Abstract
Natural selection has played an important role in establishing various phenotypes, but the molecular mechanisms of phenotypic adaptation are not well understood. The slow progress is a consequence of mutagenesis experiments in which present-day molecules were used and of the limited scope of statistical methods used to detect adaptive evolution. To fully appreciate phenotypic adaptation, the precise roles of adaptive mutations during phenotypic evolution must be elucidated through the engineering and manipulation of ancestral phenotypes. Experimental and quantum chemical analyses of dim-light vision reveal some surprising results and provide a foundation for a productive study of the adaptive evolution of various phenotypes.
Keywords: phenotypic adaptation, molecular adaptation, visual pigments, ancestral phenotypes, quantum chemistry
Nature provides extremely diverse environments, from scorching deserts to the extreme cold of Antarctica and from the tops of mountains to the bottom of the ocean. These environments are inhabited by a diverse 1.2 million known species of animals, plus a presumably much larger number of yet-to-be-described species (Mora et al. 2011). How did organisms adapt to this diversity of ecological and physiological environments? Using microorganisms such as bacteria, yeasts, and viruses, certain hypotheses and theories of evolution have been tested experimentally (Elena and Lenski 2003, Buckling et al. 2009). For vertebrates, it is extremely difficult to even establish adaptive evolution, and entirely different approaches from those used with micro-organisms are required. Independent of the organism and experimental approach we use, the answer to any evolutionary question requires all aspects of biology and the deployment of the methods developed in many scientific fields.
Visual adaptations are closely linked to organisms’ environments and behaviors, which shows that natural selection has played an important role in shaping these phenotypes (Walls 1942, Lythgoe 1979). In the 1980s, there were two critical advances in the molecular biology of vision: first, the cloning of the human opsin genes that encode visual pigments and are responsible for dim-light and color vision (Nathans and Hogness 1983, 1984, Nathans et al. 1986) and, second, the development of functional assays for visual pigments (Oprian et al. 1987).
Combining extensive data on ecology and evolution (Walls 1942, Lythgoe 1979, Yokoyama and Yokoyama 1996) and the molecular biology of visual pigments (see, e.g., Nathans and Hogness 1983, 1984, Nathans et al. 1986, Yokoyama 2000, 2008, Ebrey and Koutalos 2001, Palczewski 2006), color vision became arguably the best model system with which to study phenotypic adaptation (Carroll 2006). However, our knowledge of the evolutionary mechanisms producing various phenotypes, including color vision, is still superficial. Therefore, it is necessary to understand the causes for the slow progress and then to rectify the traditional ways of studying the molecular mechanisms of phenotypic adaptation.
Visual pigments
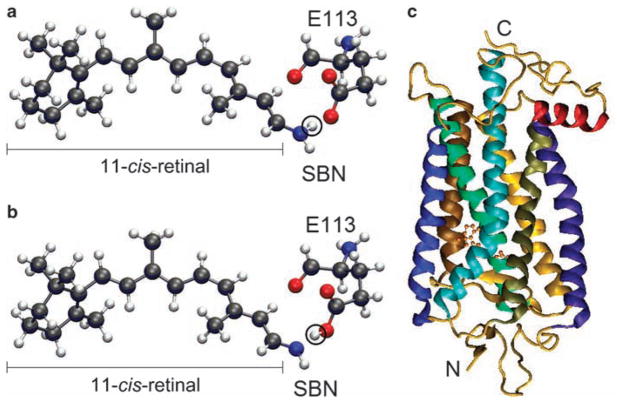
Each visual pigment consists of an opsin, encoded by a specific opsin gene, and a retinal (either 11-cis-retinal or 11-cis-3,4 dehydroretinal), which is derived from diet. The major function of each visual pigment is characterized by its wavelength of maximal absorption (λmax). In solution, the protonated Schiff base nitrogen-linked 11-cis-retinals (PSBR; figure 1a) and unprotonated Schiff base nitrogen-linked 11-cis-retinals (SBR; figure 1b) have λmax values of 440 and 365 nanometers (nm), respectively (see, e.g., Yokoyama 2000). However, by interacting with various opsins, the retinals can detect variable wavelengths between ultraviolet (λmax = 360 nm) and red (λmax = 560 nm) (Yokoyama 2008). This phenomenon is known as spectral tuning. In vertebrates, visual pigments are classified into five groups (table 1): (1) RH1 pigments that are usually expressed in rods (figure 1c), (2) RH2 pigments whose amino acid sequences are most closely related to those of RH1 pigments but that are usually expressed in cones, (3) short-wavelength-sensitive type 1 (SWS1) pigments, (4) short-wavelength-sensitive type 2 (SWS2) pigments, and (5) middle- and long-wavelength-sensitive (MWS and LWS, respectively; here collectively referred to as M/LWS) pigments. RH1 pigments containing 11-cis-retinal and 11-cis-3,4 dehydroretinal are often called rhodopsins and porphyrhopsins, respectively.
Figure 1.
The 11-cis-retinal and a visual pigment. (a) Protonated Schiff base nitrogen-linked 11-cis-retinal. The circled hydrogen atom (in white) is bound to the Schiff base nitrogen. (b) Unprotonated analog, where the circled hydrogen atom is bound to the carboxylic oxygen of E113 of the visual pigment. The black, blue, red, and white colors represent carbon, nitrogen, oxygen, and hydrogen atoms, respectively. (c) The seven transmembrane helices and 11-cis-retinal of bovine rhodopsin (pdb 1U19). The 11-cis-retinal is shown with the ball and stick model near the center, and N and C indicate the amino and carboxyl termini of the visual pigment.
Table 1.
Five groups of visual pigments generated by gene duplication.
| Visual pigment | Description | Approximate range of the wavelength of maximum absorption (in nanometers) |
|---|---|---|
| RH1 | Also called rhodopsin. These pigments are expressed mostly in rods. | 480–510 |
| RH2 | The amino acid sequences of these pigments are most closely related to those of RH1 pigments. | 450–530 |
| SWS1 | Short-wavelength-sensitive type 1 pigment. This group includes ultraviolet-, violet-, and blue-sensitive visual pigments. | 360–440 |
| SWS2 | Short-wavelength-sensitive type 2 pigment. | 400–450 |
| M/LWS | Middle- and long-wavelength-sensitive pigments. | 510–560 |
Sources: Yokoyama 1994, 2000, Ebrey and Koutalos 2001.
