebrafish bfsp2 subcloning and recombinant expression in E. coli
The zebrafish bfsp2 clone (Unigene; Dr.19486. Genbank: NM_001008633.1. MGC: 103750. Clone ID: 7074672) was obtained from Geneservice who supplied the cDNA in the cloning vector pME18S-FL3 (www.geneservice.co.uk). This cDNA was used to generate an expression construct in pET23. The oligonucleotide 5′ TCATATGCCTCTTCCAAGACG was used to engineer an NdeI site at the initiating methionine codon ATG and was PCR-amplified using the reverse primer 5′ GCATGTGTTCAGGCTGTCC and the bfsp2 clone from Geneservice (ID: 7074672). The product included a unique BstXI site present in the zebrafish cDNA. The PCR product was cloned into pGEMTeasy (Promega) and the sequence confirmed by bi-directional DNA sequencing. The pET23 expression construct was then generated by subcloning into a NdeI-NotI cut plasmid the NdeI-BstXI fragment from the sequenced pGEMTeasy vector and the BstXI-NotI fragment from the supplied Geneservice cDNA that had been cloned into pME18S-FL3. This then generated a full-length cDNA expression construct in pET23 for the zebrafish bfsp2. We refer in figures to the expressed wild-type Bfsp2 protein product as Zf WT.
In order to truncate the full-length zebrafish protein and remove the additional C-terminal tail sequences (Fig. 2A) to produce a protein product equivalent to human BFSP2, a stop codonwas introduced at residue 395. This C-terminally truncated form of zebrafish Bfsp2 we refer to in figures as Zf CTand is thus terminated at residue R394. This we achieved by PCR mutagenesis using the supplied Geneservice clone (ID: 7074672) and the two oligonucleotides 5′ ATCCGCGGAGACATCGAGC and 5′ CTAATCTGCTCTCCTCTCCGTCCAAG. The product contained a unique SacII site present in the zebrafish bfsp2 coding sequence. The amplified product was cloned into pGEMTeasy, sequenced and then subcloned into the existing pET23 expression clone by substituting the existing SacII-NotI fragment with the truncated fragment.
Fig. 2. Zebrafish Bfsp2 is present in the cytoskeletal fraction of lens extracts.
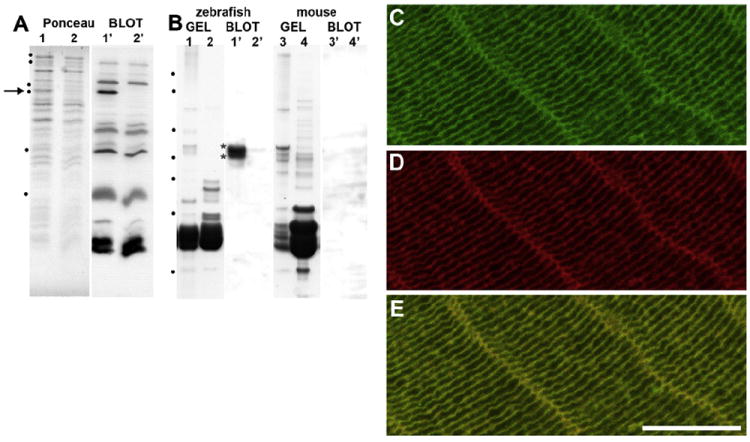
A. Characterisation of the rabbit polyclonal antibodies 2904 specific to zebrafish Bfsp2. Protein samples were separated by SDS-PAGE transferred to nitrocellulose and stained with Ponceau (Ponceau Stain), before probing with 2904 antibodies (blot). On the Ponceau stain image, lanes 1 and 2 show protein extract from the induced (lane 1) and uninduced (lane 2) recombinant Bfsp2α expression vector and the arrow in lane 1 indicates the expressed Bfsp2α. Marker proteins in order of increasing electrophoretic mobility are 116 kDa, 94 kDa, 58 kDa, 53 kDa, 29 kDa and 6.5 kDa are indicated by dots. B. Lanes 1 and 2 show protein extract from the cytoskeletal and soluble fractions respectively of the lens cortex from the lenses of 3 month old zebrafish. These samples have been separated using the 4–12% NuPAGE gel system. Lanes 1′ and 2′ is the corresponding immunoblot showing a pair of immunoreactive bands in the lens cytoskeletal fraction (blot lane 1′, asterisks). These were almost undetectable in the lens soluble fraction (blot, lane 2′). No immunoreactive bands were detected in similar fractions from mouse lenses (mouse; gel and blot). Notice the polyclonal antibodies 2904 do not cross react with lens crystallins from either mouse or zebrafish, although this antisera contains antibodies that strongly recognise bacterial proteins (A; blot lanes 1′ and 2′). Marker proteins in order of increasing electrophoretic mobility are 160 kDa, 110 kDa, 60 kDa, 50 kDa, 40 kDa, 30 kDa and 15 kDa are indicated by dots. C–E. Zebrafish C-terminal tail domain specific antibodies to zebrafish Bfsp2 (C; 1 in 2000 dilution; green channel) and TRITC-phalloidin (D; 0.5 μg/ml; red channel) were used to stain the same section from an adult zebrafish lens. Merging the green and red channels showed extensive overlap in Bfsp2 and actin as seen by the yellow signal (E). This representative image was taken from the lens cortex. The plasma membrane localisation of Bfsp2 in the zebrafish lens is apparent on both the short and long faces of the fibre cells in the lens cortex. Bar = 10 μm.
To generate human BFSP2 protein, the Geneservice clone (8322570; Unigene: Hs.659862) was obtained and the 5′ end of the cDNA PCR-amplified to contain an NdeI site at the initiating methionine, which was verified by DNA sequencing prior to subcloning into pET23.
Zebrafish (bfsp2) and human (BFSP2) constructs in the pET23 plasmids were expressed in E. coli BL21 pLysS strain as detailed previously (Der Perng et al., 2006; Perng et al., 2008, 2004b). Both human (BFSP2) and zebrafish (Bfsp2) proteins formed inclusion bodies and these were purified to homogeneity by ion exchange chromatography as described previously (Der Perng et al., 2006; Perng et al., 2008, 2004b).
2.3. Preparation of lens fractions
Lenses from approximately 2–6 adult (3–4 month) fish and mice were removed from the eye by dissection after terminal anesthesia of the animals. The lenses were collected into extraction buffer (10 mM sodium phosphate pH 7.4, 150 mM NaCl, 5 mM EDTA) and the outer lens fibre cells removed by trituration after piercing the lens capsule. The suspended cortical lens fibre cells were then homogenised on ice in 1 ml extraction buffer using a Dounce homogeniser before centrifuging for 10 min at 13,000 × g in a refrigerated Eppendorf microfuge. The pellet fraction was extracted once more with extraction buffer containing 1.5 M KCl, and washed once more in extraction buffer before solubilising the pellet directly in SDS buffer (10 mM Tris–HCl pH 7.0; 5 mM EDTA, 1% (w/v) SDS) prior to SDS-PAGE. Proteins in the supernatant fraction were precipitated using organic solvents (Wessel and Flügge, 1984) and then solubilised in SDS-PAGE sample buffer.
