Abstract
Apurinic/apyrimidinic endonuclease-1 (APE-1), a key enzyme responsible for DNA base excision repair (BER), has been linked to cancer chemoradiosensitivity. The phosphorylation of p65 plays a role in the activation of this pathway. In this study, we investigated APE-1 expression and its interaction with p65 in esophageal squamous cell carcinoma (ESCC) tissue. The expression of APE-1, p65, p65 nuclear localization sequence (p65-NLS), and monocyte chemoattractant protein-1 (MCP-1) was assessed by immunohistochemical analysis in 67 human ESCC tissue samples. Real-time PCR and western blotting were also performed. p65 siRNA was evaluated to determine the role of p65 in the regulation of APE-1 expression. We found nuclear localization of APE-1 in 89.6% (60/67) of ESCC tissue samples. We also observed the colocalization of p65-NLS and APE-1 in esophageal cancer tissue. In KYSE220 cells, pretreatment of MG-132 significantly abrogated upregulation of p65 and APE-1 levels induced by MCP-1, and treatment with 10 and 20 nM p65 siRNA significantly inhibited APE-1 mRNA expression. siRNA for p65 treatment significantly increased the apoptotic index in 5-FU-treated KYSE220 cells. We conclude that APE-1 is overexpressed and mainly localized in the nuclear compartment of cancer cells, and partly regulated by p65 in the NF-κB pathway in ESCC tissue.
Keywords: APE-1, esophageal squamous cell carcinoma, NF-κB-p65, p65-NLS, MCP-1
Introduction
Esophageal cancer is one of the most common malignancies and leading causes of death worldwide, and it has a tendency to develop quickly, metastase early, and respond poorly to chemoradiotherapy; thus, the 5-year survival rate has been estimated at a very low 10–41%.(1–3) Esophageal cancer has two primary histological types, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). The former is predominant in east countries, including Japan and China, with a proportion of more than 90% of all esophageal cancers.(4) Despite the rapid advances in adjuvant treatment of ESCC, histologic response to chemoradiotherapy is still the major determinant that is associated with survival rate.(5) Therefore, the search for a biological marker for ESCC that is predictive of chemoradiosensitivity is of clinical significance. Apurinic/apyrimidinic endonuclease-1 (APE-1) is a key enzyme responsible for DNA base excision repair (BER). Apurinic/apyrimidinic (AP) sites arise from DNA damage induced by reactive oxygen/nitrogen species (ROS), drug use or radiation. The presence of AP sites is known to block DNA synthesis or lead to mutations or genetic instability.(6) Sensitivity to chemoradiotherapy is increased through the reduction of BER function caused by the downregulation of APE-1 activity.(7) Previous studies have reported APE-1 expression in several cancer tissues.(8–15) However, there are no available reports regarding APE-1 expression in ESCC tissue.
The NF-κB signaling pathway is involved in cell proliferation, survival, and angiogenensis, and previous studies have reported that it played an important role in the genesis and development of esophageal cancer.(16) Then, a recent study revealed that inhibition of NF-κB increased the sensitivity of ESCC to chemoradiotherapy.(17) Therefore, the relationship between NF-κB and APE-1 warranted exploration. p65 is one of the key subunits of NF-κB, and it has been reported that the phosphorylation of p65 has been another pathway of activation of NF-κB that is independent of IκB.(18) Moreover, the nuclear localization sequence of p65 (p65-NLS/active p65) may be useful in monitoring overall NF-κB activity in esophageal cancer tissue.(19) Therefore, exploring the relationship between APE-1 and p65 would shed light on the regulatory mechanism of APE-1. Monocyte chemoattractant protein-1 (MCP-1/CCL2), a member of the C-C chemokine family, is involved in macrophage infiltration and cancer progression and studies have confirmed that MCP-1 activates the NF-κB pathway.(20) In our study, we also investigated the effect of MCP-1 on p65 and APE-1 in esophageal cancer in vitro and in vivo.
Materials and Methods
ESCC tissues
A total of 67 paraffin-embedded ESCC tissue samples were obtained from archived esophageal cancer patients who had undergone esophagectomies but had not received chemoradiotheraphy at Nippon Medical School Hospital. The protocol of this study was approved by the Ethics Committee of Nippon Medical School and written informed consent was obtained from all patients. Among these patients, there were 55 males and 12 females, ranging in age from 53–80 years (65.5 ± 8.2). 53 out of 67 tissues were categorized according to UICC TNM classification (2002) as follows: 4, stage 0; 5, stage I; 16, stage IIA; 15, stage III; 4, stage IVA; and 9, stage IVB. In addition, we also collected 10 adjacent non-tumor tissue samples from ESCC patients as controls to study APE-1 expression.
Immunohistochemical analysis of APE-1, p65, p65-NLS and MCP-1
The expression of APE-1, p65, p65-NLS and MCP-1 was assessed by immunohistochemical analysis. Briefly, 4-µm sections were deparaffinized, antigens were retrieved by microwaving for 5 min in 5% urea solution, and endogenous peroxidase activity was blocked with 3% H2O2 in methanol. The samples were incubated overnight at 4°C with rabbit anti-human APE-1 antibody (diluted 1:100; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-human p65 antibody (diluted 1:1000; Abcam, Cambridge, UK), mouse anti-human p65-NLS antibody (diluted 1:200; Abcam), and rabbit anti-human MCP-1 antibody (diluted 1:100; Abcam). After washing, the secondary antibody was detected by LSAB 2 kit (DAKO, Glostrup, Denmark) using diaminobenzidine as the chromogen. For the negative control, primary antibodies were replaced with isotype-matched immunoglobulin.
Colocalization of APE-1 and p65-NLS were confirmed by fluorescence double immunostaining. Briefly, the samples were incubated with primary antibodies; rabbit anti-human APE-1 (diluted 1:20; Santa Cruz) for 1 h and mouse anti-human p65-NLS (diluted 1:20; Abcam) for 1 h, then with secondary antibodies; goat anti-rabbit labeled with TRITC and goat anti-mouse labeled with FITC, followed by nuclear counterstaining with 4',6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO).