Extensive biochemical analyses of bovine rhodopsin, including a series of over 50 papers on the structure and function in rhodopsin by Khorana and his colleagues, have significantly improved our understanding of the functional properties of key amino acids in visual pigments. However, most of the mutations considered in these experiments did not occur in nature and are therefore not helpful in the understanding of the mechanism of the actual λmax shifts (or spectral tuning) in visual pigments (Yokoyama 1995). The molecular analyses of the actual mechanism of spectral tuning in visual pigments became possible when visual pigments were isolated from various species and their λmax values were determined (Yokoyama and Yokoyama 1996, Yokoyama 1997).
The problem with mutagenesis methods
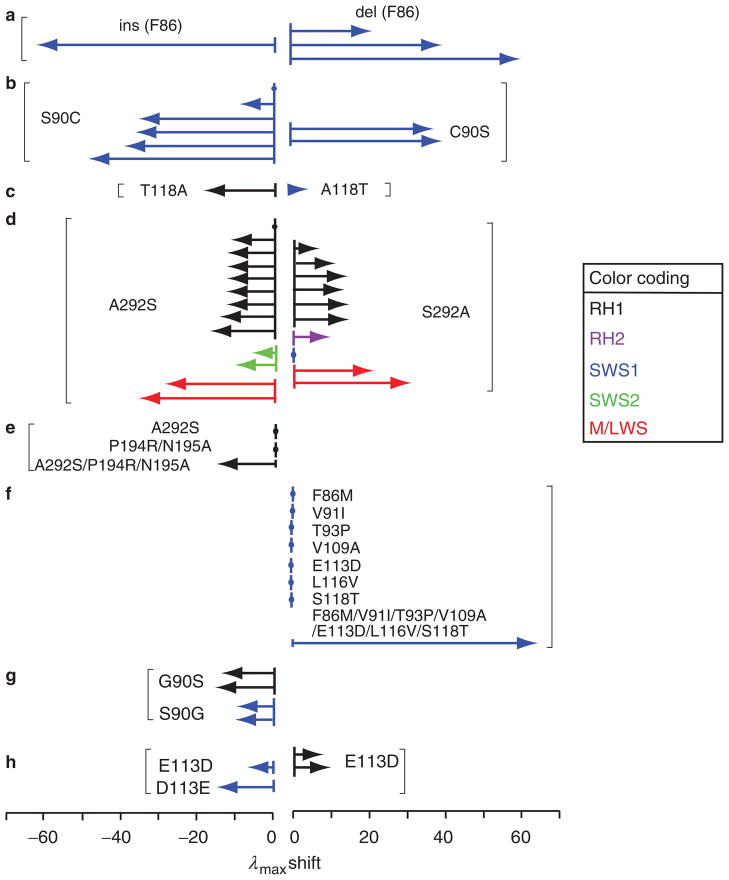
At present, certain mutations at a total of 35 amino acid residues of visual pigments are known to cause significant λmax shifts in nature (Yokoyama 2008). Most of these results have been obtained by manipulating present-day visual pigments. To interpret these mutagenesis results, two implicit assumptions are required: (1) identical amino acid changes in different pigments shift the λmax value by the same magnitude and in the same direction and (2) forward and reverse mutations shift the λmax value in opposite directions but by the same magnitude. However, these assumptions are often incorrect (Yokoyama 2008, Yokoyama et al. 2008a); in fact, not only can the same mutations in different pigments cause different magnitudes of λmax shifts (figure 2a–2f), but forward and reverse mutations can also shift the λmax values in the same direction (figure 2g, 2h). The following two examples provide further evidence for why we should not use the manipulation of modern pigments in studying the mechanisms of spectral tuning and the adaptive evolution of visual pigments.
Figure 2.
Examples of shifts in the wavelength of maximum absorption (λmax, in nanometers) affected by amino acid interactions. The black, purple, blue, green, and red colors indicate various RH1, RH2, SWS1, SWS2, MWS, and LWS pigments. The arrows reflect the λmax shifts of visual pigments from different species and are named in order from top to bottom within each example. (a) The insertion (ins) of F86 in the SWS1 pigment of scabbardfish (Lepidopus fitchi) and deletions (del) of F86 in the SWS1 pigments of a vertebrate ancestor, the lampfish (Stenobrachius leucopsarus) and the bluefin killifish (Lucania goodei) (Tada et al. 2009). (b) S90C in the SWS1 pigments of the mouse (Mus musculus), the cow (Bos taurus), an avian ancestor, the pigeon (Columba livia), the frog (Xenopus laevis), and the chicken (Gallus gallus) and C90S in the SWS1 pigments of the budgerigar (Melopsittacus undulatus) and the zebra finch (Taeniopygia guttata). (c) T118A in the RH1 pigment of the cow and A118T in the SWS1 pigment of the budgerigar. (d) A292S in three ancestral, five bovine, and one tilapia (Oreochromis niloticus) RH1 pigments; in the SWS2 pigments of the bluefin killifish and the newt (Cynops pyrrhogaster); and in the LWS pigments of two ancestral M/LWS pigments and S292A in the RH1 pigments of the scabbardfish, the coelacanth (Latimeria chalumnae), the cichlid, the thornyhead (Sebastolobus altivelis), the viperfish (Chauliodus macouni), and the conger (Conger myriaster); in the RH2 pigment of the medaka (Oryzias latipes); in the SWS1 pigment of the human (Homo sapiens); and in the M/LWS pigments of the mouse and the dolphin (Tursiops truncatus) (Yokoyama 2008, Yokoyama et al. 2008a). (e) Single and multiple mutations in the RH1 pigment of the conger. (f) Single and multiple mutations in the SWS1 pigment of a frog ancestor (Takahashi and Yokoyama 2005). (g) G90S in two bovine RH1 pigments and S90G in bovine and human SWS1 pigments. (h) E113D in the SWS1 pigments of the frog and of its ancestor and E113D in two bovine RH1 pigments. See Yokoyama (2008) and the references therein.
