Abstract
Background and purpose
Reported associations between risk of radiation-induced normal tissue injury and single nucleotide polymorphisms (SNPs) in TGFB1, encoding the pro-fibrotic cytokine transforming growth factor-beta 1 (TGF-β1), remain controversial. To overcome publication bias, the international Radiogenomics Consortium collected and analysed individual patient level data from both published and unpublished studies.
Materials and methods
TGFB1 SNP rs1800469 c.-1347T>C (previously known as C-509T) genotype, treatment- related data, and clinically-assessed fibrosis (measured at least 2 years after therapy) were available in 2782 participants from 11 cohorts. All received adjuvant breast radiotherapy. Associations between late fibrosis or overall toxicity, reported by STAT (Standardised Total Average Toxicity) score, and rs1800469 genotype were assessed.
Results
No statistically significant associations between either fibrosis or overall toxicity and rs1800469 genotype were observed with univariate or multivariate regression analysis. The multivariate odds ratio (OR), obtained from meta-analysis, for an increase in late fibrosis grade with each additional rare allele of rs1800469 was 0.98 (95% Confidence Interval (CI) 0.85–1.11). This CI is sufficiently narrow to rule out any clinically relevant effect on toxicity risk in carriers vs. non-carriers with a high probability.
Conclusion
This meta-analysis has not confirmed previous reports of association between fibrosis or overall toxicity and rs1800469 genotype in breast cancer patients. It has demonstrated successful collaboration within the Radiogenomics Consortium.
Keywords: Meta-analysis, Radiotherapy, Toxicity, Adverse effects
During the last decade, a number of studies reported associations between risk of radiation therapy related normal tissue injury and specific genetic variants [1–3]. Some of the most frequently studied were single nucleotide polymorphisms (SNPs) in TGFB1, which encodes transforming growth factor-beta 1 (TGF-β1).
TGF-β1 is a pro-fibrotic cytokine, which stimulates differentiation of fibroblasts and production of extracellular matrix and inhibits epithelial repair. It is strongly implicated in the atrophic-fibrogenic response pathway that is a main component of normal tissue injury after radiation therapy. The early phase of fibrogenesis, after irradiation, may be considered as a wound-healing response with up-regulation of pro-inflammatory cytokines, such as TGF-β1 [4]. Endothelial cell killing leading to vascular damage and macrophage activation both contribute to tissue hypoxia, which in turn perpetuates fibrosis. The role of TGF-β1 in the radiation response may be multi-factorial – related to development of fibrosis, extracellular signalling, induction of apoptosis and inhibition of proliferation in response to DNA damage [4].
TGF-β1 has been implicated in the development of radiation-related fibrosis in lung cancer patients. Some, but not all studies, reported a correlation between elevated serum TGF-β1 levels and increased fibrosis after cancer radiotherapy [5]. The hitherto largest study on SNPs in TGFB1 (with 778 participants) found no association with late radiotherapy toxicity [6]. In an accompanying meta-analysis of all published studies on late effects, where odds-ratios were reported or could be calculated, for the TGFB1 SNP c.-1347T>C (previously known as C-509T), rs1800469 [2], odds ratios of about 1.5 were reported with confidence intervals just above one. However, the possibility of publication bias was noted as the distribution of reported associations was characterised by small studies with high odds ratios, larger studies clustering around the ‘line of no effect’, and a relative absence of small studies reporting an inverse association.
The SNPs most frequently studied in TGFB1 are rs1800469 c.-1347T>C and rs1800470 (formerly rs1982073) c.29C>T (known as T+29C) encoding Leu10Pro. SNP rs1800469 is in the promoter region of TGFB1 but is not within any putative promoter regulatory elements. SNP rs1800470 is in the signal peptide. The two SNPs have been reported to act by altering the rate of secretion of TGF-β1 and hence the circulating levels of mature protein. However, not all studies have demonstrated such a relationship [7] – the presence, in the circulation, of both latent and activated forms may complicate the interpretation of studies on circulating levels.
In an attempt to weigh the evidence from multiple studies and overcome problems of publication bias, the international Radiogenomics Consortium [8] collected data from both published and unpublished studies on SNPs in TGFB1 and radiation-induced normal tissue injury. In contrast to the majority of previous studies, multivariate analysis, adjusting for patient and treatment variables (covariates) associated with the risk of normal tissue injury, was possible.