The immunostaining scores were assessed by two independent pathologists. For APE-1, immunoreactivity was determined by the proportion of positive nuclei or cytoplasm to total cell number in representative areas of the cancer tissue. Sides were evaluated by counting at least 1000 cells (mononuclear cells) in four different areas of the specimens (APE-1 positive cells were classified according to nuclear staining and cytoplasmic staining). For p65 and p65-NLS, both the distribution and the intensity of staining were assessed in a semiquantative fashion. The distribution was scored according to the number of positive cells: none (not stained), 0; focal (<1/3 of cells stained), 1; multi-focal (1/3–2/3 of cells stained), 2; and diffuse (>2/3 stained), 3. The staining intensity was scored as: none (not stained), 0; mild, 1; and strong, 2 (clearly identified by ×40 magnification). Scoring for p65 and p65-NLS was determined based on the distribution and intensity as follows; 0, negative; 1, weakly positive; 2, mildly positive; 3 or 4, moderately positive; and 5, strongly positive.(21) For MCP-1, sections with more than 30% positive cells were considered as positive, while sections with 30% or less positive cells were considered as negative, as described previously.(22)
Cell culture and drugs
The ESCC cell line KYSE220 (purchased from the National Institute of Biomedical Innovation, Osaka, Japan) was maintained in RPMI 1640 (Nikken Bio Medical Laboratory, Kyoto, Japan) supplemented with 10% fetal bovine serum (Nippon Bio-Supply Center, Tokyo, Japan) at 37°C in 5% CO2. Group 1 received no treatment and served as control, group 2 was treated with MG-132 (1 µM, 6 h), group 3 was stimulated by hr MCP-1 (0.1 µM, 24 h) without pretreatment of MG-132, and group 4 was stimulated with MCP-1 following MG-132 treatment. Then, 5-FU (0, 10, 20, 40, 80 µl/ml) were added into the KYSE220 cell cultures with or without p65 siRNA pretreatment for 48 h.
Western blot analysis of APE-1 and p65 protein expression levels in treated KYSE220 cells
Stimulated KYSE220 cells were lysed in a buffer solution containing 0.1% NP-40, 10 nM NaCl, 5 mM MgCl2, 10 nM NaH2PO4 (pH 7.4), 65 mM Na-orthovanadate (S-6508, Sigma-Aldrich, St. Louis, MO), and protease inhibitor cocktail (P-8340, 1:100; Sigma-Aldrich) and centrifuged at 1200 g for 15 min. After centrifugation, nuclei were pelleted and suspended in nuclear buffer (1 mM EDTA, 3.5% SDS, 10% glycerol, and 70 mM Tris-Cl). APE-1 and p65 proteins were visualized by western blotting. Briefly, equal amounts of protein (20 µg) were analyzed by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat milk in TBST and incubated overnight at 4°C with rabbit anti-APE-1 (diluted 1:200; Santa Cruz), mouse anti-NFκB p65 (diluted 1:200; Santa Cruz) and mouse anti-β-actin (diluted 1:1000; Sigma) antibodies, and after washing the membranes were incubated with peroxidase-conjugated secondary antibodies (diluted 1:1000; GE Healthcare, Little Chalfont, UK) for 1 h at room temperature. Electrochemiluminescence (ECL; GE Healthcare) was used for detection of APE-1 and p65 proteins. Immunoblotting results were quantitatively analyzed by scanning the image and quantitating pixels using the ImageQuant program (GE Healthcare).
Real-time PCR of APE-1 expression levels in treated KYSE220 cells
Real-time quantitative PCR was performed to detect mRNA expression of p65 and APE-1 in treated KYSE220 cells. Briefly, RNA extracted from these cells was reverse-transcribed, and subsequently cDNA was amplified using a 7500 Fast real-time PCR system (Applied Biosystems, Tokyo, Japan) with primers, dual-labeled fluorogenic probes, and a Taqman PCR Reagent kit (Applied Biosystems). The primers and probes for NF-κB-p65 (Accession numbers: HSS 109161; Invitrogen Corp., Paisley, UK), APE-1 (Accession number: Hs00172396-ml*) and β-actin (Accession number: Hs99999903-ml*) were purchased from Applied Biosystems in USA. Known concentrations of serially diluted cDNA (from KATO III) were used as standards for quantification of sample cDNA. Copy numbers of cDNA for p65, APE-1 were standardized to that of β-actin for the same sample.
RNA interference (RNAi) for p65 in KYSE220 cells
For siRNA transfection, KYSE220 cells were inoculated at a density of 6 × 104/ml in RPMI-1640 medium without 10% fetal bovine serum and antibiotics. A cell suspension of 1.0 ml per well was seeded in 24-well plates. The siRNAs were synthesized corresponding to human NF-κB-p65 (Accession numbers: HSS 109161; Invitrogen Corp.), and one nonsilencing siRNA was also supplied.
Transfection of siRNAs into KYSE220 cells was performed using Lipofectamine RNAiMAX (Invitrogen Corp.). Briefly, 6–12 pM siRNAs (final concentration 10–20 nM) and 1.5 µl of Lipofectamine were used for each well. siRNAs and Lipofectamine were first diluted in Opti-MEM I Reduced Serum Medium (Invitrogen Corp.) respectively, and then, mixed and incubated for 20 min at room temperature for complex formation. Next, the entire mixture was added to the cells in the wells. At 24 h after transfection, the cells of 24-well plates were used for real-time PCR for APE-1.
Assessment of apoptosis
To evaluate apoptosis, stimulated-KYSE220 cells were treated with terminal deoxynucleotidyl transferase (TdT) enzyme and incubated in a humidified chamber at 37°C for 1 h. Apoptosis was evaluated using terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL) assay analysis (ApopTag; Oncor, Gaithersburg, MD). To evaluate the degree of apoptosis, the number of TUNEL-positive cells was counted within a ×400 field. Four fields were selected to determine the average score count. Data were expressed as the mean percentage of total cell number (apoptotic index).
Statistical analysis
Mann-Whitney U test was used for analysis of categorical data. One-way ANOVA was used for multiple comparisons. Spearman rank correlation test was used to determine the correlation of MCP-1 with p65, p65-NLS and APE-1. Results were expressed as the mean ± SD, and a p value less than 0.05 was considered a statistically significant difference.