2.4. Purification of bovine BFSP1 and its 53-kDa fragment
Bovine BFSP1 and the BFSP1-53 kDa polypeptide (53 k) were purified from the cytoskeletal fraction of bovine lenses by a combination of anion-exchange and hydroxyapatite chromatography as described previously (Perng et al., 2004a; Quinlan et al., 1992). Fraction purity was assessed by SDS-PAGE prior to selection for in vitro assembly studies.
2.5. In vitro assembly studies
Selected fractions were dialysed into 8 M urea, 20 mM Tris–HCl pH 8.0, 2mM DTT. The urea concentrationwas sequentially reduced to 4 M, 2 M, 0 M by dialysis before finally initiating filament assembly by dialysis into 20 mM Tris–HCl pH 7.0, 50 mM NaCl, 1 mM MgCl2. All dialysis steps were carried out at 22 °C for 4 h. Protein concentrations were determined using the BCA assay and SDS-page gel densitometry. For coassembly, Bfsp2 and BFSP1 were mixed in a 1:1 mass ratio.
2.6. Sedimentation assays
In vitro sedimentation assays were carried out as described previously (Der Perng et al., 2006; Perng et al., 2008, 2004b). To investigate the extent of protein association into aggregates, assembled samples were analysed by low-speed centrifugation at 2500 × g for 15 min in a microfuge (Eppendorf, Hamburg, Germany) and by high-speed centrifugation (TLS55 rotor in a Beckman Optima max benchtop ultracentrifuge) at 80,000 × g for 30 min. The supernatant and pellet fractions were separated on 12% (w/v) polyacrylamide gels by SDS-PAGE and visualized by Coomassie Blue staining. The amount of protein in the supernatant and pellet fractions was determined by quantifying protein bands on stained gels using a luminescent image analyzer (LAS-1000plus, FujiFilm, Japan) and Image Gauge software (version 4.0, FujiFilm).
2.7. Antibodies
Polyclonal antibodies (Zf4902) specific for zebrafish Bfsp2 were generated in a rabbit using the peptide [C]-PEGPTDPTPSTSG-amide, a tridecameric peptide that corresponded to residues 407–419 in the zebrafish Bfsp2 sequence (Cambridge Research Biochemicals Ltd, Billingborough, Teeside, UK). BLAST analyses showed this peptide to be unique to the predicted zebrafish Bfsp2 protein. Primary antibodies were detected using Alexa 488 labelled mouse anti-rabbit antibodies (Jackson Labs). To detect actin, Texas Red labelled phalloidin was used.
2.8. Indirect immunofluorescence confocal light microscopy
Heads or lenses from adult zebrafish were quick frozen in isopentane at liquid nitrogen temperatures before being stored at −80 °C. Cryosections (8–10 μm) were cut using a Leica Cryostat and then fixed in 4% (w/v) paraformaldehyde (PFA) in phosphate buffered saline (PBS). The frozen sections were permeabilised with 1% (v/v) NP40, before blocking with normal goat serum (1:10). Sections were incubated with primary and secondary antibodies for 1 h at room temperature. Sections were mounted using hydromount with coverslips. Fluorescently labelled sections were imaged using a LSM 700 Confocal Microscope (Carl Zeiss Ltd, Welwyn Garden City, UK), Plan-Apochromat × 100, 1.46NA objective. Images were processed and prepared for figures using Adobe® Photoshop CS (Adobe System, USA).
2.9. SDS-PAGE and immunoblotting
SDS-PAGE was essentially as described (Perng et al., 2008) using 12% (w/v) polyacrylamide gels to separate the protein samples. Immunoblotting was performed using the semi-dry blotting method (Khyse-Anderson, 1984) according to the manufacturer’s specifications (Bio-Rad Laboratories, UK) and modified as described (Der Perng et al., 2006). In some cases precast Novex NuPAGE 4–12% (w/v) Bis-Tris polyacrylamide gels were used according to the manufacturers instructions (Invitrogen, USA).
2.10. Electron microscopy
Bfsp2 was diluted in assembly buffer to 5 μg/ml and negatively stained with 1% (w/v) uranyl acetate (Agar Scientific, UK). Samples were prepared using the Valentine method (Valentine et al., 1968) and the copper grids examined in an Hitachi H-7600 transmission electron microscope (Hitachi High-Technologies Corporation, Japan). An accelerating voltage of 100 kV was used. Images were acquired using a CCD camera (Advanced microscopy Technology, Danvers, MA). Width measurements of filaments were made on these images using ImageJ 1.44 (http://rsb.info.nih.gov/ij). 50 width measurements were made using three different images to give a total of 150 width measurements for each sample. Measurements were made at 100 nm intervals along the length of the filaments in randomly selected areas. Average and standard deviations of sample filament width were then calculated. Images for presentation were processed in Adobe® Photoshop CS (Adobe System, San Jose, CA).
2.11. Data mining and sequence analysis programmes
Isoelectric points and molecular weights of the protein products from the cDNA transcripts were calculated using the web resource at ExPASY (http://www.expasy.ch/cgi-bin/pi_tool). Database mining used TBLASTN 2.2.21 (Altschul et al., 1997) and NBLAST (Zhang et al., 2000). Sequence alignments were made using the clustalW2 package (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and Jalview (http://www.jalview.org/; (Waterhouse et al., 2009)). Unless otherwise stated default settings were used on all programmes.
3. Results and discussion
3.1. Teleost Bfsp2 contains a C-terminal non-α-helical tail domain
The publication of the trout bfsp2 sequence was the first to identify a C-terminal non-α-helical domain in a vertebrate Bfsp2. Further data mining of the genomic and EST databases (www.ncbi.nlm.nih.gov) identified cDNA clones for other fish species (stickleback, medaka, green-spotted puffer fish, gold bluetip cichlid) with C-terminal non-α-helical tail domains, including zebrafish. Interestingly the gene structure for these fish bfsp2 (stickleback, medaka, green-spotted puffer fish, zebrafish) revealed an exon–intron structure similar to, for instance, the type III intermediate filament vimentin (Fig. 1A) with the presence of two exons encoding this C-terminal tail domain. No database evidence for an alternatively spliced second exon to extend helix 1B of Bfsp2 in birds by 49 residues was found (Table 2). In zebrafish, however, two distinct splice variants (Bfsp2α, Bfsp2β) were identified that differed in the last ten residues of the C-terminus. The last exon (exon 7) contains a splice donor site, which is alternatively spliced as confirmed from the available EST and cDNA evidence (Table 2) to generate a product with a distinct C-terminus.
Fig. 1. Conservation of Bfsp2 from fishes to man.