Oprian and his colleagues (Asenjo et al. 1994) attempted to explain the λmax difference between the human LWS pigment (λmax = 563 nm; denoted as HUMAN LWS) and the human MWS pigment (λmax = 532 nm; HUMAN MWS). When they replaced serine at site 180 by alanine (S180A), tyrosine at site 277 by phenylalanine (Y277F), and tyrosine at site 285 by alanine (T285A) in HUMAN LWS, the three mutant pigments decreased λmax by 33 nm in total, which fully explains the λmax value of HUMAN MWS. However, the λmax value of HUMAN MWS with all three reverse changes (A180S, F277Y, and A285T) increased by 23 nm, which explains only 74% of the 31-nm difference. After an extensive search, it has been found that an additional four amino acid changes (Y116S, T230I, S233A, and F309Y) are required to achieve the desired λmax value of HUMAN LWS. Therefore, HUMAN LWS with three amino acid changes showed a 33-nm λmax shift, but HUMAN LWS was not convertible with the three amino acid reversals. Clearly, depending on which pigment is chosen for the mutation, the molecular results of spectral tuning in the two pigments can be different.
The CONGER A pigment (λmax = 486 nm) of the conger eel (Conger myriaster) provides another important example. When A292S was introduced into its engineered ancestral pigment (AncCONGER A; λmax = 501 nm), λmax was unchanged, but when S292A was introduced into CONGER A, λmax increased by 12 nm (Yokoyama et al. 2008b). The latter result alone leads to the erroneous conclusion that CONGER A evolved from AncCONGER A simply by the A292S change. In fact, the actual functional divergence of CONGER A occurred through three amino acid substitutions: P194R, N195A, and A292S (Yokoyama et al. 2008b).
The solution
How can these discrepancies be resolved? The example from the conger eel shows that the problem disappears if we recapitulate the actual evolutionary change from AncCONGER A to CONGER A. The same principle also works for the HUMAN LWS and HUMAN MWS problem. Indeed, when S180A, Y277F, and T285A were introduced into the engineered ancestral mammalian pigment (AncMAMMALIAN LWS), the mutant pigment decreased the λmax value by 28 nm, which explains the λmax value of HUMAN MWS (Yokoyama et al. 2008a). Obviously, the seven amino acid changes used in manipulating HUMAN MWS are artificial, not natural. We have seen that S180A, Y277F, and T285A decreased the λmax value of HUMAN LWS by 33 nm (Asenjo et al. 1994). After 100 million years of HUMAN LWS evolution, its amino acid composition has been modified from that of AncMAMMALIAN LWS. The 5-nm difference between the results of the two experiments in which AncMAMMALIAN LWS and HUMAN LWS pigments were used is significant. If the amino acid interactions in HUMAN LWS were much stronger, the molecular basis of spectral tuning in HUMAN LWS could have been explained by more than three amino acid changes. Therefore, the HUMAN LWS result is also considered invalid.
As these examples demonstrate, the mechanisms of spectral tuning and the adaptive evolution of dim-light and color vision must be studied together by manipulating engineered ancestral pigments (Yokoyama and Radlwimmer 2001, Shi and Yokoyama 2003). Amino acid interactions are certainly not limited to visual pigments. In fact, the importance of reconstructing and manipulating ancestral molecules has also been stressed by Thornton and his colleagues (Harms and Thornton 2010).
The glucocorticoid receptor (GR) and the mineralo-corticoid receptor (MR), which are hormone-regulated transcription factors in vertebrates, descended from the ancestral corticoid receptor (AncCR) some 470 million years ago. The MR is activated by aldosterone and deoxy-corticosterone to control such functions as electrolyte homeostasis and kidney and colon function, whereas the GR is activated by the adrenal steroid cortisol and regulates such functions as stress response and glucose homeostasis. The engineered AncCR shows the MR-like sensitivity to aldosterone, deoxycorticosterone, and cortisol, which demonstrates that the function of GR was derived from that of AncCR, whereas the function of the ancestral receptor has been maintained in MR (Bridgham et al. 2006). When S106P and L111Q were introduced into AncCR (Bridgham et al. 2006) and into a more recent ancestral receptor (Ortlund et al. 2007), the function of GR was achieved. However, when the same two mutations were introduced into the contemporary human MR, the mutant receptor became nonfunctional (Ortlund et al. 2007), which shows that a single amino acid change in different proteins can produce very different phenotypes.
To genetically engineer ancestral molecules, their amino acid sequences must be inferred by using statistical methods, and they are always associated with uncertainty or possible estimation errors. In studying ancestral pigments, therefore, we have to deal with the statistical uncertainties of the inferred amino acids. For example, the Bayesian posterior probabilities of inferred amino acids can be used to check the reliabilities of the statistical predictions (Yang et al. 1995, Zhang and Nei 1997). Fortunately, most inferred amino acids of the five groups of visual pigments in the vertebrate ancestor have posterior probabilities of more than .95 and so are highly reliable (Yokoyama and Radlwimmer 2001, Shi and Yokoyama 2003). However, if we go a little lower in a phylogenetic tree and consider the common ancestor of all five groups of visual pigments, most inferred amino acids have posterior probabilities of much less than .90, and the inference is no longer reliable. Therefore, it seems biologically meaningful to engineer ancestral pigments only within each of the five visual pigment groups, not in their common ancestor.