Results
Expression of APE-1, MCP-1, p65, p65-NLS in ESCC tissue, and of APE-1 in adjacent non-tumor tissue
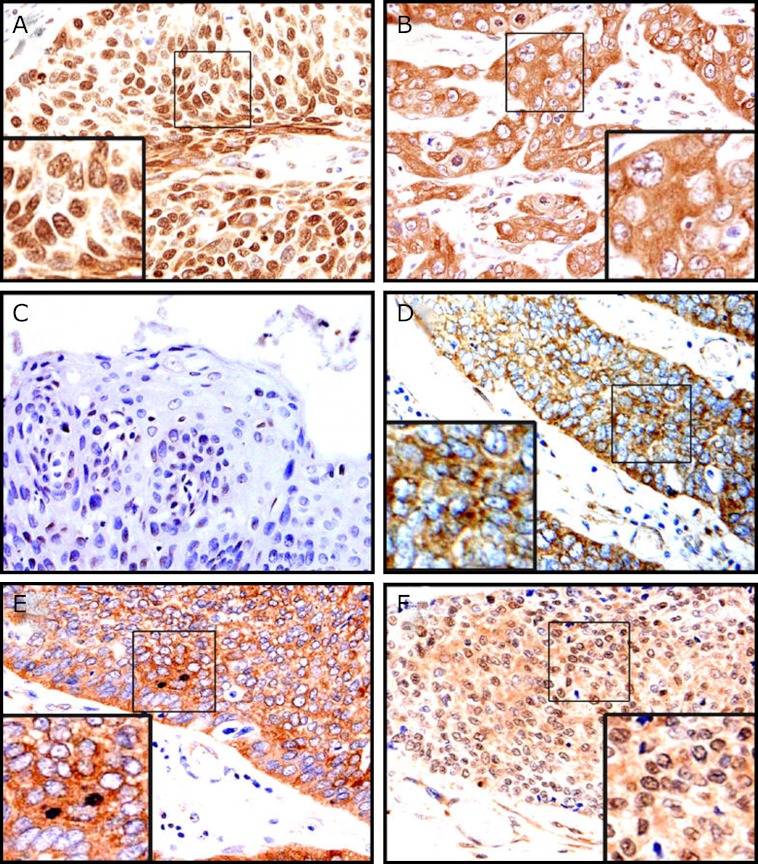
In human ESCC tissue samples, the nuclear and cytoplasmic localization of APE-1 were observed in 89.6% (60/67) and 11.4% (7/67) of all tissue samples, respectively (Fig. 1A and B). In contrast, in adjacent non-tumor tissue, APE-1 was weakly expressed and localized in nuclei of squamous epithelium (Fig. 1C). MCP-1 was found exclusively in the cytoplasmic compartment of cancer cells (Fig. 1D), while p65 was found in both the cytoplasmic (Fig. 1E) and nuclear compartments of cancer cells (p65-NLS, Fig. 1F).
Fig. 1.
Representative immunohistochemical findings in ESCC tissue (×200). APE-1 was found primarily in cancer cells with nuclear localization (A). APE-1 was found in cancer cells with cytoplasmic localization (B). In paracancerous tissues, APE-1 was weak (C). MCP-1 was found in cancer cells exclusively with cytoplasmic localization (D). p65 was found in cancer cells with both cytoplasmic localization and nuclear localization (NLS) (E, F).
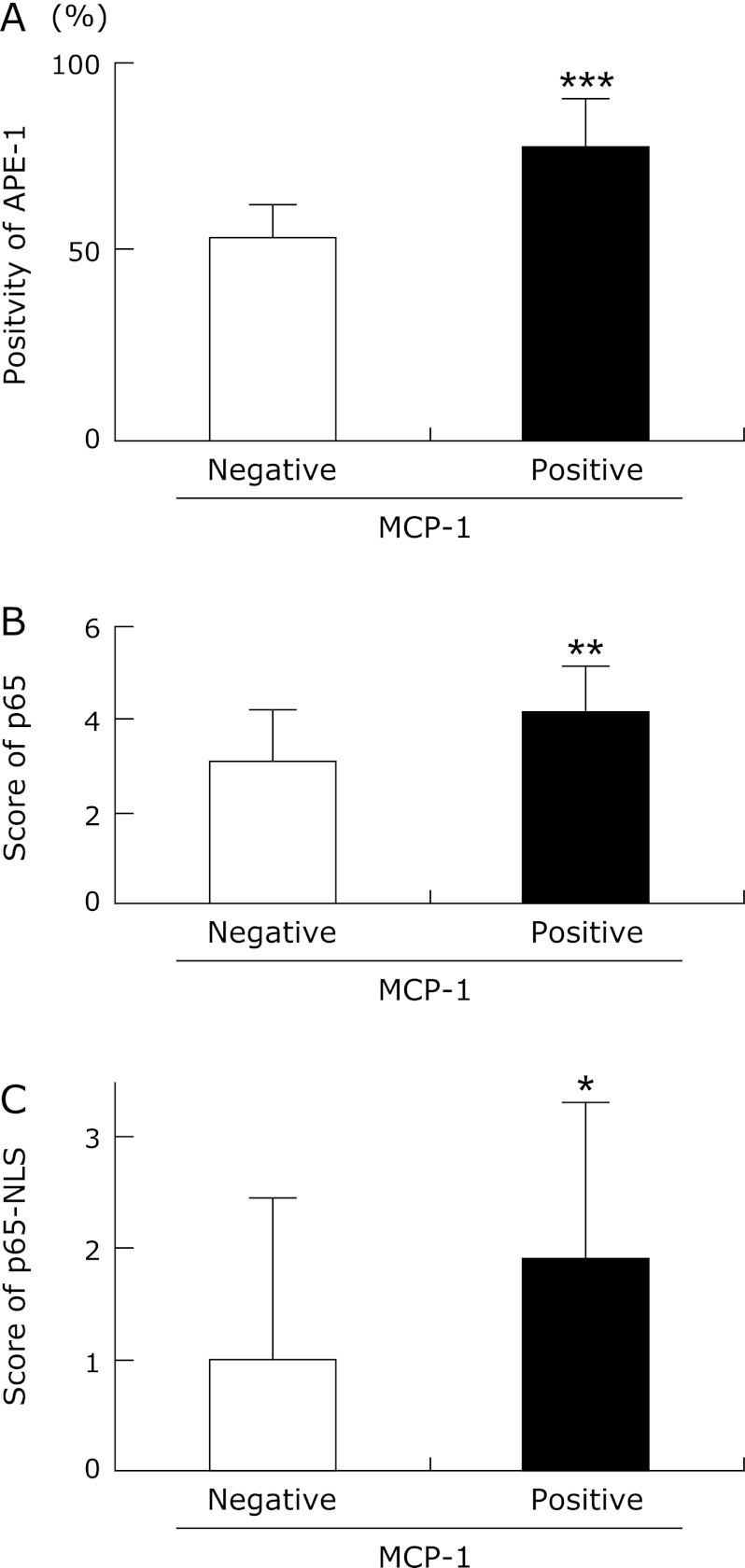
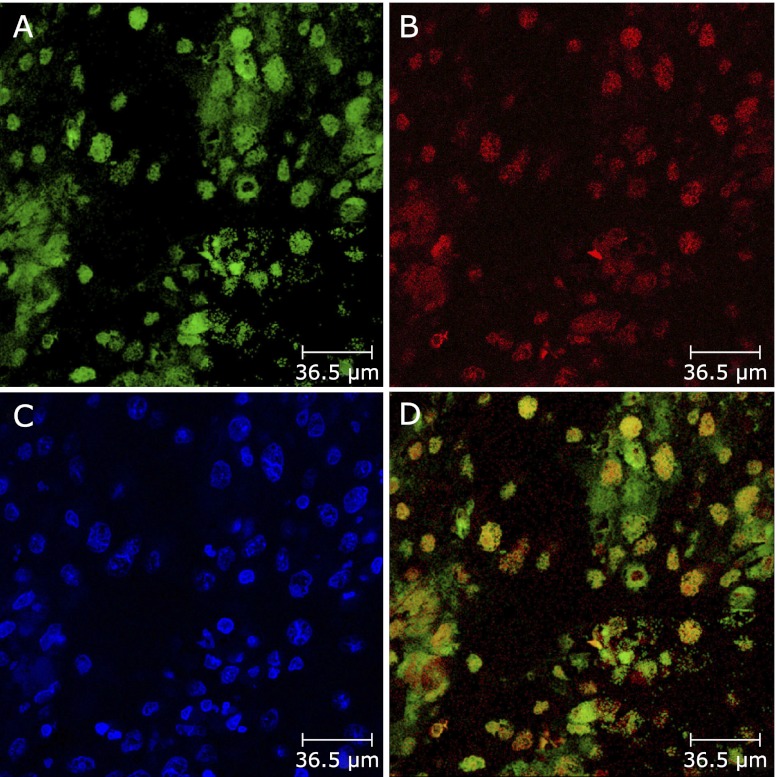
The tissue samples were divided into two groups according to MCP-1 positivity or negativity. The levels of APE-1, p65 and p65-NLS expression in the MCP-1-positive tissue samples were significantly higher than in the MCP-1-negative tissue samples (p<0.001; p = 0.001; p<0.05, respectively) (Fig. 2A, B and C). In addition, we found that MCP-1 correlated with p65, p65-NLS and APE-1 expression (p = 