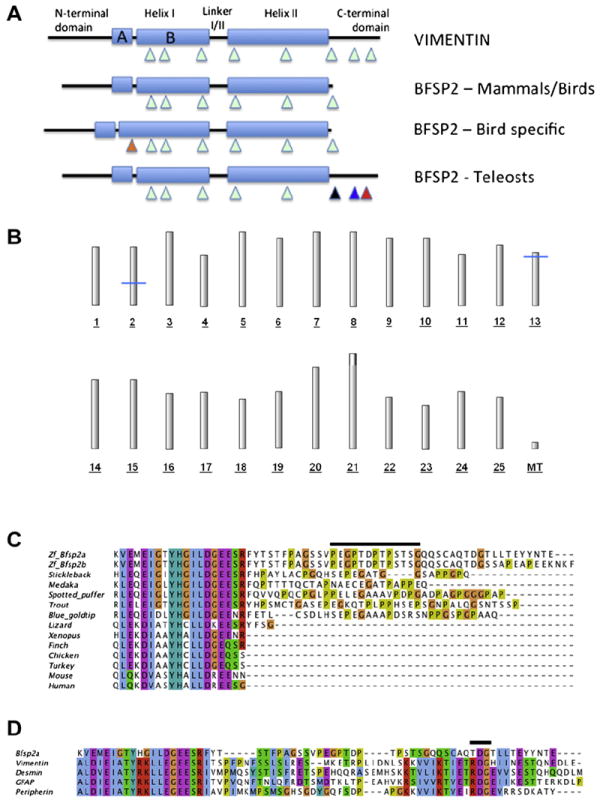
A. Schematic highlighting the major gene and protein differences between vertebrate BFSP2 genes. Exon locations are indicated (triangles) relative the major secondary sequence feature of the predicted protein. This contains two major α-helical domains (Helix I and II) punctuated by a linker region (Linker I/II), which is collectively called the rod domain. This is flanked on the N-terminal side by the non-α-helical head domain and on the C-terminus by the non-α-helical head tail domain. Compared to other mammalian intermediate filament proteins, such as vimentin, BFSP2 is distinct in that it misses a C-terminal tail domain. This feature is preserved in amphibian and avian orthologues. Interestingly, some birds (chicken and turkey) have a different feature, namely an alternatively spliced exon (orange triangle) that extends helix IB (BFSP2 – bird specific), which as first discovered for chicken BFSP2 (Wallace et al., 1998). In contrast to Xenopus, mammals and birds, BFSP2 genes of bony fishes have retained a non α-helical head C-terminal tail domain. This domain is usually specified by two exons (blue and black triangles). The position of the exon boundary immediately flanking helix 2 in avian bfsp2 (black triangle), is different to those of mammals. In zebrafish, the last exon is alternatively spliced (red triangle). B. Radiation hybrid mapping of the zebrafish beaded filament genes. Position of the zebrafish genes bfsp2 (CP49) and bfsp1 (filensin) on the T51 radiation hybrid map. The approximate positions of the genes are marked with blue lines; bfsp2 is located at a distance of approximately 47 cM from top of chromosome 2; bfsp1 is located approximately 7 cM from top of chromosome 13. These data confirm that there is a single bfsp2 gene and combining this with the data above suggest two transcripts (Bfsp2α, Bfsp2β) are produced by alternative splicing. C. CLUSTALW alignment of the C-terminal tail and TYRKLLEGE-motif sequences from the rod domain for various vertebrate BFSP2. Notice the sequence identity within the first 19 residues, which span the highly conserved TYRKLLEGE-motif at the C-terminal end of helix II. The sequence of the C-terminal tail domain in fish Bfsp2 appears not to be highly conserved between species, except to note that the region is rich in glycine and proline residues (lime and orange shaded residues respectively). Data for this Figure was mined from the ENSEMBL and GENBANK databases. Transcript predictions were confirmed independently by BLAST searches of the EST and nr databases. The presence of STOP codons was independently confirmed for the stickleback, xenopus and zebra finch BFSP2 sequences. The zebrafish gene (Danio rerio, ENSDARG00000011998) and its two predicted protein products (Bfsp2α, ENSDARP00000025416; Bfsp2β, ENSDARP00000111147) and genes for the other fishes, namely stickleback (Gasterosteus aculeatus, ENSGACG00000002441), medaka (Oryzias latipes, ENSORLG00000007668) and the spotted green puffer fish tetraodon (Tetraodon nigroviridis, ENSTNIG00000003071), were all analysed for evidence of alternative splicing in the C-terminal tail domain. The full-length cDNA sequence for trout (Oncorhynchus mykiss) was previously published (Binkley et al., 2002) and is supported by other cDNA data (Genbank: U071847.1 and FP319084.1), but no genomic data are available. The sequence for the blue goldtip cichlid (Ophthalmotilapia ventralis; (GenBank: JL564127.1; (Baldo et al., 2011)). These sequences were compared to transcripts derived from BFSP2 of representative tetrapods, namely human (Homo sapiens, ENSG00000170819), mouse (Mus musculus, ENSMUSG00000032556), chick (Gallus gallus, ENSGALG00000011712), turkey (ENSMGAG00000011532), zebra finch (Taeniopygia guttata, ENSTGUG00000004473) and finally Xenopus (Xenopus tropicalis, ENSXETG00000033786). The black line above the zebrafish Bfsp2α sequence identifies the peptide sequence used to generate polyclonal antibodies. This sequence is present in both splice variants (α and β) of zebrafish Bfsp2. D. Comparison of zebrafish Bfsp2 C-terminal tail sequence and neighbouring TYRKLLEGE-motif sequences to those of type III intermediate filament proteins, vimentin, GFAP, desmin and peripherin. The junction between the rod domain and the C-terminal tail domain in type III intermediate filament proteins, like Bfsp2, contains proline residues. The comparison of zebrafish Bfsp2α sequences with those from desmin (desma-201, ENSDARP00000075991/desmb-201, ENSDARP00000065355), GFAP (ENSDARP00000029607), peripherin (Prph, ENSDARP00000108086) and vimentin (Vim-201, ENSDARP00000016261) reveals additional similarities, including conservation of the RDG-motif and hydrophobic residue (I, L, V). This region also locates the last exon boundary. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Summary of bfsp2/Bfsp2 sequences resulting from database mining to illustrate the extent of the conservation of the helix IB extension and the presence of C-terminal tail domains.