Materials and methods
Patient data
Genotype of SNPs in TGFB1 and late toxicity data on 5555 patients from 21 different cohorts were collected from members of the international Radiogenomics Consortium, including 3257 patients with breast cancer, 78 with cervical cancer, and 2220 with prostate cancer (Table 1 and Supplementary Fig. 1). The current analysis includes data from the breast cancer patients. Data from patients with other tumour types will be investigated in subsequent analysis. In addition, data on patient- and treatment-related factors were also collected from each cohort. Patient-related factors included age, smoking status, body mass index (BMI), breast volume and the presence of co-morbidity such as diabetes mellitus and hypertension. Treatment-related factors included total dose, number of fractions, use of a radiotherapy boost, chemotherapy, hormone-therapy, acute toxicity, post-operative infection and surgical cosmesis. Due to the retrospective nature of our meta-analysis not all covariates were available in all studies.
Table 1.
The cohorts and covariates included in the meta-analysis of genotype at SNP rs1800469 and the presence of late fibrosis after radiotherapy to the breast.
| Trial | n | Genotyping platform | Fibrosis endpoint | Covariates included for fibrosis residual | Endpoints in STAT | Covariates included for STAT residual | Length of follow-up (years) | Refs. |
|---|---|---|---|---|---|---|---|---|
| RAPPER | 1010 | Fluidigm | Breast shrinkage (photos) | Age Breast volume Chemotherapy Boost |
Breast shrinkage (photos) Telangiectasia Oedema Pigmentation Pain Oversensitivity |
Acute toxicity week 3 Boost Smoker status DM Breast volume Actual volume 107% Post-op infection Surgical cosmesis |
2 | [12] |
| LeND wide Local excision | 493 | SNPlex | LS fibrosis | Age Chemotherapy Boost EQD2 Gy |
LS pain LS fibrosis LS telangiectasia LS oedema LS atrophy |
Boost Smoker DM Chemotherapy Hypertension EQD2 Gy |
1.5–7 | [14,15] |
| MARIERAD study | 389 | MassArray iPlex | RTOG fibrosis | Age BMI Boost EQD2 Gy |
Telangiectasia Fibrosis in operation site |
Boost dose Smoker DM BMI EQD2 Gy |
>5 | [15] |
| Post-mastectomy DBCG2 | 234 | ABI TaqMan | LS Fibrosis | Age EQD2 Gy Chemotherapy (concurrent) |
Variable | [16] | ||
| RACE | 190 | ABI 3730 automated sequencer | Breast shrinkage (photos) | Age Boost Estimated breast volume Chemotherapy EQD2 Gy |
Breast shrinkage (photos) Telangiectasia Oedema Breast symptoms (QoL) |
Boost Chemotherapy Estimated breast volume Surgical deficit EQD2 Gy |
2 & 5 | [17] |
| LeND mastectomy | 134 | SNPlex | LS fibrosis | Age Chemotherapy EQD2 Gy |
LS pain LS fibrosis LS telangiectasia |
Smoker DM Chemotherapy EQD2 Gy |
1.5–7 | [14,15] |
| Gene-PARE | 92 | DHPLC | RTOG palpation | Age EQD2 Gy |
RTOG visual exam RTOG palpation |
Acute toxicity week 5 Smoker DM EQD2 Gy |
>2 | [18] |
| TGFB prospective study in Sant Pau Hospital, Barcelona | 78 | Sanger sequencing | RTOG subcutaneous tissue | Age BMI Chemotherapy Boost EQD2 Gy |
RTOG skin RTOG subcutaneous tissue |
Acute toxicity Boost Smoker DM Chemotherapy BMI Total dose Surgical cosmesis |
2 | |
| Hamburg Fibrosis breast | 69 | RFLP | LS fibrosis | Age Chemotherapy EQD2 Gy |
2 | [7,21] | ||
| Post-mastectomy DBCG1 | 41 | ABI SNaPshot | LS fibrosis | Age EQD2 Gy |
Variable | [19] | ||
| Post-lumpectomy pre-START trial | 52 | ABI SNaPshot/Taqman | Induration | Age Breast size Chemotherapy Boost EQD2 Gy |
1–5 | [20] |
Abbreviations: ABI, Applied Biosystems™; RFLP, Restriction Fragment Length Polymorphism; DHPLC, Denaturing High-Performance Liquid Chromatography; LS, LENT-SOM (Late effects on Normal Tissues-Subjective Objective Management); RTOG, Radiation Therapy Oncology Group; DM, diabetes mellitus; BMI, body mass index; QoL, Quality of Life; Post-op, post-operative; EQD2 Gy, equivalent dose in 2 Gy fractions; RAPPER, Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy; LeND, Leicester Nottingham Derby; MARIERAD, Mammakarzinom-Risikofaktoren-Erhebung” – Mammary Carcinoma Risk Factor Investigation Radiotherapy; DBCG, Danish Breast Cancer Group; RACE, Radiation Complications and Epidemiology; Gene-PARE, genetic predictors of Adverse Radiotherapy Effects; START, Standardisation of Breast Radiotherapy.