0.001; p<0.05; p<0.001, by Spearman rank correlation test, respectively). Moreover, we also investigated the colocalization of APE-1 and p65-NLS by fluorescence double immunostaining. FITC labelled (green) cells in ESCC tissue samples in Fig. 3A show p65 immunoreactivity. Fig. 3B shows APE-1 positive cells labelled with Texas red for the same section. Fig. 3C shows DAPI staining. Double immunostaining (yellow cells) for p65-NLS and APE-1 could be seen in the nuclei of cancer cells (Fig. 3D).
Fig. 2.
APE-1, p65 and p65-NLS expression in MCP-1 negative and MCP-1 positive ESCC tissue. Expression levels of APE-1 (A), p65 (B) and p65-NLS (C) in MCP-1-positive tissue were significantly increased compared to MCP-1-negative tissue (p<0.001; p = 0.001; p<0.05, respectively). Data are expressed as mean ± SD. (***p<0.001, **p = 0.001, *p<0.05).
Fig. 3.
Colocalization of p65-NLS and APE-1 in ESCC tissue. p65-NLS was found in the nuclear compartment of cancer cells (green cells, A) (original magnification, ×200), APE-1 was primary found in the nuclear compartment of cancer cells (red cells, B) (original magnification, × 200). 4',6-diamidino-2-phenylindole staining (blue staining, C) (original magnification, × 200). p65-NLS-positive and APE-1-positive cells were shown in the same section (yellow cells, D) (original magnification, × 200).
p65 and APE-1 mRNA and protein levels of expression in MCP-1-stimulated KYSE220 cells with or without MG-132 pretreatment
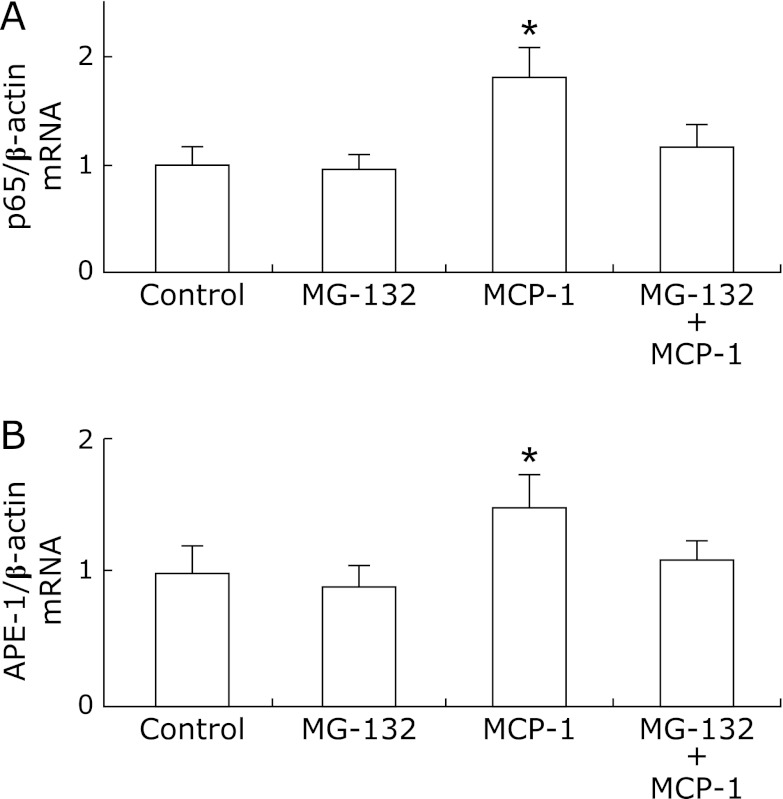
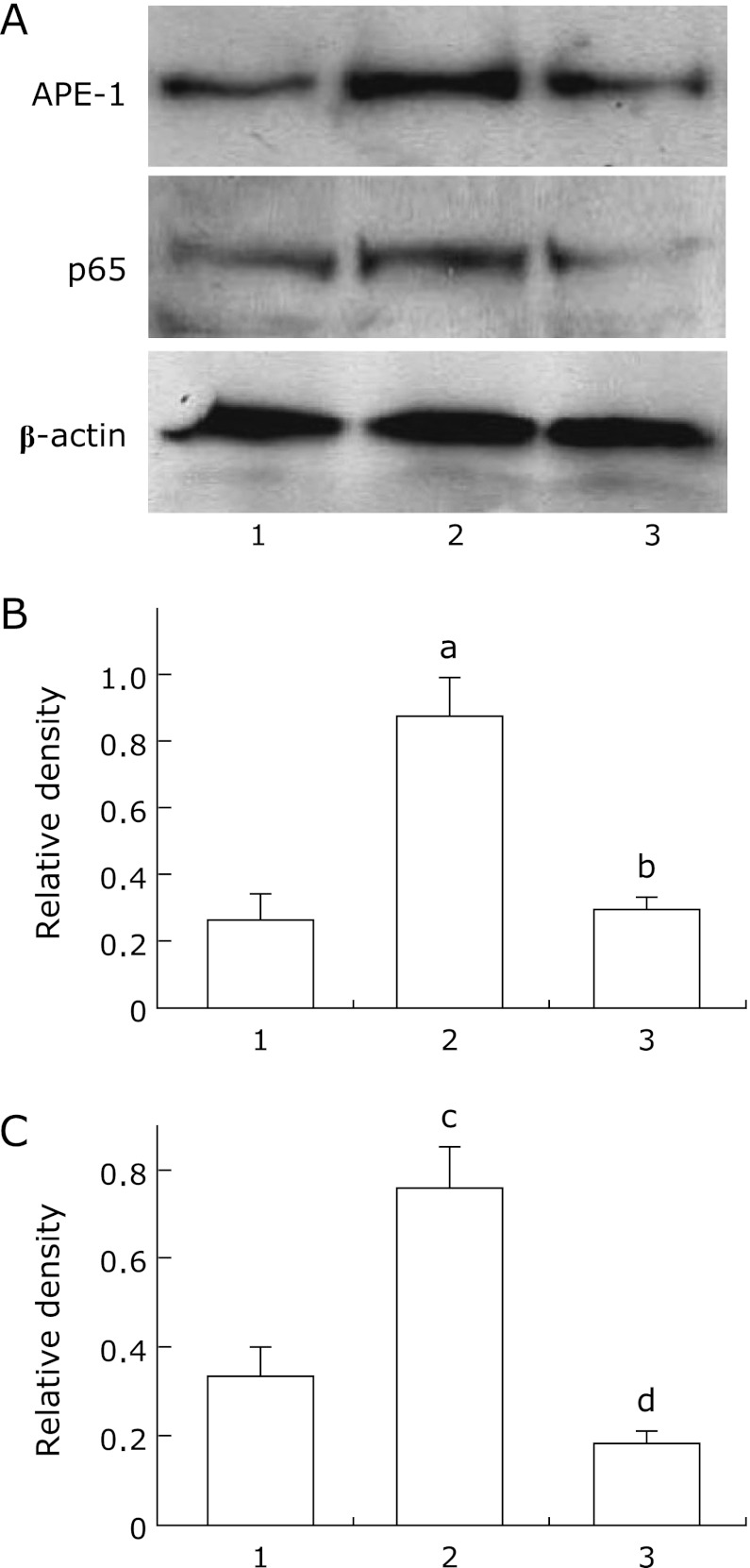