In engineering an ancestral pigment at a particular node in an evolutionary tree, we may select the amino acid sequence that has the highest posterior probability. This sequence still contains amino acids whose posterior probabilities are much lower than .95. At these “ambiguous” sites, amino acids with the highest posterior probabilities may be replaced either individually by those with the second-highest probabilities or collectively through the construction of chimeric pigments (Yokoyama and Radlwimmer 2001, Shi and Yokoyama 2003). Then, the effects of these “ambiguous” amino acids on the λmax shift can be evaluated. When no functional change is detected, we may conclude that the “ambiguous” amino acid sites are inconsequential. In addition, visual pigments in rods (Yokoyama et al. 2008b), SWS1 pigments (Shi and Yokoyama 2003), and M/LWS pigments (Yokoyama and Radlwimmer 2001) in various vertebrate ancestors have identical λmax values, which suggests that a majority of amino acid substitutions do not shift the value of λmax
Adaptive evolution of dim-light vision
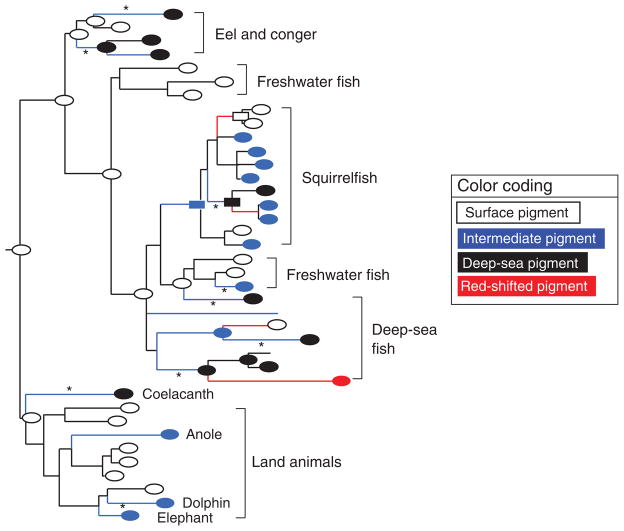
One of the critical times for the survival of animals in shallow water and on land is at twilight, when most abundant visible light has wavelengths shorter than 550 nm (Munz and McFarland 1973). Dim-light vision is controlled mostly by RH1 pigments in rod photoreceptor cells. On the basis of the λmax values of RH1 pigments and their ecological or physiological environments, the dim-light vision of a wide range of vertebrates has been classified into four groups (figure 3): (1) the surface vision (λmax = 500–507 nm; the white ovals in figure 3) of animals living on land and in shallow water; (2) the deep-sea vision (λmax ≅480–485 nm; the black ovals in figure 3) of deep-sea fishes, which reflects the narrow distribution of wavelengths (approximately 480 nm) of light that reach deep water; (3) the intermediate vision (λmax ≅ 490–495 nm; the blue ovals in figure 3) of animals living at intermediate depths; and (4) the red-shifted vision (λmax = 526 nm; the red oval in figure 3) of the intriguing case of the shiny loosejaw (Aristostomias scintillans), a deep-sea fish that seems to be able to detect its own far-red bioluminescence (O’Day and Fernandez 1974).
Figure 3.
The composite maximum likelihood tree of 38 representative RH1 pigments in vertebrates. The black, blue, white, and red ovals indicate the experimentally determined deep-sea (wavelength of maximum absorption [λmax] = 480–485 nanometers [nm]), intermediate (λmax = 490–495 nm), surface (λmax = 500–507 nm), and red-shifted (λmax = 526 nm) pigments, respectively, and the rectangles indicate λmax values estimated theoretically (Yokoyama et al. 2008b). The black, blue, and red branches show that no shift, blue shifts, and red shifts in λmax, respectively, occurred. Nine A292S substitutions are indicated by asterisks (★).
The mutagenesis analyses of a total of 11 engineered ancestral and extant RH1 pigments have revealed four major evolutionary characteristics (Yokoyama et al. 2008b). First, the evolutionary switches among the four vision groups have occurred on at least 18 separate occasions, which highlights the adaptive evolution of dim-light vision. Second, a total of 15 amino acid substitutions (D83N, Y96V, Y102F, E122I, E122M, E122Q, M183F, P194R, N195A, M253I, F261Y, T289G, A292S, S292I, and M317I) at 12 amino acid sites have undergone adaptive evolution individually or interactively, whereas 191 sites cause no λmax shift and are most likely to have undergone neutral evolution. Third, dim-light vision reversed the direction of evolutionary changes in λmax in several lineages. Fourth, the substitutions D83N (seven times), A292S (nine times), and F261Y (five times) have occurred multiple times independently.
Under dim-light conditions, animals also use ultraviolet and violet vision. Inhabiting a violet-light-rich environment, scabbardfish (Lepidopus fitchi) achieved the violet-sensitive extant SCABBARD pigment by deleting a functionally important amino acid site (F86) of its immediate ancestor (AncSCABBARD; Tada et al. 2009). In comparison, despite inhabiting much deeper depths than scabbardfish, where ultraviolet light does not reach, the lampfish (Stenobrachius leucopsarus) has retained ultraviolet vision. However, lampfish feed on ultraviolet-reflecting copepods near the surface at twilight, when perception of ultraviolet light, rather than higher wavelengths of light, is important (Tada et al. 2009). Therefore, the acquisition of violet vision in scabbardfish and the retention of ultraviolet vision by lampfish are adaptive to their foraging behaviors—consequences of adaptive evolution.
Statistical methods for detecting positive selection: Controversy
Over the last two decades, many biologists have used statistical methods for detecting adaptive mutations (e.g., Nei 2005, Nei et al. 2010). These statistical methods are based on the fundamental principle of molecular adaptation (dn > ds or ω = dn/ds > 1), where dn and ds are the number of nonsynonymous nucleotide substitutions per nonsynonymous site and that of synonymous substitutions per synonymous site, respectively (Hughes and Nei 1988). This approach has become so popular that “thousands of papers have been published every year claiming evidence of adaptive evolution on the basis of computational analyses alone” (Hughes 2008, p. 13,193). Adaptive amino acid substitutions have frequently been inferred individually using the site-prediction method (Nielsen and Yang 1998, Suzuki and Gojobori 1999, Yang et al. 2000, Suzuki et al. 2001, Yang 2007), whereas individual adaptive amino acid substitutions or positively selected genes have been identified along a predetermined branch on a phylogenetic tree using the branch-site method (Yang and Nielsen 2002, Yang et al. 2005, Zhang et al. 2005).