Genotype data
Amongst the 20 cohorts included in this meta-analysis, the SNPs most frequently studied in TGFB1 were rs1800469 c.-1347T>C (previously known as C-509T) and rs1800470 (formerly rs1982073) c.29C>T (known as T+29C) encoding Leu10Pro. The two SNPs are in strong linkage disequilibrium (LD) with each other such that the minor T allele of c.-1347T>C and the minor C allele of c.29C>T, encoding proline Pro10, tend to be inherited together. An association was therefore sought between c.-1347T>C (rs1800469) and the development of late radiotherapy toxicity. Table 1 shows the genotyping platforms used in each cohort as well as selected study characteristics. Supplementary Table 1 gives the distribution of genotypes in each study and the minor allele frequency (MAF). In all studies, the genotype distributions did not differ from those expected under Hardy Weinberg Equilibrium.
Toxicity data
The toxicity endpoints recorded in each study are listed in Supplementary Table 2. Some endpoints were graded using identical scaling systems, whereas others used slightly different scoring scales. Some studies reported the same endpoint at multiple time points after therapy. For each of the breast patients, the endpoint was selected which was judged to reflect breast fibrosis most closely. As late toxicity can progress up to 10 years following radiotherapy, where toxicity was recorded longitudinally, data from the latest time point available were used. The incidence of ≥ grade 2 late fibrosis ranged from 1.3% to 56.1% among the studies and the incidence across all studies was 25.2%.
The primary investigation was a univariate analysis of fibrosis. A Z-score was obtained for an individual patient (k) for the fibrosis endpoint for which that patient had a valid (non-missing) score, sk.
where Mean and Standard Deviation are taken over all cases in the study population where fibrosis data were available. Converting individual toxicity scores to Z-scores eliminates the problem of grades for one toxicity item not being directly comparable with grades for another item. Z-scores define, for a particular endpoint, whether a patient’s score is high or low relative to the distribution of the scores of other patients in the population.
In addition, a secondary analysis of overall toxicity, reported by the Standardised Total Average Toxicity (STAT) score [9], and genotype was performed. To obtain a STAT score for an individual patient (k), a standardised Z-score, Zk,i, is derived for each toxicity endpoint (i) for which that patient has a valid (non-missing) score, sk,i:
The STAT score for patient k, STATk, is simply the average of all non-missing Z scores for that patient:
Statistical analysis
Stata version 11.0 was used. Univariate analysis (UVA) was initially performed to look for an association between genotype at rs1800469 and either fibrosis or overall toxicity using the non-parametric Spearman’s rank correlation coefficient. UVA was also performed on each cohort to determine the non-genetic factors that influenced both fibrosis and overall toxicity. Spearman’s rank correlation coefficient was used to test for associations between fibrosis or STAT scores and continuous covariates. The Mann–Whitney two-sample statistic was used for binary covariates.
In order to quantify the prescribed radiation dose, and to account for differences in dose per fraction used in the participating cohorts, the equivalent dose in 2 Gy fractions was calculated in each patient using the formula:
where D = total dose, d = dose per fraction and the α/β ratio was assumed to be 3.4 Gy, which was the value estimated for late change in breast appearance (photographic) in the meta-analysis of START trial A and the pilot RMH/GOC trial [10].
In addition, ordinal logistic regression was used to look for an association between late fibrosis and genotype. This was considered appropriate as late fibrosis was scored using a graded scale in all studies. No numeric relationship is assumed between these grades; it is only assumed that lower grades correspond to milder reactions. Patients are assumed to have a decreasing risk of developing a specific grade of reaction when going from rare allele homozygous to heterozygous and to common allele homozygous. This form of regression effectively looks for a trend both down and across a table of toxicity score against genotype and has greater power than simple dichotomization of the toxicity endpoints, e.g. <grade 2 or ≥ grade 2. Univariate and multivariate analyses were performed, including factors that were associated with fibrosis with P-value <0.1 in each data set (Supplementary Table 3). The analyses were also repeated including any factors that were associated with fibrosis in the largest dataset with P-value <0.1. The inclusion of these additional factors did not significantly change the results of the analysis (data not shown).
The resulting univariate and multivariate odds ratios for each study were meta-analysed and Forest plots created, using the Der-Simonian–Laird random-effects model. This model assumes heterogeneity between the studies; i.e., it assumes that the true effect can be different for each study. It is assumed that the individual-study true effects are distributed around an overall true effect, but the model makes no assumptions about the form of the distribution of either the within-study or the between-studies effects. Meta-analysis was also repeated with a fixed effect model which assumes no heterogeneity between studies.