To determine whether MCP-1 could induce p65 and APE-1 expression, we evaluated p65 and APE-1 mRNA and protein levels in MCP-1-stimulated KYSE220 cells with or without MG-132 pretreatment. p65 and APE-1 mRNA levels were significantly upregulated in MCP-1-stimulated KYSE220 cells compared to unstimulated KYSE220 cells or MG-132-treated KYSE220 cells (p<0.001; p<0.001, respectively). In addition, pretreatment with MG-132 abrogated the upregulation of p65 and APE-1 mRNA levels induced by MCP-1 in KYSE220 cells (p<0.001; p<0.001, respectively; Fig. 4A and B). In similar tendency, p65 and APE-1 protein levels were significantly upregulated in MCP-1-stimulated KYSE220 cells compared to unstimualted KYSE220 cells (Fig. 5). Then, pretreatment with MG-132 significantly reduced the upregulation of p65 and APE-1 proetin levels induced by MCP-1 in KYSE 220 cells (Fig. 5).
Fig. 4.
The relative expression of p65 mRNA and APE-1 mRNA by real-time PCR in KYSE220 stimulated by MCP-1 with or without pretreatment of MG-132. Control: unstimulated KYSE 220 cells; MG-132: KYSE220 cells treated with MG-132; MCP-1: MCP-1 stimulated KYSE 220 cells; MCP-1 + MG-132: MCP-1 stimulated KYSE 220 cells pretreated with MG-132. MCP-1 (0.1 µM) significantly induced the upregulation of p65 mRNA, which was abrogated by pretreatment of MG-132 (1 µM) (p<0.001; p<0.001, respectively; A). MCP-1 (0.1 µM) significantly induced the upregulation of APE-1 mRNA, which was abrogated by pretreatment of MG-132 (1 µM) (p<0.001; p<0.001, respectively; B). The results are representative of three separate experiments. Data are expressed as mean ± SD. (*p<0.001).
Fig. 5.