Recently, the theoretical bases and reliability of these statistical methods have been debated intensely among statistical evolutionary geneticists (Nozawa et al. 2009a, 2009b, Yang et al. 2009, Yang and dos Reis 2011). One major point of the controversy is the distribution of the type I error value (p) associated with branch-site method. Using computer simulations, it has been found that the distribution can have peaks in the region of .05 > p > .95. Possibly because of this and other reasons, using Fisher’s exact test, Nozawa and colleagues (2009a, 2009b) identified only 5 positively selected genes out of the 13,888 that they examined, whereas 154 positively selected genes were identified by Bakewell and colleagues (2007), using the branch-site method. However, Yang and dos Reis (2011) claimed that the levels of false positives were within twice the standard deviation of the expected 5% and that the branch-site method is reliable. Moreover, whereas the former group (Nozawa et al. 2009b) claimed that it is meaningless to evaluate the power of branch-site method because of the peculiar distribution of p, the latter group (Yang and dos Reis 2011) claimed otherwise.
Fortunately, the two groups agree on one thing; that is, the final answer can be obtained only by subjecting the statistical results to some form of experimental test (Nozawa et al. 2009a, 2009b, Yang et al. 2009). It is of considerable interest, therefore, to see how the site-prediction method and the branch-site method perform when they are applied to amino acid sites that are shown through mutagenesis experiments to cause adaptive or functional changes.
To examine the reliability of the statistical methods, it is of utmost importance to realize that the fundamental principle of molecular adaptation is an assumption without any experimental support. Therefore, it would be necessary to evaluate whether this assumption really holds in nature. To date, only visual pigments have been used for such purposes. It turns out that the actual ω value evaluated for the 12 adaptive sites of the RH1 pigments (figure 3) was 0.4 and that the corresponding ω values for the functionally important amino acid sites for the remaining four groups of visual pigments varied between 0.31 and 0.67 (Nozawa et al. 2009a). These results cast serious doubt on the fundamental principle (dn > ds or ω > 1) that has been widely used by evolutionary and molecular biologists for nearly 25 years.
When the site-prediction method was applied to the experimental data on opsin genes (Yokoyama et al. 2008b) and odorant receptor genes (Zhuang et al. 2009), over 90% of the estimates were false positives or false negatives. Moreover, when it was applied to opsin genes, the branch-site method generated equally high rates of false positives and false negatives (Nozawa et al. 2009b).
Although it is not widely recognized, the site-prediction results are also strongly affected by the number of DNA sequences compared. To illustrate the point, we may apply the Bayes empirical Bayes inference and the naive empirical Bayes methods of the phylogenetic analysis by maximum likelihood (version 4; Yang 2007) to the 11 squirrelfish RH1 genes in figure 3. The statistical analyses then predict 8 adaptive sites but identify none of the 12 experimentally determined adaptive sites, including site 292, where A292S occurred on nine separate occasions (figure 3).
The real problem starts to emerge when the other fish sequences in figure 3 are added into the same statistical tests. For the expanded data set, only two, rather than eight, adaptive sites are predicted. Furthermore, when all 38 opsin genes (figure 3) are considered, no adaptive site can be found, which provides a third answer to the same question. However, the parsimony method (Suzuki et al. 2001) could not identify any adaptive sites. This was expected, because 38 sequences is too few for the 12 adaptive sites to be detected (Nei 2005).
The site-prediction method has also been applied to the M/LWS opsin genes. When the method was applied to 14 primate M/LWS pigments, amino acid sites 180 and 285, together with four other sites, were predicted to be adaptive. When the data were expanded to 40 vertebrate M/LWS pigments, the same method then identified only site 275 as adaptive (Nozawa et al. 2009a). However, this site does not seem to shift λmax (Asenjo et al. 1994). Similarly, 10 adaptive sites were predicted for six mammalian SWS1 pigments by using the same statistical method, but all of them disappeared when the data were expanded to 21 orthologous vertebrate pigments (Nozawa et al. 2009a).
All of these observations show that the validity of the fundamental principle of molecular adaptation is highly questionable and that the performances of the currently available statistical tests for detecting adaptive evolution are not reliable. The poor performance of the statistical methods may have arisen from unrealistic mathematical models and from the unwarranted underlying assumption that most nonsynonymous nucleotide substitutions were positively selected (Nei et al. 2010). As was alluded to previously (Hughes 2007), the relaxation of purifying selection and the fluctuation of population sizes, which reduces the effective population size and, consequently, increases the effect of random genetic drift, may also make the statistical inference ambiguous.
To study the possibility of the adaptive evolution of red–green color vision, various approaches have been taken. In mammals, red–green color vision is controlled by M/LWS pigments. The genetic engineering of these pigments in vertebrate ancestors shows that the early ancestral pigments had λmax values of approximately 560 nm and extant M/LWS pigments have been achieved by various combinations of five amino acid substitutions (S180A, H197Y, Y277F, T285A, and A308S), where the amino acid sites were standardized by those of the orthologous pigments in humans (Yokoyama and Radlwimmer 2001, Yokoyama et al. 2008a). Among these, H197Y and A308S occurred only along the rodent lineages, and A308S occurred only along the dolphin (Tursiops truncatus) lineage. Interestingly, the three amino acid substitutions (S180A, Y277F, and T285A) occurred independently along cavefish (Astyanax fasciatus), gecko (Gekko gecko), deer (Odocoileus virginianus), human, some New World monkeys (e.g., the squirrel monkey [Saimiri boliviensis]), and wallaby (Macropus eugenii) lineages. It is easily imagined that the chance of the three identical amino acid substitutions occurring on six separate occasions is extremely small, which implies that they have undergone adaptive evolution (Yokoyama and Takenaka 2005).