Multivariate linear regression of STAT score with genotype at rs1800469 was then performed on each cohort, including covariates associated with overall toxicity on univariate analysis of that cohort with P-value <0.1 or of the largest cohort in the data set. Linear regression was considered appropriate as STAT scores follow an approximately normal distribution. Supplementary Table 3 shows the results of univariate analysis of association between covariates and overall late toxicity for the 11 studies. After multivariate analysis (MVA), residuals were calculated for each patient, for the STAT score (R-STAT), to quantify the overall toxicity not explained by available patient and treatment related factors (Table 1) [11]. A residual is the difference between the observed and the estimated toxicity score. Patients with residuals of zero have toxicity entirely accounted for by these factors. Patients with negative or positive residuals have less or greater toxicity respectively than is explained by known factors. Residuals were used in the analysis as clinical data were available in some patients from the cohorts who did not have genotyped data. This meant that the clinical factors that influence toxicity could be determined in all patients to maximise power and the resulting factors used in the calculation of residuals in patients for whom genetic data were available.
Results
The minimally required data: SNP rs1800469 genotype, treatment variables, and clinically-assessed fibrosis (measured 2 years or later after therapy) were available in 2782 participants who had received adjuvant radiotherapy to the breast, from 11 cohorts. Supplementary Fig. 1 shows the CONSORT diagram for this meta-analysis. Table 1 shows the number of patients included from each cohort, the endpoints used to score fibrosis and calculate overall toxicity reported by the STAT score as well as the covariates included in multivariate analysis.
On initial univariate analysis there was no evidence of association between rs1800469 genotype and either fibrosis or overall toxicity. Spearman’s correlation coefficient of Z scores for fibrosis and genotype was 0.02 (P = 0.38) and the correlation between overall toxicity and genotype was 0.005 (P = 0.80). Meta-analysis of the univariate odds ratios obtained from ordinal logistic regression in all data sets gave an OR = 1.015 (95% CI 0.89, 1.14), with no significant evidence of heterogeneity in this effect size between studies (P = 0.67).
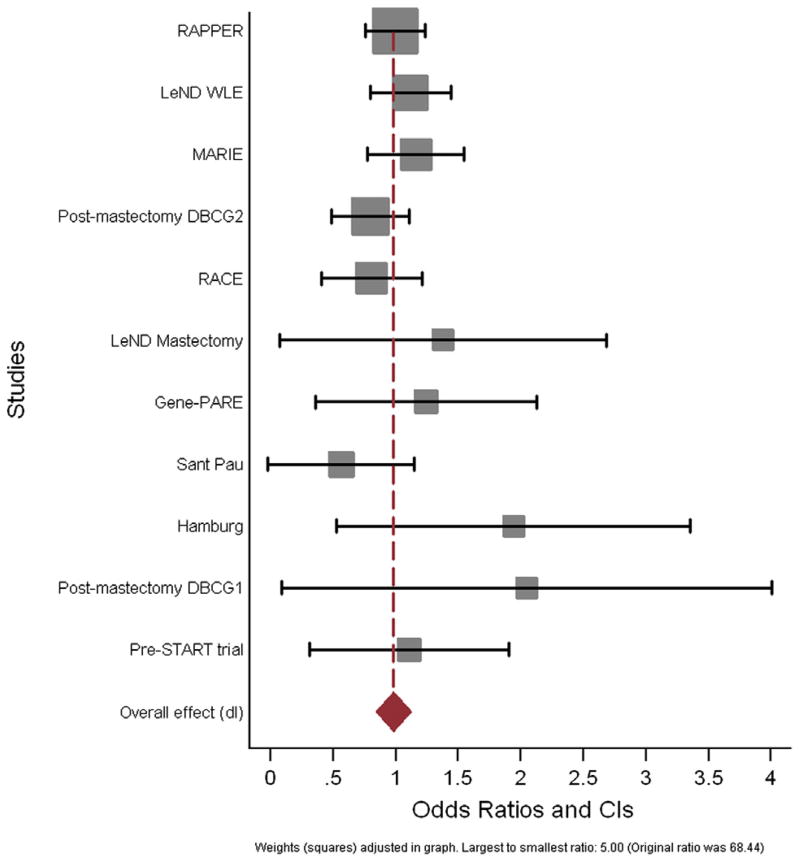
Multivariate analysis was next performed, adjusting for all variables found to be associated with late toxicity. Fig. 1 shows the Forest plot of odds ratios obtained from multivariate ordinal logistic regression of fibrosis by genotype for all 11 cohorts included in the meta-analysis, with the corresponding confidence intervals. Smaller trials have larger confidence intervals, reflecting the decreased power to detect an association between genotype and the development of late fibrosis of the breast. The multivariate OR obtained from meta-analysis, for an increase in late fibrosis grade with each additional rare allele of rs1800469 was 0.98 (95% CI 0.85–1.11). The corresponding 99% CI are 0.81–1.13. Thus, the null hypothesis that the OR equals 1 could not be rejected, i.e. there was no association between the SNP and risk of subcutaneous fibrosis in breast cancer patients. Table 2 shows the univariate and multivariate odds ratios and standard errors obtained for the ordinal regression of fibrosis against genotype at rs1800469. The results of meta-analysis did not change when a fixed-effect model was used (data not shown).