| Species (splice variant) | Exon No | C-terminal Sequence (3′ end Exon 6 – Exon7 –Exon 8). Residues on the exon boundaries indicated in contrasting colours. | Exon 6/7 end phase | cDNA/EST evidence (GenBank ref. no) | ENSEMBL reference (UniGENE) |
|---|---|---|---|---|---|
| Danio rerio (Bfsp2α) | 7 | KVEMEIGTYHGILDGEESRFYTSTFPAGSSVPEGPTDPTPSTSGQQSC AQTDGTLLTEYYNTE* | 2 | YES (NM_001008633.1 and BC165202.1) | ENSDART00000004712 (UGID:112945; zgc:103750) |
| Danio rerio (Bfsp2β) | 8 | KVEMEIGTYHGILDGEESRFYTSTFPAGSSVPEGPTDPTPSTSGQQSC AQTDGSSAPEAPEEKNKF* | 2 and 1 | YES (EH478706.1, EH477771.1, EH477419.1, EH469985.1, EH469616.1, EH463619.1, EH463390.1) | ENSDART00000125889 |
| Gasterosteus aculeatus | 7a | HLEQEIGIYHGILDGEESRFHPAYLACPGQHSEPEGATGGSAPPGPQ* | 2 | YES (DW608316.1, DW604438.1, DW600648.1, DW593699.1, DN691906.1, DN692062.1, DW597869.1, DW597286.1, DN723004.1, DW600502.1, DN690742.1, DN685169.1, DW603481.1, DW600649, DN705778.1, DN723005.1) | ENSGACT00000003202 (UGID:1671045) |
| Oryzias latipes | 7 | KLEQEIGLYHGILDGEESRFQPTTTTQCTAPNAECEGATPAPPEQ* | 2 | YES (AM373876.1, AM354986.1, AM372182.1, BJ739079.1, FS537903.1, FS513580.1, FS538891.1, FS514567.1, FS540557.1, FS516248.1) | ENSORLT00000009608b |
| Tetraodon nigroviridisc | 7 | RLEQEIGLYHGILDGEESRFQVVQPQCPGLPPELEGAAAVPDPGA DPAGPGGGPAP* | 2 | YES (CR693727.1, CR688948.2, CR691352.2, CR704350.2, CR702307.2, CR689298.2, CR703338.1, CR694039.2, CR693777.2, CR682294.2, CR702596.2, CR682354.2, CR697322.2, CR703892.1, CR685998.2, CR693576.2, CR701067.2, CR699002.2) | ENSTNIT00000005791 |
| Oncorhynchus mykissd | NAe | RLELEIGTYHGILDGEESRYHPSMCTGASEPEGKQTPLPPHSEPSG NPALQGSNTSSP* | NA | YES (CU071847.1, FP319084.1) | NA |
| Ophthalmotilapia ventralis | NAe | f RLEQEIDLYHGILDGEENRFETLCSDLHSEPEGAAAPDSRSNPPG SPGPAAQ | NA | YES (JL564127.1) | NA |
| Xenopus tropicalisa | 7 | HLEKDIAAYHAILDGEENR* | 2 | YES (ES687881.1, ES680166.1) | ENSXETT00000060324 |
| Taeniopygia guttataa | 7 | QQLEKDIAAYHCLLDGEQSR* | 2 | YES (DV953306.1, FE712239.1) | ENSTGUT00000004669 |
| Gallas gallusa | 8h | QLEKDIAAYHCLLDGEQSS* | 2 | YES (NM_205218, X84807) | ENSGALG00000011712 andh ENSGALT00000019140. (UGID:117124) |
| Meleagris gallopavog | 8h | QLEKDIAAYHCLLDGEQSS* | 2 | NO | ENSMGAG00000011532 |
| Mus musculus | 7 | QLQKDVASYHALLDREENN* | 2 | YES (NM_001002896) | ENSMUSG00000032556 (UGID:1134439) |
| Home sapiens | 7 | QLQKDVASYHALLDREESG* | 2 | YES (NM_003571) | ENSG00000170819 (UGID:2729180) |
| Predicted sequence for a reptilian Bfsp2 | |||||
| Anolis carolinensisi | 7 | QLEKDIATYHCLLDKEESRYFSG* | 2 | NO | ENSACAG00000004403 |
Incomplete information in the ENSEMBL database, but Exon 7 containing an inframe STOP codon was deduced from available EST data.
Two transcripts given in the ENSEMBL database, but exon prediction for ENSORLT00000009605 is defies convention.
Two transcripts in the database, but there is no cDNA or EST evidence for ENSTNIT00000002586.
cDNA and predicted protein sequence published by Binkley et al.
Exon structure can not be confirmed.
The published cDNA sequence does not include an STOP codon.
In birds there is an extra exon that is alternatively spliced, which lengthens helix 1 by 49 residues. This exon was first identified in chicken. There is high sequence identity between G.gallus and M. gaaopavo and a similar sequence (VSYLSQEPRGSHRRRKTTLWELRRARRRQEQQVFHGSLRRTTLKRTANM) is present in intron 1 of T. guttata Bfsp2. There are issues affecting the predicted protein sequences for the all three ENSEMBL database entries. The insert sequences need to be formally proven for both turkey and zebra finch.
The predicted exon pattern for this splice variant claims 9 exons by splitting Exon 1 which accounts for this discrepancy.
This is predicted from an analysis of the genome sequence. As found for all other Bfsp2, the end phase of the penultimate exon is 2, and occurs just before an in frame STOP codon.
This presented the possibility that zebrafish possesses two bfsp2 copies as seen for desmin, which has genes located on chromosomes 6 (desmb; ENSDARG00000005221) and 9 (desma: ENSDARG00000058656). Coincidentally the desmin gene on zebrafish chromosome 9 is also alternatively spliced, and this too alters the very end of the C-terminal non-α-helical domain by inserting 15 residues before the last exon (cf ENSDARP00000075991 and ENSDARP00000075994). We confirmed that zebrafish possess only one bfsp2 by radiation hybrid mapping (Fig. 1B), in contrast to desmin and other single copy mammalian genes that are duplicated in zebrafish due to a genome wide duplication event (Postlethwait et al., 2004). The same holds for bfsp1 (Fig. 1B), the other major structural protein of beaded filaments and coassembly partner for Bfsp2. There appears therefore not to be any paralogues, only orthologues, to the human beaded filament genes in zebrafish.
Translation of the bfsp2 coding sequence for the one full-length cDNA (Unigene; Dr.19486. Genbank: NM_001008633.1. MGC: 103750. Clone ID: 7074672) revealed a C-terminal non-α-helical tail domain (Fig. 1C) that extended beyond the highly conserved helix termination motif (TYHGILDGEE). This splice variant we have designated Bfsp2α. The other splice variantof the zebrafish bfsp2 is longer by 3 residues and has replaced the last 10 residues of Bfsp2α with a newexon.We designate this Bfsp2β (see Fig. 1B). All mammalian, avian and the Xenopus orthologues thus far described lack this C-terminal domain ((Perng et al., 2004a, 2007); Fig. 1C). The available avian, mammalian and Xenopus sequences have only a two residue extension beyond the helix termination motif, whereas the anole lizard is predicted to have a six residue extension, which is more similar to the position of the exon 6 donor site in the fish gene sequences (Fig. 1C). Whilst all the available fish Bfsp2 sequences possess C-terminal sequences that extend beyond the helix termination motif, no consensus sequence is readily apparent, although proline and glycine residues are well represented (Fig. 1C).
Comparison of the Bfsp2α sequence to vimentin, desmin, GFAP and peripherin sequences of zebrafish (Fig. 1D) highlighted two features. Firstly, the type III proteins also contained prolines and sometimes glycines (peripherin) adjacent to the helix termination motif. Secondly, type III proteins contain a RDG-motif in the C-terminal tail domain and this feature is partially maintained in Bfsp2α (Fig. 1D). These data suggest that the tail domain of zebrafish Bfsp2α is also important to filament assembly like the C-terminal tail domain in mammalian type III proteins.