Interestingly, these positively selected MWS pigments are found not only in animals with red–green color vision (e.g., cavefish, New World monkeys, humans) but also in those with red–green color blindness (e.g., geckos, deer, wallabies) and lacking LWS pigments. This means that red–green color vision and red–green color blindness have undergone adaptive evolution independently in different lineages. This observation contradicts a widely accepted notion that animals with red–green color vision have a selective advantage over those with color blindness but is compatible with the observation that the majority of mammals and many other species are red–green color blind. Evidence is scant and sometimes controversial, but several observations are consistent with the idea that animals with color blindness can have a selective advantage over those with red–green color vision, including the possibility of decoding color camouflages (for such a discussion, see Yokoyama and Takenaka 2005). To resolve the issue, more behavioral analyses are needed.
Neutral mutations can become adaptive
In molecular evolution, mutations are often classified as adaptive or neutral, which has been the source of the controversy between the so-called selectionists and neutralists (Nei 2005, Nei et al. 2010). However, neutral mutation can, in combination with subsequent mutations, become adaptive or vice versa. We have seen that during the evolution of CONGER A from AncCONGER A, the λmax value decreased by about 15 nm when the three changes—A292S, P194R, and N195A—became combined. Interestingly, however, neither A292S nor the combination of P194R and N195A can shift the λmax value of AncCONGER A (figure 2e). Therefore, the neutral mutations (A292S and the paired P194R and N195A) became adaptive only in combination. Indeed, selectively neutral alleles can have a latent potential for selection in different environments (Dykhuizen and Hartl 1980). This phenomenon has been called the Dykhuizen–Hartl effect (Kimura 1983), and the three amino acid substitutions (P194R, N195A, and A292S) in CONGER A perfectly demonstrate it.
The FROG SWS1 pigment (λmax = 423 nm) of the African clawed frog (Xenopus laevis) evolved from its ancestral ultraviolet pigment (AncFROG SWS1; λmax = 359 nm) by seven specific amino acid changes (F86M, V91I, T93P, V109A, E113D, L116V, and S118T; Takahashi and Yokoyama 2005). However, none of these mutations alone can shift the λmax value of AncFROG SWS1 (figure 2f). Therefore, any one of these seven mutations could have originally been neutral and could have became adaptive during the frog’s evolution after it had accumulated multiple mutations. Similarly, the λmax value of the MOUSE SWS1 pigment (λmax = 359 nm) in the mouse (Mus musculus) can be converted into that of the HUMAN SWS1 pigment (λmax = 414 nm) by T46F, L49F, F52T, L86F, P93T, G114A, and T118S, but once again, none of these mutations individually causes any λmax shift (Shi and Yokoyama 2003). Here, the ancestral pigment has not been modified, but it is likely that the neutral mutants became adaptive during HUMAN SWS1 evolution. The preliminary analyses of the ancestral ultraviolet-sensitive pigment (λmax = 359 nm) also suggest the switch of neutral mutations into adaptive mutations.
The quantum chemistry of SWS1 pigments
At present, in studying molecular adaptation, the best thing we can do is to identify functionally critical amino acid changes. However, even if all adaptive mutations are identified using experimental methods, their actual roles in the functional differentiation of proteins are still unclear. Fortunately, the precise roles of amino acid changes in the adaptive evolution of visual pigments can be studied using quantum mechanical and molecular mechanical calculations (see, e.g., Altun et al. 2011). This method uses the homology models based on the crystal structure of bovine rhodopsin (Okada et al. 2004) as a template (figure 1c). In elucidating the mechanisms of spectral tuning in visual pigments, we have seen that the molecular analyses of present-day visual pigment are misleading. The same argument holds for quantum chemical analyses as well. If amino acid interactions are important in understanding the structure–function relationship of certain proteins, ancestral molecules—rather than the extant forms—must be analyzed.
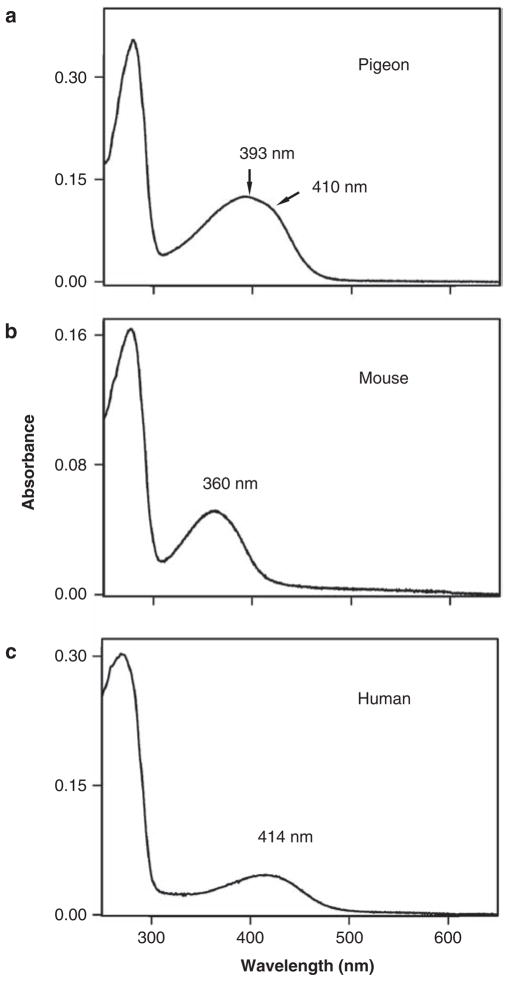
So far, I have probably given the impression that each visual pigment has a single absorption spectrum. Strictly speaking, this is not true, even for visual pigments constructed using the in vitro assay. For example, each SWS1 pigment consists of a mixture of two groups: the PSBR group and the SBR group (figure 1a, 1b). The major shift in the λmax value of a visual pigment is caused by differences in the ground-state energies of the SBR group (ESBR) and the PSBR group (EPSBR). Experiments show that the absorption spectra of several SWS1 pigments consist of a major peak at around 390 nm and a minor peak at 410 nm (Shi et al. 2001, Fasick et al. 2002, Shi and Yokoyama 2003, Takahashi and Yokoyama 2005). This occurs when the difference in energy (ΔE = EPSBR − ESBR) is near zero, as is exemplified by the pigeon pigment (ΔE = −0.3 kilocalories per mole [kcal/mol]; figure 4a), and the two peaks are generated as a result of the overlapped signals from the SBR and PSBR groups (Altun et al. 2011).