Fig. 1.
Forest plot of odds ratios obtained from multivariate ordinal logistic regression of fibrosis by genotype for all 11 cohorts included in the meta-analysis, with the corresponding confidence intervals.
Table 2.
The univariate and multivariate odds ratios (OR) and standard errors (SE) obtained from ordinal regression of fibrosis against genotype at rs1800469.
| Study | n (UVA) | OR (UVA) | SE (UVA) | n (MVA) | OR (MVA) | SE (MVA) |
|---|---|---|---|---|---|---|
| RAPPER | 786 | 0.99 | 0.11 | 773 | 1.00 | 0.12 |
| LeND WLE | 480 | 1.13 | 0.17 | 480 | 1.12 | 0.16 |
| MARIE | 389 | 1.24 | 0.21 | 389 | 1.16 | 0.20 |
| Post-mastectomy DBCG 2 | 234 | 1.00 | 0.17 | 234 | 0.80 | 0.16 |
| RACE | 161 | 0.75 | 0.18 | 145 | 0.81 | 0.21 |
| LeND mastectomy | 132 | 1.45 | 0.72 | 132 | 1.38 | 0.67 |
| Gene-PARE | 92 | 1.24 | 0.45 | 92 | 1.24 | 0.45 |
| Sant Pau | 78 | 0.67 | 0.32 | 78 | 0.56 | 0.30 |
| Hamburg | 69 | 1.94 | 0.72 | 69 | 1.94 | 0.72 |
| Post-mastectomy DBCG 1 | 41 | 1.19 | 0.49 | 41 | 2.05 | 1.00 |
| Pre-START trial | 52 | 1.13 | 0.41 | 52 | 1.11 | 0.41 |
UVA, univariate analysis; MVA, multivariate analysis; OR, odds ratio; SE, standard error; RAPPER, Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy; LeND, Leicester Nottingham Derby; MARIERAD, Mammakarzinom-Risikofaktoren-Erhebung” – Mammary Carcinoma Risk Factor Investigation Radiotherapy; DBCG, Danish Breast Cancer Group; RACE, Radiation Complications and Epidemiology; Gene-PARE, Genetic Predictors of Adverse Radiotherapy Effects; START, Standardisation of Breast Radiotherapy.
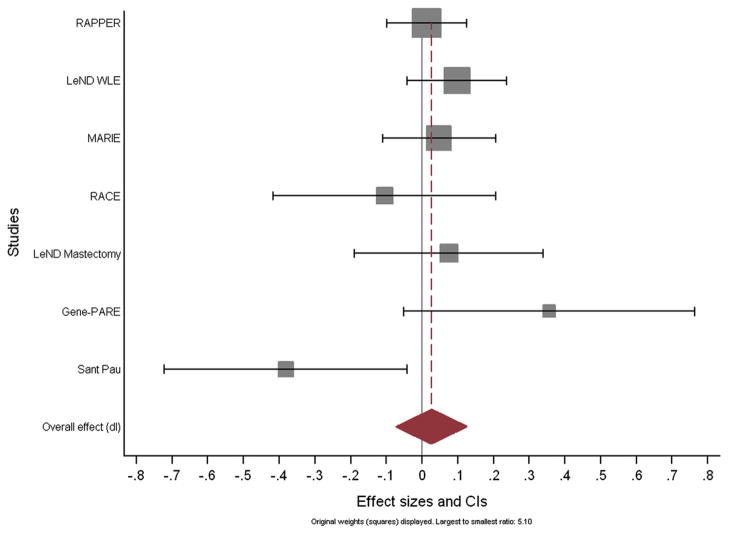
Multivariate linear regression of overall toxicity with genotype at rs17800469 was performed to obtain residuals. The residuals therefore quantify the toxicity not explained by available patient and treatment related factors, i.e. they are the difference between the observed and the estimated toxicity score. Fig. 2 shows the Forest plot of the regression coefficients for residuals of overall toxicity and genotype for each of the seven cohorts with more than one toxicity endpoint available, with the corresponding confidence intervals. Meta-analysis reveals that each additional rare allele of rs1800469 corresponds to a 0.026 increase in STAT score (95% CI −0.074 to 0.126). Thus, the null hypothesis that this coefficient of regression equals 0 could not be rejected, i.e. there was no significant association between the SNP and overall toxicity in breast cancer patients.