APE-1 and p65 protein expression levels in MCP-1-stimulated KYSE220 cells. Representative western blots in MCP-1-stimulated KYSE220 cells for APE-1, p65 and β-actin. Lane 1: Unstimulated KYSE220 cells; lane 2: MCP-1 stimulated KYSE 220 cells; lane 3: MCP-1 stimulated KYSE 220 cells pretreated with MG-132 (A). Relative densities of APE-1 and p65 protein bands. Quantitation of APE-1 (B) and p65 (C) expression relative to β-actin expression was determined by densitometric analysis of each group using 3 separate experiments. Data are means ± SE. ap<0.05 vs unstimulated KYSE220 cells, bp<0.05 vs MCP-1-stimulated KYSE 220 cells, cp<0.05 vs unstimulated KYSE220 cells, dp<0.05 vs MCP-1-stimulated KYSE 220 cells.
mRNA Expression of APE-1 in KYSE 220 cells Treated with p65 siRNA
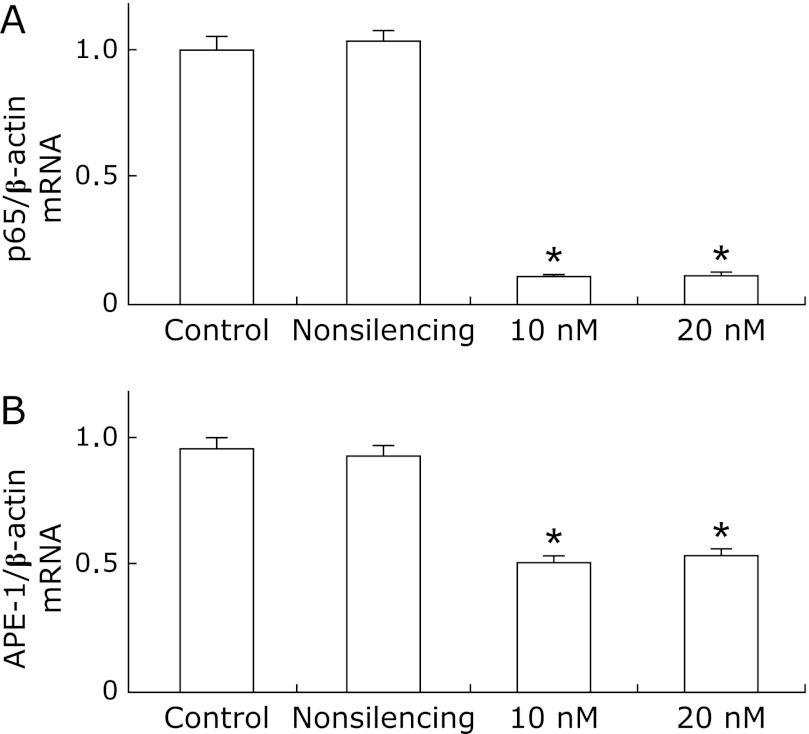
To determine the role of p65 in the regulation of APE-1 expression, we evaluated APE-1 mRNA in KYSE220 cells treated with p65 siRNA. Treatment with p65 siRNA nearly eliminated p65 mRNA expression. Also, 10 nM and 20 nM p65 siRNA significantly inhibited APE-1 mRNA expression compared to unstimulated cells or cells treated with nonsilencing siRNA (p<0.001; p<0.001, Fig. 6A and B).
Fig. 6.
The relative expression of APE-1 mRNA by real-time PCR in KYSE 220 cells treated with p65 siRNA. Control: unstimulated KYSE 220 cells; Nonsilencing: KYSE220 cells treated with nonsilencing siRNA. 10 nM: KYSE220 cells treated with 10 nM p65 siRNA. 20 nM: KYSE220 cells treated with 20 nM p65 siRNA. 10 nM and 20 nM p65 siRNA nearly knocked down p65 mRNA expression (p<0.001; p<0.001, respectively; A). 10 nM and 20 nM p65 siRNA significantly inhibited APE-1 mRNA (p<0.001; p<0.001, respectively; B). The results are representative of three separate experiments. Data are expressed as mean ± SD. (*p<0.001).
Apoptotic Index in 5-FU-incubated Esophageal Cancer Cell-lines Treated with p65 siRNA
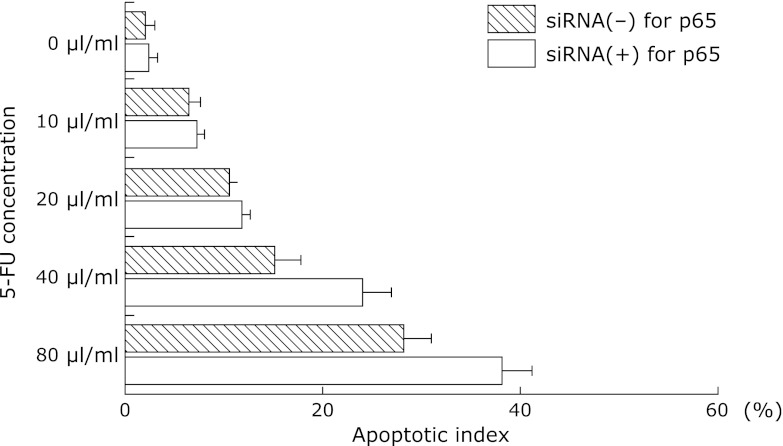
To clarify whether the APE-1-p65 pathway was associated with sensitivity to chemotherapy, we investigated the apoptotic index in 5-FU-incubated KYSE220 cells with or without p65 siRNA. p65 siRNA treatment significantly increased the apoptotic index in 5-FU (40 and 80 µl/ml)-treated KYSE220 cells compared to cells without p65 siRNA (Fig. 7).
Fig. 7.
Apoptotic index in 5-FU-incubated esophageal cancer cell-lines treated with siRNA for p65. siRNA for p65 treatment significantly (p<0.05; p<0.05, respectively) increased apoptotic index in 5-FU (40 and 80 µl/ml)-incubated KYSE220 cells compared to those without siRNA for p65. Data are means ± SE. The results are representative of four separate experiments. *p<0.05 vs siRNA (–) for p65, **p<0.05 vs siRNA (–) for p65.
Discussion
In this study, we aimed to investigate the expression of APE-1 in ESCC tissues and to determine whether APE-1 was induced by activation of NF-κB in ESCC cells. Our major findings are: (1) APE-1 was overexpressed and found primarily with nuclear localization in ESCC tissue; (2) APE-1 and p65-NLS were colocalized in the nuclei of cancer cells in ESCC tissue; (3) MCP-1 induced a significant increase in p65 and APE-1 mRNA and protein expression levels in KYSE220 cells; (4) Inhibition of NF-κB with MG-132 significantly abrogated the upregulation of p65 and APE-1 induced by MCP-1 in KYSE220 cells; and (5) Knockdown of p65 by siRNA significantly inhibited APE-1 mRNA expression and significantly increased the apoptotic index in 5-FU-treated KYSE220 cells.