Figure 4.
The absorption spectra of SWS1 pigments in (a) the pigeon, (b) the mouse, and (c) the human. Abbreviation: nm, nanometers.
When ΔE is positive, the SBR group is energetically more stable than the PSBR group, and consequently, the pigment consists of mostly SBR-type pigment and is sensitive to ultraviolet wavelengths, whereas when ΔE is negative, the pigment consists of mostly PSBR-group pigment and is sensitive to violet light. MOUSE SWS1 and HUMAN SWS1 represent the SBR (figure 4b) and PSBR (figure 4c) pigments, respectively.
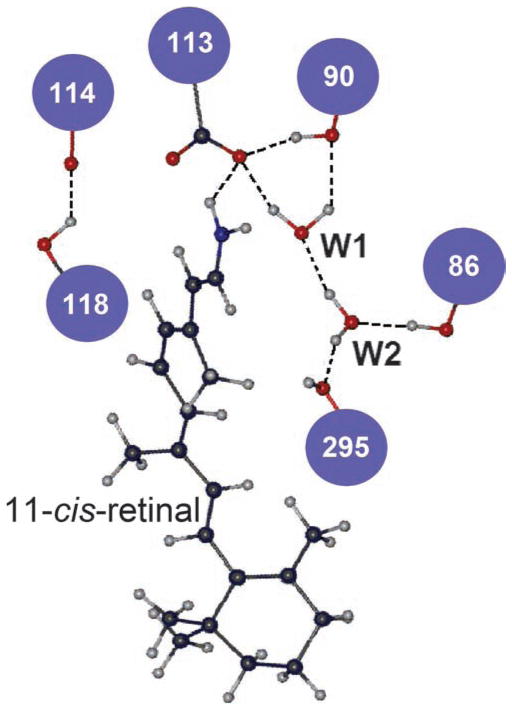
Returning to the SCABBARD example on dim-light vision, its immediate ancestor (AncSCABBARD; ΔE = 3.2 kcal/mol) is ultraviolet sensitive, whereas AncSCABBARD with an F86 deletion (ΔE = −6.2 kcal/mol) and SCABBARD (ΔE = −1.3 kcal/mol) are violet sensitive (Tada et al. 2009). Here, the number of hydrogen bonds of the SBR group decreased from five to three, and the number of hydrogen bonds of the PSBR group increased from three to five, which makes the PSBR group the majority in SCABBARD. More generally, by studying the switches between ultraviolet and violet pigments at various stages of vertebrate evolution, it has been shown that the evolution of SWS1 pigments is mediated by the hydrogen-bond network connected by amino acid sites 86, 90, 113, 114, 118, and 295 and by two water molecules (figure 5; Altun et al. 2011).
Figure 5.
The hydrogen-bond network of bovine rhodopsin (pdb 1U19), connected by six amino acid sites and two nearby water molecules (W1 and W2).
Experimental and quantum chemical analyses of the ultraviolet pigment in the vertebrate ancestor (AncSWS1) reveal another fascinating feature of phenotypic adaptation (Tada et al. 2009). That is, when F86 is deleted from AncSWS1, ΔE does not change much (from 6.2 to 5.8 kcal/mol), and the experimentally determined λmax value of the mutant pigment is 380 nm, which is still near ultraviolet sensitive. Clearly, ultraviolet-sensitive AncSWS1 and its descendant, AncSCABBARD, can respond to the same amino acid change very differently, and, again, depending on which ancestral pigments are chosen to be mutated, the outcomes can be very different.
Conclusions
Because of the biologically unwarranted fundamental principle of molecular adaptation (dn > ds or ω > 1) and high rates of false positives and false negatives, statistically detected adaptive amino acid changes must eventually be examined through experimental means. Today, only a small number of genetic systems have been explored in which experimental molecular biology and evolutionary biology were synthesized (Golding and Dean 1998, Dean and Thornton 2007). One hopes that more evolutionary biologists will find such experimental approaches necessary in studying phenotypic adaptation. As the analyses of dim-light vision indicate (Tada et al. 2009), such experiments will undoubtedly uncover some fascinating features of adaptive evolution.
Should experimental approaches be taken, the relationship between genotype and phenotype must first be established unambiguously. Then, by linking different phenotypes to organisms’ environments, adaptive phenotypic changes can be specified. To identify adaptive amino acid changes, proper ancestral phenotypes must be engineered and manipulated. Finally, to fully understand the molecular mechanism of phenotypic adaptation, the precise roles of adaptive mutations in the process of modifying the chemical structures of proteins and their functions must be clarified. To achieve this goal, quantum chemical methods can be used in the vision system, and equivalent analyses must be explored in other genetic systems.
Acknowledgments
I thank Ahmet Altun, Phil Dunham, Masafumi Nozawa, Marty Tracey, and Ruth Yokoyama for their comments. This work was supported by National Institutes of Health grant no. R01EY016400 and by Emory University.