3.2. Bfsp2 is expressed in the zebrafish lens and is present in the cytoskeletal fraction
In order to demonstrate that the single bfsp2 gene gives rise to a gene product containing the extra C-terminal tail sequences, a rabbit polyclonal antibody was raised against a tridecameric peptide in this region (Fig.1C, black line). The characterization of this antibody used two different approaches (Fig. 2A, B). In the first, the zebrafish bfsp2 coding sequence was inserted into a bacterial expression vector based on pET23. Induction of protein expression (Fig. 2A Ponceau, lane 1) produced a single immunoreactive band (Fig. 2A blot; lane 1′) compared to an uninduced culture (Fig. 2A blot, lane 2′). In the second approach, soluble and cytoskeletal fractions from zebrafish lenseswere prepared. The zebrafish Bfsp2 C-terminal specific antibodies detected two immunoreactive bands in the cytoskeletal fraction of the zebrafish lens extract (Fig. 2B blot, lane 1′). These were absent from the soluble fraction (Fig. 2B blot; lane 2′). Note too that the antibodies do not cross-react with any other zebrafish lens proteins, despite the abundance of the lens crystallins in the lower half of the gel (Fig. 2B blot; lanes 1′ and 2′). The antibodies failed to recognize proteins in similar fractions prepared from mouse lenses (Fig. 2B, mouse). These data show that we have generated a polyclonal antibody reagent capable of selectively detecting zebrafish Bfsp2 and specifically an epitope in the C-terminal tail domain. Moreover, these data also show that this protein resides primarily in the cytoskeletal fraction of lens extracts.
Using these antibodies, confocal immunofluorescence light microscopy was performed on frozen sections of adult zebrafish lenses to locate Bfsp2 (Fig. 2C) in the lens fibre cells and counterstaining the same sections for actin (Fig. 2D). Bfsp2 was seen to locate primarily to the plasma membranes of lens fibre cells (Fig. 2C) supporting the biochemical fractionation data (Fig. 2B). Phalloidin, a label for F-actin, also highlights the plasma membrane profiles of the fibre cells (Fig. 2D) and from the merged green and red signals (Fig. 2E), both actin and Bfsp2 are seen to concentrate at the plasma membrane of the cortical fibre cells. The zebrafish Bfsp2 signal located to both the short and the long faces (Fig. 2C) of the fibre cell profiles. These data demonstrate that in zebrafish lenses, Bfsp2 containing the C-terminal tail sequences is present in the cytoskeletal fraction and predominantly locates to fibre cell plasma membranes in the cortical regions, just as has been reported for mammalian Bfsp2 (Sandilands et al., 1995).
3.3. Functional consequences of the C-terminal tail domain of zebrafish Bfsp2 – effects upon protein assembly in vitro
The biochemical and immunolocalisation data suggest that the presence of the extra C-terminal tail domain in zebrafish Bfsp2 does not prevent its correct compartmentalisation to the cytoskeletal fraction of lens fibre cell extracts as seen for mammalian orthologues (Sandilands et al., 1995). We pursued this further by determining the effect of the zebrafish Bfsp2 C-terminal non-α-helical domain upon filament formation in vitro. We assembled recombinant Bfsp2 proteins with an assembly partner, BFSP1 (reviewed in (Song et al., 2009)). Both BFSP1 and its 53 kDa fragment, BFSP1-53 k (53 k) have been shown previously to be assembly competent. Mixing purified BFSP2 with BFSP1 leads to the formation of 10 nm filaments under appropriate in vitro assembly conditions (Carter et al., 1995; Goulielmos et al., 1996). Therefore native bovine BFSP1 and its 53 kDa fragment were selected as surrogate assembly partners for the recombinantly produced BFSP2/Bfsp2α proteins in this study. The ability of these beaded filament protein combinations to assemble into filaments was then assessed by sedimentation assay (Fig. 3A, B) and electron microscopy (Fig. 3C). The two sedimentation assays measure different aspects of protein assembly. In the high-speed assay all the assembled filaments will be sedimented, whilst in the low-speed assay, only filaments that self-associate will be sedimented. The former therefore gives an estimation of the assembly efficiency of the protein, whilst the latter assesses the self-association properties of the assembled material.
Fig. 3. In vitro assembly of zebrafish Bfsp2 with BFSP1.
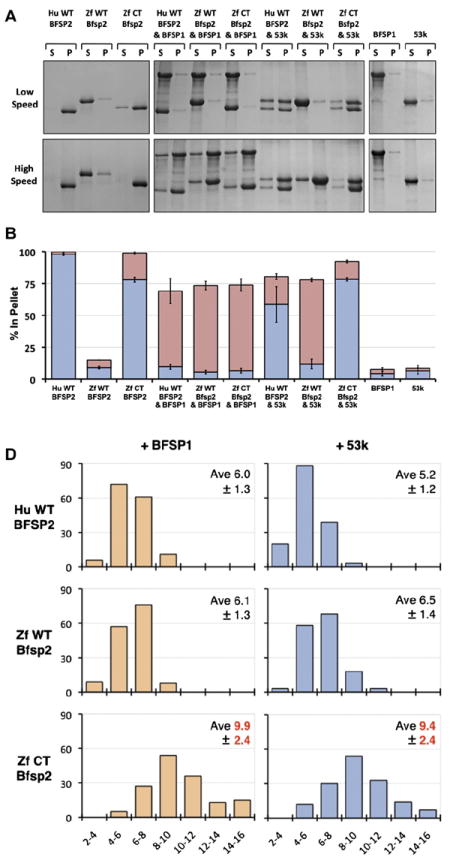
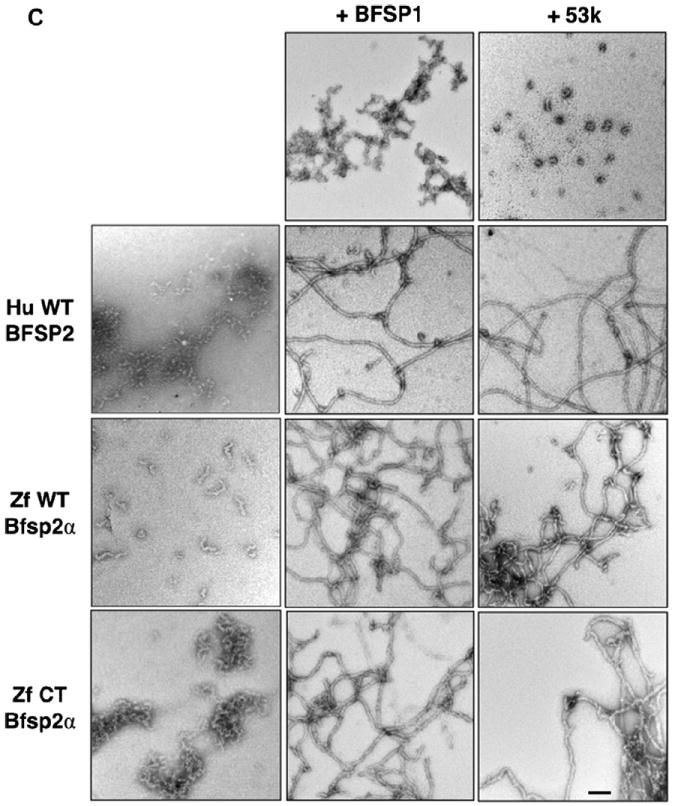
A Assembly of human (Hu WT) and zebrafish (wild-type (Zf WT) and C-terminally truncated (Zf CT)) Bfsp2 proteins with bovine native BFSP1 or bovine native BFSP1-53 kDa (53 k). Prior to assembly, the named protein combinations were mixed in 1:1 mass ratios. Assembly was analysed by low- and high-speed sedimentation assays. Representative examples from supernatant (s) and pellet (p) fractions obtained after low- and high-speed sedimentation assays are shown. By sedimentation assay, both bovine BFSP1 and its 53 kDa (53 k) fragment remained mostly (>90%) in the soluble fractions when assembled alone. Only when these proteins were combined with an appropriate assembly partner were they found in the pellet fractions. Notice that wild-type zebrafish Bfsp2α was as efficient as human wild-type BFSP2 in shifting BFSP1 and its 53 kDa fragment to the pellet fraction in the high-speed sedimentation assay (Zf WT Bfsp2α & BFSP1, red bar; Zf WT Bfsp2α & 53 k, red bar). In fact, coassembly with BFSP1 also shifted wild-type zebrafish Bfsp2α to the pellet fraction indicative of a compatible partnership for coassembly. B. Densitometric quantification of low- (blue) and high-speed (red) sedimentation data from ‘A’. The filaments formed by a combination of zebrafish wild-type Bfsp2α and bovine BFSP1-53 kDa fragment (ZfWT Bfsp2α & 53 k) self-associate less than those formed when C-terminally truncated Bfsp2α or human wild-type BFSP2 were combined with the 53 kDa fragment of BFSP1 (Zf CT Bfsp2α & 53 k and HuWT BFSP2 & 53 k respectively), as shown by the low-speed sedimentation assay. Filaments formed by a combination of zebrafish Bfsp2α and bovine BFSP1 self-associate less then those formed by a combination of zebrafish Bfsp2α and BFSP1-53 kDa (cf blue columns for Zf