Fig. 2.
Forest plot of the regression coefficients for residuals of overall toxicity and genotype for each of the seven cohorts with more than one toxicity endpoint available, with the corresponding confidence intervals.
Discussion
This meta-analysis did not confirm previously reported associations between TGFB1 SNP c.-1347T>C (rs1800469) and increased risk of radiation-induced fibrosis in breast cancer patients. After adjustment for covariates which influence the development of radiation fibrosis, this meta-analysis enables the exclusion of an odds ratio >1.13 associated with the T allele, with >99% confidence. We can translate this into a confidence interval for the increased risk (around the overall average risk) of fibrosis in carriers vs. non-carriers. Assuming an incidence of ≥ grade 2 fibrosis of 25.2% after adjustment for covariates, we can exclude an incidence of ≥ grade 2 fibrosis greater than 28.5% for carriers of the rare allele of rs1800469 with >99% confidence. Thus, the statistical power of the present analysis is sufficient to rule out a clinically relevant difference in risk of moderate/severe fibrosis between carriers and non-carriers of the rs1800469 SNP with a high probability.
This is the first unbiased evaluation, with sufficient statistical power and adjustment for covariates, on a SNP that has been reported to be associated with radiation-induced normal tissue injury. The strengths of the current study include its large size, the use of both published and unpublished data and the availability of non-genetic factors that influence toxicity.
SNP rs1800469 is in the promoter region of TGFB1, but its effect on secretion and hence circulating levels of the cytokine remains unclear [7,22]. In addition the development of radiation fibrosis, a complex multi-factorial process, involving atrophy of normal tissues and excess collagen deposition, involves complex interactions between many proteins. Therefore it is possible that genetic variation in many proteins will influence the development of radiotherapy toxicity in a complex fashion.
Radiotherapy toxicity is a complex phenotype involving a variety of different pathological mechanisms, and these different processes lead to a variety of different clinical end-points. Despite a recognised need for a standardised approach for reporting toxicity, a variety of scoring systems are used and toxicity remains generally under-reported. In this meta-analysis, an attempt was made to minimise these problems by studying a single clinical endpoint (fibrosis) in breast cancer patients. Patients from clinical trials or dedicated radiogenomics studies were included. In each study the toxicity endpoint selected was judged to reflect breast fibrosis most closely. Note, that in a meta-analysis all effect estimates are within a specific study rather than between studies. Thus, it is not necessary to assume consistency in grading and use of scales across studies. It is also noteworthy that the toxicity scoring used in most cases had face validity in the sense that factors known to influence toxicity were found to have a significant impact on the toxicity scores. The current analysis corrects for total dose and dose fractionation, which is one of the most important non-genetic risk factors. However, due to the retrospective design of the analysis, it was not possible to include volume-related parameters. Whenever possible, future prospective studies should aim to include sufficient information on both dose and volume related parameters. The more accurate the information included on the non-genetic factors that influence late toxicity, the greater the chance of determining the genetic factors that determine radiotherapy toxicity. However, sample size is paramount. Most published GWAS have shown that a large sample size, even if the data are “noisy”, is more effective for detecting associations than a smaller sample size with very clean data.
The odds ratios and standard errors obtained from UVA and MVA of each data set were similar (Table 2). This means that in these studies the adjustment for covariates did not significantly alter the magnitude of the effect size of genotype at rs1800469. This implies that for SNPs that are not significantly associated with radiotherapy toxicity, i.e. true negative results, the adjustment for covariates does not have an undue influence on the results. It could be hypothesised that the effect of genotype may be constant despite increases in dose or the addition of other treatments, such as chemotherapy. However, it is possible that for a SNP that is truly associated with toxicity, adjustment for covariates would have a significant effect on the magnitude of the odds ratio. The relative importance of the inclusion of non-genetic factors will be demonstrated once univariate and multivariate analyses are performed on SNPs that are found to be truly associated with late radiotherapy toxicity.
The Radiogenomics Consortium was established in Manchester in 2009 [8] to provide a collaborative link between the major groups performing genotyping studies and existing cancer epidemiology cohorts in radiogenomics. It provides a route for sharing and developing expertise and quality assurance procedures; developing best practices for data collection; pooling data, addressing methodological challenges associated with radiogenomics and carrying out large, sufficiently powered studies. This meta-analysis of both published and unpublished studies demonstrates the Consortium’s ability to achieve its collaborative goals and has successfully overcome a publication bias problem.