References cited
- Altun A, Morokuma K, Yokoyama S. H-bond network around retinal regulates the evolution of ultraviolet and violet vision. ACS Chemical Biology. 2011;6:775–780. doi: 10.1021/cb200100f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Bakewell MA, Shi P, Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proceedings of the National Academy of Sciences. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1681/01.asn.0000926836.46869.e5. [DOI] [PubMed] [Google Scholar]
- Buckling A, Craig Maclean R, Brockhurst MA, Colegrave N. The Beagle in a bottle. Nature. 2009;457:824–829. doi: 10.1038/nature07892. [DOI] [PubMed] [Google Scholar]
- Carroll SB. The Making of the Fittest: DNA and the Ultimate Forensic Record of Evolution. Norton; 2006. [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: The functional synthesis. Nature Reviews Genetics. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen D, Hartl DL. Selective neutrality of 6PGD allozymes in E. coli and the effects of genetic background. Genetics. 1980;96:801–817. doi: 10.1093/genetics/96.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrey T, Koutalos Y. Vertebrate photoreceptors. Progress in Retinal and Eye Research. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nature Reviews Genetics. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Fasick JI, Applebury ML, Oprian DD. Spectral tuning in the mammalian short-wavelength sensitive cone pigments. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- Golding GB, Dean AM. The structural basis of molecular adaptation. Molecular Biology and Evolution. 1998;15:355–369. doi: 10.1093/oxfordjournals.molbev.a025932. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Current Opinion in Structural Biology. 2010;20:360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: The misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The origin of adaptive phenotypes. Proceedings of the National Academy of Sciences. 2008;105:13193–13194. doi: 10.1073/pnas.0807440105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge University Press; 1983. [Google Scholar]
- Lythgoe JN. The Ecology of Vision. Clarendon; 1979. [Google Scholar]
- Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. How many species are there on Earth and in the ocean? PLOS Biology. 2011;9:art. e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz FW, McFarland WN. The significance of spectral position in the rhodopsins of tropical marine fishes. Vision research. 1973;13:1829–1874. doi: 10.1016/0042-6989(73)90060-6. [DOI] [PubMed] [Google Scholar]
- Nathans J, Hogness DS. Isolation, sequence analysis, and intronexon arrangement of the gene encoding bovine rhodopsin. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Nathans J, Hogness DS. Isolation and nucleotide sequence of the gene encoding human rhodopsin. Proceedings of the National Academy of Sciences. 1984;81:4851–4855. doi: 10.1073/pnas.81.15.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: The genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Nei M. Selectionism and neutralism in molecular evolution. Molecular Biology and Evolution. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annual Review of Genomics and Human Genetics. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Suzuki Y, Nei M. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proceedings of the National Academy of Sciences. 2009a;106:6700–6705. doi: 10.1073/pnas.0901855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Suzuki Y, Nei M. Response to Yang et al.: Problems with Bayesian methods of detecting positive selection at the DNA sequence level. Proceedings of the National Academy of Sciences. 2009b;106:E96. doi: 10.1073/pnas.0906089106. [DOI] [Google Scholar]
- O’Day WT, Fernandez HR. Aristostomias scintillans (Malacosteidae): A deep-sea fish with visual pigments apparently adapted to its own bioluminescence. Vision Research. 1974;14:545–550. doi: 10.1016/0042-6989(74)90044-3. [DOI] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. Journal of Molecular Biology. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proceedings of the National Academy of Sciences. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: Evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. G protein-coupled receptor rhodopsin. Annual Review of Biochemistry. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yokoyama S. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proceedings of the National Academy of Sciences. 2003;100:8308–8313. doi: 10.1073/pnas.1532535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Radlwimmer FB, Yokoyama S. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proceedings of the National Academy of Sciences. 2001;98:11731–11736. doi: 10.1073/pnas.201257398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Gojobori T. A method for detecting positive selection at single amino acid sites. Molecular Biology and Evolution. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Gojobori T, Nei M. ADAPTSITE: Detecting natural selection at single amino acid sites. Bioinformatics. 2001;17:660–661. doi: 10.1093/bioinformatics/17.7.660. [DOI] [PubMed] [Google Scholar]
- Tada T, Altun A, Yokoyama S. Evolutionary replacement of UV vision by violet vision in fish. Proceedings of the National Academy of Sciences. 2009;106:17457–17462. doi: 10.1073/pnas.0903839106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Yokoyama S. Genetic basis of spectral tuning in the violet-sensitive visual pigment of African clawed frog, Xenopus laevis. Genetics. 2005;171:1153–1160. doi: 10.1534/genetics.105.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls GL. The Vertebrate Eye and Its Adaptive Radiation. Cranbrook Institute of Science; 1942. [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, dos Reis M. Statistical properties of the branch-site test of positive selection. Molecular Biology and Evolution. 2011;28:1217–1228. doi: 10.1093/molbev/msq303. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Molecular Biology and Evolution. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen A-MK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N. In defense of statistical methods for detecting positive selection. Proceedings of the National Academy of Sciences. 2009;106:E395. doi: 10.1073/pnas.0904550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S. Gene duplications and evolution of the short wavelength– sensitive visual pigments in vertebrates. Molecular Biology and Evolution. 1994;11:32–39. doi: 10.1093/oxfordjournals.molbev.a040090. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Amino acid replacements and wavelength absorption of visual pigments in vertebrates. Molecular Biology and Evolution. 1995;12:53–61. doi: 10.1093/oxfordjournals.molbev.a040190. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular genetic basis of adaptive selection: Examples from color vision in vertebrates. Annual Review of Genetics. 1997;31:315–336. doi: 10.1146/annurev.genet.31.1.315. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Progress in Retinal and Eye Research. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Evolution of dim-light and color visual pigments. Annual Review of Genome and Human Genetics. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Takenaka N. Statistical and molecular analyses of evolutionary significance of red–green color vision and color blindness in vertebrates. Molecular Biology and Evolution. 2005;22:968–975. doi: 10.1093/molbev/msi080. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Yokoyama R. Adaptive evolution of photoreceptors and visual pigments in vertebrates. Annual Review of Ecology and Systematics. 1996;27:543–567. [Google Scholar]
- Yokoyama S, Yang H, Starmer WT. Molecular basis of spectral tuning in the red- and green-sensitive (M/LWS) pigments in vertebrates. Genetics. 2008a;179:2037–2043. doi: 10.1534/genetics.108.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proceedings of the National Academy of Sciences. 2008b;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nei M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. Journal of Molecular Evolution. 1997;44 (suppl 1):S139–S146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular Biology and Evolution. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhuang H, Chien M-S, Matsunami H. Dynamic functional evolution of an odorant receptor for sex-steroid-derived odors in primates. Proceedings of the National Academy of Sciences. 2009;106:21247–21251. doi: 10.1073/pnas.0808378106. [DOI] [PMC free article] [PubMed] [Google Scholar]