First, immunohistochemical analysis of ESCC tissue showed that APE-1 was weakly expressed in adjacent non-tumor tissue whereas nuclear localization of APE-1 expression was observed in 89.6% of ESCC tissue samples, indicating that APE-1 is overexpressed in ESCC tissue. Esophageal carcinogenesis induced by ethanol and smoking is closely related to the metabolism of ethanol, acetaldehyde and acetate, and the carcinogens in smoking, such as polycyclic aromatic hydrocarbons and N-nitrosamines, respectively. These factors generate cytokines, prostaglandins, and ROS in the esophageal tissue, which damage biomolecules including DNA, leading to activation of oncogenes and/or inactivation of tumor suppressor genes, and finally to tumor development by creating a local microenvironment that facilitates neoplastic transformation and potentiates the progression of cancer.(23,24) The host response to free radical-induced damage includes the induction of DNA repair enzymes, including APE-1. The overexpression of APE-1 in ESCC tissues may be associated with chemoradiotherapy resistance and a poor outcome.(8) Considering that p65 siRNA treatment significantly reduced APE-1 mRNA levels while increasing the apoptotic index in 5-FU-treated KYSE220 cells, the APE-1-p65 pathway might be associated with apoptosis in 5-FU-treated esophageal cancer cells. In addition, the cytoplasmic localization of APE-1 was observed in 10.4% of ESCC tissue samples, but not in adjacent non-tumor tissue. The inability of APE-1 to translocate into the nuclei has been frequently observed in the tumorigenesis of some malignancies. Kakolyris et al.(25) found APE-1 expression was cytoplasmic in 11 of 30 adenomas (37%) and 22 of 44 carcinomas (50%). Wu et al.(26) recently demonstrated that cytoplasmic APE-1 expression promoted lung tumor aggressiveness via NF-κB activation. Nuclear APE-1 functions as DNA repair, and cytoplasmic APE-1 is involved in the interaction with certain transcription factors through its redox activity, such as p53, hypoxia-inducible factor-1α, NF-κB and so on.(27–29) Therefore, the localization of APE-1 in the cytoplasm found in ESCC tissues may prompt the aggressiveness, metastasis, and angiogenesis of cancer.
We then investigated the interaction of p65, one of the key subunits of NF-κB, with APE-1. In this study, p65 siRNA treatment significantly inhibited APE-1 expression in KYSE220 cells. In double immunostaining, we could find the colocalization of p65-NLS and APE-1 in esophageal cancer tissue. Tian et al.(17) confirmed that p65 had DNA-binding activity when it was translocated to the nucleus. In addition, γ-H2AX is a sensitive marker for the quantification of DNA damage and repair processes, and a study showed γ-H2AX decreased after 2 h of DNA damage in p65-positive cells, while this signal persisted for 12 h in cells lacking p65, indicating p65 might bind to DNA repair genes and regulate its expression.(30) Further studies will be needed to clarify the precise interaction between p65 and APE-1 in esophageal cancer tissue.
In our immunohistochemical analysis data, we found that MCP-1 positivity correlated with p65, p65-NLS and APE-1 expression. Subsequently, in KYSE220 cells, MCP-1 increased both p65 and APE-1 mRNA and protein expression levels. In addition, in our study, MG-132, an NF-κB inhibitor, abrogated the upregulation of p65 and APE-1 mRNA and protein levels induced by MCP-1. Ohta et al.(22) have reported that MCP-1 expression was also linked to poor prognosis in esophageal cancer. Viedt et al.(20) have reported that MCP-1 was linked to the activation of NF-κB with stimulating binding activity of NF-κB. Considering our data and a previous study, the downregulation of MCP-1 might also be an important strategy in preventing the development of esophageal cancer. Finally, previous studies have revealed that alcohol and smoking could induce MCP-1 expression in cancer tissues.(31,32) Therefore, we speculate that alcohol and smoking induces the release of MCP-1 in tissue, which in turn activates p65 in the cytoplasm. After upregulation, phosphorylated p65 is translocated into the nucleus, binding to the promoter, and ultimately regulating the transcription of APE-1 or other target genes.
In conclusion, our study demonstrates that APE-1 is overexpressed with primary nuclear localization in ESCC tissues. As a downstream target in the NF-κB signaling pathways, APE-1 is partly regulated by p65 in human ESCC cells. MCP-1 plays a role in the activation of NF-κB and APE-1. Our study suggests that both MCP-1 and the APE-1-p65 pathway are potential therapeutic targets for the treatment of ESCC. However, the precise mechanism regulating the expression of APE-1 via p65 needs further clarification.