Genome-wide association studies have highlighted the limitations of our current understanding of the genetic basis of most complex traits and diseases. Radiosensitivity is a complex phenotype and positive findings from candidate gene studies have proved difficult to replicate [12]. Several Stage 1 GWAS of radiogenomics are underway [13], the future meta-analysis of which should identify true causative variants. The large amount of data collected for this meta-analysis (Supplementary Fig. 1) highlight the need for standardisation between studies of radiogenomics in the Consortium.
In conclusion, this relatively large meta-analysis, found no clinically relevant association between the frequently-studied candidate SNP rs1800469 in TGFB1 and the development of fibrosis or other late radiotherapy toxicity. The meta-analysis demonstrates successful collaboration of groups included in the Radiogenomics Consortium.
Supplementary Material
Acknowledgments
Funding Support
Dr. Gillian Barnett is funded the National Institute of Health Research (NIHR) and previously by a fellowship from Cancer Research UK and The Royal College of Radiologists [C26900/A8740]. She also received funding from Addenbrooke’s Charitable Trust. Dr. Charlotte Coles and Prof. Neil Burnet are supported by the Cambridge NIHR Biomedical Research Centre. Dr. Alison Dunning is funded by Cancer Research UK [C8197/A10865] and the Joseph Mitchell Trust. Prof. Catharine West is supported by Cancer Research UK and Experimental Cancer Medicine Centre (ECMC) funding. The collaborative group (RAPPER) is funded by Cancer Research UK. The LeND cohort was funded by a grant from the Breast Cancer Campaign to Prof. R. Paul Symonds and Dr. Chris Talbot. The MARIE study was funded in part by the Dietmar Hopp Stiftung and the Deutsche Krebshilfe e.V. (project numbers 108253 and 108419). Dr. Sara Gutiérrez-Enríquez was supported by a Grant of the “Fondo de Investigación Sanitaria” from the Spanish Health Ministry [FIS 05/2181] and is currently funded by a Miguel Servet contract of the Instituto de Salud Carlos III. Prof. Barry Rosenstein and Dr. Sarah Kerns are funded by Grants RSGT-05-200-01-CCE from the American Cancer Society, PC074201 from the Department of Defense and 1R01CA134444 from the National Institutes of Health. The RACE study was funded by Cancer Research UK, and Prof. John Yarnold is supported by the Royal Marsden and ICR NIHR Biomedial Research Centre. Christian Nicolaj Andreassen and Jan Alsner received funding from the Danish Cancer Society, the Danish Council for Strategic Research, and CIRRO – The Lundbeck Foundation Centre for Interventional Research in Radiation Oncology. Prof. Soren Bentzen acknowledges support from the National Cancer Institute Grant No. 2P30 CA 014520.34.
The authors wish to acknowledge the Trial Management Groups of the component trials including:
The Cambridge Breast IMRT Trial Management Group.
Charlotte E. Coles, Gillian C. Barnett, Jennifer Wilkinson, Anne M. Moody, Charles B. Wilson, Nicola Twyman, Andrew Hoole, Gordon C. Wishart, Neil G. Burnet.
The LeND study.
Prabir Chakraborti, Steve Chan, Kwok-Leung Cheung, Ahmed Osman, Irene Peat, George Tanteles.
The Manchester Prospective Study.
Catharine West, Vivek Misra.
The MARIERAD study:
Irmgard Helmbold, Ursula Eilber, Christina Krieg.
The TGFB prospective study in Sant Pau Hospital of Barcelona (Spain) is grateful to David Fisas for the design and maintenance of the database.
The RACE Trial Management Group.
John Yarnold, Mark Sydenham, Judith Bliss, Jo Haviland, Lone Gothard, Richard Houlston, Nazneen Rahman and Mike Stratton (all Institute of Cancer Research Sutton), Roger Owen (Gloucestershire Oncology Centre, Cheltenham).
The DBCG post-mastectomy study.
Marie Overgaard, Jens Overgaard, Jørgen Johansen.
The Hamburg/Berlin Prediction Group.
Ulrike Höller, Kerstin Borgmann.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.radonc.2012.10.017.
Footnotes
Conflicts of interest statement
None declared.