WT Bfsp2α & BFSP1 and Zf WT Bfsp2α & 53 k). C. Electron microscopy characterisation of the in vitro assembled combinations of zebrafish wild-type Bfsp2α (Zf WT Bfsp2α), C-terminally truncated zebrafish Bfsp2 (Zf CT Bfsp2α), human BFSP2 (Hu WT BFSP2) with either BFSP1 (BFSP1) or BFSP1-53 kDa fragment (53 k). The left hand column shows the Bfsp2 proteins assembled alone. The centre and right hand columns show the Bfsp2 proteins assembled with BFSP1 or BFSP1-53 kDa fragment respectively. The first row shows BFSP1 and BFSP1-53 kDa assembled alone. The same magnification was used for all micrographs. Bar = 100 nm. D. Effect of the zebrafish C-terminal tail sequences upon the width of in vitro assembled filaments. The left and right hand columns show filament width distributions when the Bfsp2 proteins are assembled with BFSP1 or BFSP1-53 kDa respectively. The x axis show filament width catagories (nm) and the y axis show measurement frequency. The average (Ave) and standard deviation (±) of filament widths are also shown. A total of 150 width measurements were made for each sample. Filaments formed by Zf CT Bfsp2α are significantly larger and more varied in width than those formed by Hu WT BFSP2 or Zf WT Bfsp2α. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We analysed first the assembly properties of zebrafish Bfsp2α alone and compared this to human BFSP2. Human BFSP2 sediments in both the low- and high-speed assays (Fig. 3A, B; Hu WT BFSP2), but the wild-type zebrafish Bfsp2α (Fig. 3A, B; Zf WT Bfsp2α) remained soluble in both assays. In fact, nearly 90% of the zebrafish protein remained soluble whereas all of human BFSP2 pelletted (Fig. 3A, B). Negatively stained samples of the assembled material (Fig. 3C) confirmed these sedimentation characteristics as the filamentous material formed by the human BFSP2 tended to selfassociate. In contrast, zebrafish Bfsp2α formed short fibrils that did not self-associate (Fig. 3C). We reasoned that the additional C-terminal sequences were the likely cause of this significant change in solubility for zebrafish Bfsp2α. Therefore we also generated a zebrafish equivalent to the human Bfsp2 (see Fig. 1C) by removing the C-terminal 44 residues by site directed mutagenesis before expressing and purifying this protein (Zf CT Bfsp2α) for in vitro assembly studies. Now the in vitro assembly properties of the zebrafish Bfsp2α was dramatically changed by the deletion of the C-terminal tail domain as seen by the very significant shift is sedimentation properties of Zf CT Bfsp2α in both the high- and low-speed assay (Fig. 3A, B).
Human (BFSP2) and both the wild-type and C-terminally truncated zebrafish Bfsp2α recombinant proteins formed filaments when coassembled with either bovine BFSP1 (Fig. 3C, ZfWT Bfsp2α & BFSP1) or its 53 kDa fragment (Fig. 3C, ZfWT Bfsp2α & 53 k). These data show that the zebrafish Bfsp2α is indeed assembly competent when combined with an appropriate assembly partner, such as bovine BFSP1. These data also confirm that zebrafish Bfsp2 is indeed a functional orthologue of mammalian BFSP2, at least in this in vitro assembly assay. The high-speed centrifugation assay showed zebrafish Bfsp2α to be as efficient as human BFSP2 in forming filaments with both bovine BFSP1 and its assembly competent 53 kDa fragment ((Carter et al., 1995); Fig. 3A, B). Interestingly, filament aggregation (Fig. 3B blue bars) was significantly greater for coassembly assays involving the BFSP1-53 kDa fragment assembled with either human BFSP2 (Fig. 3A, B blue bars; HuWT BFSP2 & 53 k) or the zebrafish C-terminally truncated Bfsp2 (Fig. 3A, B blue bars; Zf CT Bfsp2α & 53 k) when compared to wild-type zebrafish Bfsp2α (Fig. 3A, B blue bars; ZfWT Bfsp2α & 53 k). Filament self-association was detected by the low-speed centrifugation assay and so these data (Fig. 3B blue bars) therefore suggest that the C-terminal sequences of zebrafish Bfsp2α help prevent filament self-association. Indeed the data from this low-speed assay also suggest that the C-terminal tail of BFSP1 contains sequences that also prevent filament–filament interactions, as seen from the human wild-type BFSP2 and bovine BFSP1 combination (Fig. 3A, B; Hu WT BFSP2 & BFSP1) compared with the human wild-type and BFSP1-53 kDa combination (Fig. 3 A, B; HuWT BFSP2 & 53 k). Closer scrutiny of the morphology of the assembled filaments (Fig. 3C) also suggested that the presence of the C-terminal sequences of Bfsp2α produced filaments with BFSP1 and its 53 kDa fragment that were more uniform in width (Fig. 3D). Therefore we suggest that these C-terminal sequences of Bfsp2α not only help prevent filament aggregation, but also contribute directly to the in vitro assembly process as seen for other intermediate filament proteins by helping regulate filament width (reviewed in (Herrmann and Aebi, 2004; Herrmann et al., 1996)). On the evidence presented here, the C-terminal non-α-helical tail domain of Bfsp2α is not redundant within the context in its in vitro assembly properties.
3.4. Implications for the lens function and the evolution of intermediate filaments
Zebrafish Bfsp2α has distinct assembly properties when compared to the human BFSP2 orthologue. The study here shows that the additional C-terminal sequences of zebrafish Bfsp2α has significant effects on its in vitro assembly as it regulates both the filament–filament association properties and width control of the assembled filaments in vitro. Although the human BFSP2 is functionally equivalent to zebrafish Bfsp2α in being able to regulate filament width when coassembled with BFSP1, its role in regulating filament–filament associations is not entirely equivalent. Our data show that the C-terminal non-α-helical tail sequences of BFSP1 can also contribute to filament–filament associations in vitro (Fig. 3A, B), which is not the case when BFSP1 is coassembled with Bfsp2α (Fig. 3A, B). Therefore, the loss of the C-terminal non-α-helical tail domain in human BFSP2 is not without some consequences for its in vitro assembly properties (Fig. 3) and our data suggest some compensation by the C-terminal non-α-helical tail domain in BFSP1, particularly with respect to regulating filament–filament interactions (Fig. 3A, B, low-speed).