Abbreviations
- APE-1
apurinic/apyrimidinic endonuclease-1
- ESCC
esophageal squamous cell carcinoma
- MCP-1
monocyte chemoattractant protein-1
- MG-132
carbobenzoxy-Lleucyl-L-leucyl-L-leucinal
- NF-κB
nuclear factor-kappa B
- p65-NLS
p65 nuclear localization sequence
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Xiao ZF, Yang ZY, Liang J, et al. Value of radiotherapy after radical surgery for esophageal carcinoma: a report of 495 patients. Ann Thorac Surg. 2003;75:331–336. doi: 10.1016/s0003-4975(02)04401-6. [DOI] [PubMed] [Google Scholar]
- 2.Shimizu K, Hihara J, Yoshida K, Toge T. Clinical evaluation of low-dose cisplatin and 5-fluorouracil as adjuvant chemoradiotherapy for advanced squamous cell carcinoma of the esophagus. Hiroshima J Med Sci. 2005;54:67–71. [PubMed] [Google Scholar]
- 3.Hagymási K, Tulassay Z. Risk factors for esophageal cancer, and possible genetic background. Orv Hetil. 2009;150:407–413. doi: 10.1556/OH.2009.28558. [DOI] [PubMed] [Google Scholar]
- 4.Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 2009;24:729–735. doi: 10.1111/j.1440-1746.2009.05824.x. [DOI] [PubMed] [Google Scholar]
- 5.Meredith KL, Weber JM, Turaga KK, et al. Pathologic response after neoadjuvant therapy is the major determinant of survival in patients with esophageal cancer. Ann Surg Oncol. 2010;17:1159–1167. doi: 10.1245/s10434-009-0862-1. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 8.Koukourakis MI, Giatromanolaki A, Kakolyris S, et al. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int J Radiat Oncol Biol Phys. 2001;50:27–36. doi: 10.1016/s0360-3016(00)01561-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Xiang DB, Yang XQ, et al. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer. 2009;66:298–304. doi: 10.1016/j.lungcan.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Futagami S, Hiratsuka T, Shindo T, et al. Expression of apurinic/apyrimidinic endonuclease-1 (APE-1) in H. pylori-associated gastritis, gastric adenoma, and gastric cancer. Helicobacter. 2008;13:209–218. doi: 10.1111/j.1523-5378.2008.00605.x. [DOI] [PubMed] [Google Scholar]
- 11.Di Maso V, Avellini C, Crocè LS, et al. Subcellular localization of APE1/Ref-1 in human hepatocellular carcinoma: possible prognostic significance. Mol Med. 2007;13:89–96. doi: 10.2119/2006-00084.DiMaso. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Zhou S, Sandusky GE, Kelley MR, Fishel ML. Reduced expression of DNA repair and redox signaling protein APE1/Ref-1 impairs human pancreatic cancer cell survival, proliferation, and cell cycle progression. Cancer Invest. 2010;28:885–895. doi: 10.3109/07357907.2010.512816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obtulowicz T, Swoboda M, Speina E, et al. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis. 2010;25:463–471. doi: 10.1093/mutage/geq028. [DOI] [PubMed] [Google Scholar]
- 14.Tanner B, Grimme S, Schiffer I, et al. Nuclear expression of apurinic/apyrimidinic endonuclease increases with progression of ovarian carcinomas. Gynecol Oncol. 2004;92:568–577. doi: 10.1016/j.ygyno.2003.10.037. [DOI] [PubMed] [Google Scholar]
- 15.Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res. 2004;24:2127–2134. [PubMed] [Google Scholar]
- 16.Li B, Li YY, Tsao SW, Cheung AL. Targeting NF-κB signaling pathway suppresses tumor growth, angiogenesis, and metastasis of human esophageal cancer. Mol Cancer Ther. 2009;8:2635–2644. doi: 10.1158/1535-7163.MCT-09-0162. [DOI] [PubMed] [Google Scholar]
- 17.Tian F, Zang WD, Hou WH, Liu HT, Xue LX. Nuclear factor-κB signaling pathway constitutively activated in esophageal squamous cell carcinoma cell lines and inhibition of growth of cells by small interfering RNA. Acta Biochim Biophys Sin (Shanghai) 2006;38:318–326. doi: 10.1111/j.1745-7270.2006.00166.x. [DOI] [PubMed] [Google Scholar]
- 18.Vermeulen L, De Wilde G, Notebaert S, Vanden Berghe W, Haegeman G. Regulation of the transcriptional activity of the nuclear factor-κB p65 subunit. Biochem Pharmacol. 2002;64:963–970. doi: 10.1016/s0006-2952(02)01161-9. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins GJ, Mikhail J, Alhamdani A, et al. Immunohistochemical study of nuclear factor-κB activity and interleukin-8 abundance in oesophageal adenocarcinoma; a useful strategy for monitoring these biomarkers. J Clin Pathol. 2007;60:1232–1237. doi: 10.1136/jcp.2006.043976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viedt C, Dechend R, Fei J, Hänsch GM, Kreuzer J, Orth SR. MCP-1 induces inflammatory activation of human tubular epithelial cells: involvement of the transcription factors, nuclear factor-κB and activating protein-1. J Am Soc Nephrol. 2002;13:1534–1547. doi: 10.1097/01.asn.0000015609.31253.7f. [DOI] [PubMed] [Google Scholar]
- 21.Charalambous MP, Lightfoot T, Speirs V, Horgan K, Gooderham NJ. Expression of COX-2, NF-κB-p65, NF-κB-p50 and IKKα in malignant and adjacent normal human colorectal tissue. Br J Cancer. 2009;101:106–115. doi: 10.1038/sj.bjc.6605120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohta M, Kitadai Y, Tanaka S, et al. Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human esophageal squamous cell carcinomas. Int J Cancer. 2002;102:220–224. doi: 10.1002/ijc.10705. [DOI] [PubMed] [Google Scholar]
- 23.Rieder F, Biancani P, Harnett K, Yerian L, Falk GW. Inflammatory mediators in gastroesophageal reflux disease: impact on esophageal motility, fibrosis, and carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2010;298:G571–G581. doi: 10.1152/ajpgi.00454.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdel-Latif MM, Duggan S, Reynolds JV, Keller D. Inflammation and esophageal carcinogenesis. Curr Opin Pharmacol. 2009;9:396–404. doi: 10.1016/j.coph.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Kakolyris S, Kaklamanis L, Engels K, et al. Human apurinic endonuclease 1 expression in a colorectal adenoma-carcinoma sequence. Cancer Res. 1997;57:1794–1797. [PubMed] [Google Scholar]
- 26.Wu HH, Cheng YW, Chang JT, et al. Subcellular localization of apurinic endonuclease 1 promotes lung tumor aggressiveness via NF-κB activation. Oncogene. 2010;29:4330–4340. doi: 10.1038/onc.2010.178. [DOI] [PubMed] [Google Scholar]
- 27.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley MR, Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid Redox Signal. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- 29.Ando K, Hirao S, Kabe Y, et al. A new APE1/Ref-1-dependent pathway leading to reduction of NF-κB and AP-1, and activation of their DNA-binding activity. Nucleic Acids Res. 2008;36:4327–4336. doi: 10.1093/nar/gkn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sedelnikova OA, Rogakou EP, Panyutin IG, Bonner WM. Quantitative detection of (125)IdU-induced DNA double-strand breaks with γ-H2AX antibody. Radiat Res. 2002;158:486–492. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Xu M, Li F, et al. Ethanol promotes mammary tumor growth and angiogenesis: the involvement of chemoattractant factor MCP-1. Breast Cancer Res Treat. 2012;133:1037–1048. doi: 10.1007/s10549-011-1902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazar M, Sullivan J, Chipitsyna G, et al. Induction of monocyte chemoattractant protein-1 by nicotine in pancreatic ductal adenocarcinoma cells: role of osteopontin. Surgery. 2010;148:298–309. doi: 10.1016/j.surg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]