References
- 1.Alsner J, Andreassen CN, Overgaard J. Genetic markers for prediction of normal tissue toxicity after radiotherapy. Semin Radiat Oncol. 2008;18:126–35. doi: 10.1016/j.semradonc.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Andreassen CN. Searching for genetic determinants of normal tissue radiosensitivity – are we on the right track? Radiother Oncol. 2010;97:1–8. doi: 10.1016/j.radonc.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Barnett GC, West CM, Dunning AM, et al. Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer. 2009;9:134–42. doi: 10.1038/nrc2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentzen SM. Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat Rev Cancer. 2006;6:702–13. doi: 10.1038/nrc1950. [DOI] [PubMed] [Google Scholar]
- 5.Evans ES, Kocak Z, Zhou SM, et al. Does transforming growth factor-beta1 predict for radiation-induced pneumonitis in patients treated for lung cancer? Cytokine. 2006;35:186–92. doi: 10.1016/j.cyto.2006.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett GC, Coles CE, Burnet NG, et al. No association between SNPs regulating TGF-beta1 secretion and late radiotherapy toxicity to the breast: results from the RAPPER study. Radiother Oncol. 2010;97:9–14. doi: 10.1016/j.radonc.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reuther S, Metzke E, Bonin M, Petersen C, Dikomey E, Raabe A. No effect of the transforming growth factor beta1 promoter polymorphism C-509T on TGFB1 gene expression, protein secretion, or cellular radiosensitivity. Int J Radiat Oncol Biol Phys. 2012 doi: 10.1016/j.ijrobp.2012.01.090. [DOI] [PubMed] [Google Scholar]
- 8.West C, Rosenstein BS, Alsner J, et al. Establishment of a Radiogenomics Consortium. Int J Radiat Oncol Biol Phys. 2010;76:1295–6. doi: 10.1016/j.ijrobp.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 9.Barnett GC, West CM, Coles CE, et al. Standardized Total Average Toxicity score: a scale- and grade-independent measure of late radiotherapy toxicity to facilitate pooling of data from different studies. Int J Radiat Oncol Biol Phys. 2011;82:1065–74. doi: 10.1016/j.ijrobp.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Bentzen SM, Agrawal RK, Aird EG, et al. The UK Standardisation of Breast Radiotherapy (START) Trial A of radiotherapy hypofractionation for treatment of early breast cancer: a randomised trial. Lancet Oncol. 2008;9:331–41. doi: 10.1016/S1470-2045(08)70077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentzen SM, Overgaard J. Patient-to-patient variability in the expression of radiation-induced normal tissue injury. Semin Radiat Oncol. 1994;4:68–80. doi: 10.1053/SRAO00400068. [DOI] [PubMed] [Google Scholar]
- 12.Barnett GC, Coles CE, Elliott RM, et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol. 2012;13:65–77. doi: 10.1016/S1470-2045(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 13.Kerns SL, Ostrer H, Stock R, et al. Genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with the development of erectile dysfunction in African-American men after radiotherapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2010;78:1292–300. doi: 10.1016/j.ijrobp.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giotopoulos G, Symonds RP, Foweraker K, et al. The late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causes. Br J Cancer. 2007;96:1001–7. doi: 10.1038/sj.bjc.6603637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talbot CJ, Tanteles GA, Barnett GC, et al. A replicated association between polymorphisms near TNFalpha and risk for adverse reactions to radiotherapy. Br J Cancer. 2012 doi: 10.1038/bjc.2012.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreassen CN, Alsner J, Overgaard M, Sorensen FB, Overgaard J. Risk of radiation-induced subcutaneous fibrosis in relation to single nucleotide polymorphisms in TGFB1, SOD2, XRCC1, XRCC3, APEX and ATM – a study based on DNA from formalin fixed paraffin embedded tissue samples. Int J Radiat Biol. 2006;82:577–86. doi: 10.1080/09553000600876637. [DOI] [PubMed] [Google Scholar]
- 17.Martin S, Sydenham M, Haviland J, et al. Test of association between variant tgbeta1 alleles and late adverse effects of breast radiotherapy. Radiother Oncol. 2010;97:15–8. doi: 10.1016/j.radonc.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Ho AY, Fan G, Atencio DP, et al. Possession of ATM sequence variants as predictor for late normal tissue responses in breast cancer patients treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2007;69:677–84. doi: 10.1016/j.ijrobp.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Andreassen CN, Alsner J, Overgaard M, Overgaard J. Prediction of normal tissue radiosensitivity from polymorphisms in candidate genes. Radiother Oncol. 2003;69:127–35. doi: 10.1016/j.radonc.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Andreassen CN, Alsner J, Overgaard J, et al. TGFB1 polymorphisms are associated with risk of late normal tissue complications in the breast after radiotherapy for early breast cancer. Radiother Oncol. 2005;75:18–21. doi: 10.1016/j.radonc.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Zschenker O, Raabe A, Boeckelmann IK, et al. Association of single nucleotide polymorphisms in ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with clinical and cellular radiosensitivity. Radiother Oncol. 2010;97:26–32. doi: 10.1016/j.radonc.2010.01.016. R&O. [DOI] [PubMed] [Google Scholar]
- 22.Dunning AM, Ellis PD, McBride S, et al. A transforming growth factor beta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003;63:2610–5. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.