The eye lens is believed to be under significant selection pressure given its key role in the visual process (Kappe et al., 2010; Kröger et al., 2009). Could there be a functional explanation for such a difference between mammalian and fish BFSP2? The fish lens does not accommodate, but there is a huge variation of functional requirements even amongst teleosts to be met by the lens proteins. Large size differences exist as well as lenses with short or long normalized focal lengths, that are monofocal or multifocal is different teleosts (Kröger et al., 2009). In cichlids, the lens has the property of changing its refractive power of the lens cortex in phase with light–dark cycles (Schartau et al., 2010). Interestingly, phosphorylation of Bfsp2 is a mechanism to alter the subcellular distribution of beaded filaments (Ireland et al., 1993) and so Bfsp2 and beaded filaments as well as lens crystallins (Kappe et al., 2010) are potential candidates to elicit such a change in lens refractive power. Thus far, however, it is only zebrafish that have evidence of alternative splicing to further increase Bfsp2 diversity (see Table 1), but where this occurs in mammalian intermediate filament proteins it can change the assembly properties (Perng et al., 2008) and alter function (Langbein et al., 2010).
The studies here provide further evidence (Perng et al., 2007; Wallace et al., 1998) of the flexibility tolerated in domain structure when it comes to the in vitro assembly of beaded filament proteins. It is remarkable that vertebrate Bfsp2 orthologues exhibit such distinctive sequence features that include an extended helix 1B and the loss of the C-terminal tail domain. Both are very unusual amongst the intermediate filament protein family for cytoplasmic proteins in vertebrates (Herrmann and Strelkov, 2011), but offer a unique opportunity to detail specific events in the evolution of the intermediate filament protein family. The studies here suggest that the C-terminal non-α-helical tail domain is functional for fish Bfsp2 proteins. Birds appear to be the only vertebrates to possess an extended helix 1B, albeit in a splice variant that is a minor component of the lenticular cytoskeleton (Wallace et al., 1998). In cephalopods, lens specific intermediate filament proteins share both these features (Tomarev et al., 1993), which is typical of the cytoplasmic intermediate filament proteins in invertebrates (Herrmann and Strelkov, 2011). The studies here suggest that the loss of the C-terminal tail occurred between fish and tetrapods. The Bfsp2 sequence predicted for the anole lizard (Fig. 1C) possibly indicates a transition point as the splice site location for the last exon is more fish-like than mammalian-like, suggesting the loss of the C-terminal domain is a multi-step process. We present evidence that one of these could involve reducing the coding sequence of the last exon to complete the penultimate exon and add a STOP codon. Another is the moving the exon splice site closer to the helix termination motif. The studies here demonstrate that zebrafish Bfsp2α is a functional orthologue of human BFSP2 and therefore offers the possibility to explore the functional implications of Bfsp2 sequence differences in terms of lens function by genetic complementation in animal models.
Acknowledgments
RAQ, LYZ and ZQJ acknowledge the financial support for the Royal Society for the International Collaborative Grant that supported these studies. RAQ was supported by a Leverhulme research leave fellowship (0546) and support of the National Eye Institute (EY04542, to JIC). AL is supported by a BBSRC CASE award. Financial assistance to QB and YzL by National Natural Science Foundation of China (Grant No. 30973277), the Natural Science Foundation (Grant No. 7001571) and the Science and Technology Planning Project of Guangdong Province, China (Grant No. 2007B060401050) is gratefully acknowledged. We thank Terry Gibbons for technical support.
Abbreviations
- Bfsp1
beaded filament structural protein 1
- Bfsp2
beaded filament structural protein 2
- WT
wild-type
- CT
C-terminally truncated
- dpf
days post fertilization
- hpf
hours post fertilization
- MWt
molecular weight
References
- Alexander LE. An experimental study of the role of optic cup and overlying ectoderm in lens formation in the chick embryo. J Exp Zool. 1937;75:41–73. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Santos ME, Salzburger W. Comparative transcriptomics of eastern African cichlid fishes shows signs of positive selection and a large contribution of untranslated regions to genetic diversity. Genome Biol Evol. 2011;3:443–455. doi: 10.1093/gbe/evr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkley PA, Hess J, Casselman J, FitzGerald P. Unexpected variation in unique features of the lens-specific type I cytokeratin CP49. Invest Ophthalmol Vis Sci. 2002;43:225–235. [PubMed] [Google Scholar]
- Bousquet O, Ma L, Yamada S, Gu C, Idei T, Takahashi K, Wirtz D, Coulombe PA. The nonhelical tail domain of keratin 14 promotes filament bundling and enhances the mechanical properties of keratin intermediate filaments in vitro. J Cell Biol. 2001;155:747–754. doi: 10.1083/jcb.200104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JM, Hutcheson AM, Quinlan RA. In vitro studies on the assembly properties of the lens beaded filament proteins: co-assembly with α-crystallin but not with vimentin. Exp Eye Res. 1995;60:181–192. doi: 10.1016/s0014-4835(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Dahm R, Geisler R, Nüsslein-Volhard C. Zebrafish (Dario rerio) genome and genetics. In: Meyers RA, editor. Encyclopedia of Molecular Cell Biology and Molecular Medicine. Wiley-VCH; Weinheim: 2005. pp. 593–626. [Google Scholar]
- Der Perng M, Su M, Wen SF, Li R, Gibbon T, Prescott AR, Brenner M, Quinlan RA. The Alexander disease-causing glial fibrillary acidic protein mutant, R416W, accumulates into Rosenthal fibers by a pathway that involves filament aggregation and the association of alpha B-crystallin and HSP27. Am J Hum Genet. 2006;79:197–213. doi: 10.1086/504411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler R. Mapping and cloning. In: Nüsslein-Volhard C, Dahm R, editors. Zebrafish – A Practical Approach. Oxford University Press; Oxford: 2002. pp. 175–212. [Google Scholar]
- Goulielmos G, Gounari F, Remington S, Muller S, Haner M, Aebi U, Georgatos SD. Filensin and phakinin form a novel type of beaded intermediate filaments and coassemble de-novo in cultured-cells. J Cell Biol. 1996;132:643–655. doi: 10.1083/jcb.132.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Haner M, Brettel M, Muller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Strelkov SV. History and phylogeny of intermediate filaments: now in insects. BMC Biol. 2011;9:16. doi: 10.1186/1741-7007-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland ME, Klettner C, Nunlee W. Cyclic AMP-mediated phosphorylation and insolubilization of a 49-kDa cytoskeletal marker protein of lens fiber terminal differentiation. Exp Eye Res. 1993;56:453–461. doi: 10.1006/exer.1993.1058. [DOI] [PubMed] [Google Scholar]
- Ireland ME, Wallace P, Sandilands A, Poosch M, Kasper M, Graw J, Liu A, Maisel H, Prescott AR, Hutcheson AM, Goebel D, Quinlan RA. Up-regulation of novel intermediate filament proteins in primary fiber cells: an indicator of all vertebrate lens fiber differentiation? Anat Rec. 2000;258:25–33. doi: 10.1002/(SICI)1097-0185(20000101)258:1<25::AID-AR3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Kappe G, Purkiss AG, van Genesen ST, Slingsby C, Lubsen NH. Explosive expansion of betagamma-crystallin genes in the ancestral vertebrate. J Mol Evol. 2010;71:219–230. doi: 10.1007/s00239-010-9379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khyse-Anderson J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Meth. 1984;10:203–211. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Kim S, Coulombe PA. Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev. 2007;21:1581–1597. doi: 10.1101/gad.1552107. [DOI] [PubMed] [Google Scholar]
- Kröger RH, Fritsches KA, Warrant EJ. Lens optical properties in the eyes of large marine predatory teleosts. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2009;195:175–182. doi: 10.1007/s00359-008-0396-1. [DOI] [PubMed] [Google Scholar]
- Langbein L, Eckhart L, Rogers MA, Praetzel-Wunder S, Schweizer J. Against the rules: human keratin K80: two functional alternative splice variants, K80 and K80.1, with special cellular localization in a wide range of epithelia. J Biol Chem. 2010;285:36909–36921. doi: 10.1074/jbc.M110.161745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier JF, Kas J, Hartwig J, Vegners R, Janmey PA. Mechanical effects of neurofilament cross-bridges. Modulation by phosphorylation, lipids, and interactions with F-actin. J Biol Chem. 1996;271:15687–15694. doi: 10.1074/jbc.271.26.15687. [DOI] [PubMed] [Google Scholar]
- Lin YC, Broedersz CP, Rowat AC, Wedig T, Herrmann H, Mackintosh FC, Weitz DA. Divalent cations crosslink vimentin intermediate filament tail domains to regulate network mechanics. J Mol Biol. 2010;399:637–644. doi: 10.1016/j.jmb.2010.04.054. [DOI] [PubMed] [Google Scholar]
- Lowrie DJ, Jr, Stickney JT, Ip W. Properties of the nonhelical end domains of vimentin suggest a role in maintaining intermediate filament network structure. J Struct Biol. 2000;132:83–94. doi: 10.1006/jsbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Merdes A, Gounari F, Georgatos SD. The 47-kD lens-specific protein phakinin is a tailless intermediate filament protein and an assembly partner of filensin. J Cell Biol. 1993;123:1507–1516. doi: 10.1083/jcb.123.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Jorgensen AL. Structural and functional characterization of the zebrafish gene for glial fibrillary acidic protein, GFAP. Gene. 2003;310:123–132. doi: 10.1016/s0378-1119(03)00526-2. [DOI] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Tao GZ, Toivola DM, Liao J. “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem Sci. 2006;31:383–394. doi: 10.1016/j.tibs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Parry DA. Microdissection of the sequence and structure of intermediate filament chains. Adv Protein Chem. 2005;70:113–142. doi: 10.1016/S0065-3233(05)70005-X. [DOI] [PubMed] [Google Scholar]
- Perng MD, Quinlan RA. Seeing is believing! The optical properties of the eye lens are dependent upon a functional intermediate filament cytoskeleton. Exp Cell Res. 2005;305:1–9. doi: 10.1016/j.yexcr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Perng MD, Sandilands A, Kuszak J, Dahm R, Wegener A, Prescott AR, Quinlan RA. The intermediate filament systems in the eye lens. Methods Cell Biol. 2004a;78:597–624. doi: 10.1016/s0091-679x(04)78021-8. [DOI] [PubMed] [Google Scholar]
- Perng MD, Wen SF, Gibbon T, Middeldorp J, Sluijs JA, Hol EM, Quinlan RA. GFAP filaments can tolerate the incorporation of assembly-compromised GFAP-{delta}, but with consequences for filament Organization and {alpha}B-crystallin association. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-03-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng MD, Wen SF, vanden IP, Prescott AR, Quinlan RA. Desmin aggregate formation by R120G alphaB-crystallin is caused by altered filament interactions and is dependent upon network status in cells. Mol Biol Cell. 2004b;15:2335–2346. doi: 10.1091/mbc.E03-12-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng MD, Zhang Q, Quinlan RA. Insights into the beaded filament of the eye lens. Exp Cell Res. 2007;313:2180–2188. doi: 10.1016/j.yexcr.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Quinlan RA, Carter JM, Hutcheson AM, Campbell DG. The 53 kDa polypeptide component of the bovine fibre cell cytoskeleton is derived from the 115 kDa beaded filament protein: evidence for a fibre cell specific intermediate filament protein. Curr Eye Res. 1992;11:909–921. doi: 10.3109/02713689209033488. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Prescott AR, Carter JM, Hutcheson AM, Quinlan RA, Richards J, FitzGerald PG. Vimentin and CP49/Filensin form distinct networks in the lens which are independently modulated during lens fibre cell differentiation. J Cell Sci. 1995;108:1397–1406. doi: 10.1242/jcs.108.4.1397. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Prescott AR, Wegener A, Zoltoski RK, Hutcheson AM, Masaki S, Kuszak JR, Quinlan RA. Knockout of the intermediate filament protein CP49 destabilises the lens fibre cell cytoskeleton and decreases lens optical quality, but does not induce cataract. Exp Eye Res. 2003;76:385–391. doi: 10.1016/s0014-4835(02)00330-5. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Wang X, Hutcheson AM, James J, Prescott AR, Wegener A, Pekny M, Gong X, Quinlan RA. Bfsp2 mutation found in mouse 129 strains causes the loss of CP49 and induces vimentin-dependent changes in the lens fibre cell cytoskeleton. Exp Eye Res. 2004;78:109–123. doi: 10.1016/j.exer.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Schartau JM, Kroger RH, Sjogreen B. Dopamine induces optical changes in the cichlid fish lens. PLoS One. 2010;5:e10402. doi: 10.1371/journal.pone.0010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Landsbury A, Dahm R, Liu Y, Zhang Q, Quinlan RA. Functions of the intermediate filament cytoskeleton in the eye lens. J Clin Invest. 2009;119:1837–1848. doi: 10.1172/JCI38277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak PC, Lane EB. Sequence of cDNA coding for human keratin 19. Nucl Acids Res. 1987;15:10058. doi: 10.1093/nar/15.23.10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomarev SI, Zinovieva RD, Piatigorsky J. Primary structure and lens-specific expression of genes for an intermediate filament protein and a beta-tubulin in cephalopods. Biochim Biophys Acta. 1993;1216:245–254. doi: 10.1016/0167-4781(93)90151-3. [DOI] [PubMed] [Google Scholar]
- Valentine RC, Shapiro BM, Stadtman ER. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry. 1968;7:2143–2152. doi: 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]
- Wallace P, Signer E, Paton IR, Burt D, Quinlan R. The chicken CP49 gene contains an extra exon compared to the human CP49 gene which identifies an important step in the evolution of the eye lens intermediate filament proteins. Gene. 1998;211:19–27. doi: 10.1016/s0378-1119(98)00117-6. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]


