Abstract
Discovery of novel antigens associated with infectious diseases is fundamental to the development of serodiagnostic tests and protein subunit vaccines against existing and emerging pathogens. Efforts to predict antigenicity have relied on a few computational algorithms predicting signal peptide sequences (SignalP), transmembrane domains, or subcellular localization (pSort). An empirical protein microarray approach was developed to scan the entire proteome of any infectious microorganism and empirically determine immunoglobulin reactivity against all the antigens from a microorganism in infected individuals. The current database from this activity contains quantitative antibody reactivity data against 35,000 proteins derived from 25 infectious microorganisms and more than 30 million data points derived from 15,000 patient sera. Interrogation of these data sets has revealed ten proteomic features that are associated with antigenicity, allowing an in silico protein sequence and functional annotation based approach to triage the least likely antigenic proteins from those that are more likely to be antigenic. The first iteration of this approach applied to Brucella melitensis predicted 37% of the bacterial proteome containing 91% of the antigens empirically identified by probing proteome microarrays. In this study, we describe a naïve Bayes classification approach that can be used to assign a relative score to the likelihood that an antigen will be immunoreactive and serodiagnostic in a bacterial proteome. This algorithm predicted 20% of the B. melitensis proteome including 91% of the serodiagnostic antigens, a nearly twofold improvement in specificity of the predictor. These results give us confidence that further development of this approach will lead to further improvements in the sensitivity and specificity of this in silico predictive algorithm.
Introduction
Our lab has developed an approach to construct and probe protein microarrays on a genome-wide scale. We have applied this approach to more than 25 medically important infectious microorganisms, including Mycobacterium tuberculosis [1], Plasmodium falciparum [2– 4], Plasmodium vivax, Brucella melitensis [5], Chlamydia trachomatis [6], Francisella tularensis [7] [8], Burkholderia pseudomallei [9] [10], Coxiella burnetii [11] [12], Borrelia burgdorferi [13], Salmonella enterica serovar Typhi, Rickettsia prowazekii, Rickettsia rickettsii, Orientia tsutsugamushi, Bartonella henselae [14], Leptospira interrogans, Toxoplasma gondii [15], Candida albicans [16], and Schistosoma mansoni [17], and viruses, including vaccinia [18 – 21], monkeypox, herpes type 1 and 2, varicella zoster, human papillomavirus (HPV) [22], HIV, dengue, influenza, West Nile, and Chikungunya. In total, we have now made more than 35,000 plasmids, printed the encoded proteins on 25,000 microarrays, and probed the arrays with 15,000 serum specimens, to determine disease-associated antibody profiles in people infected with each agent. The individual proteins printed on these arrays capture antibodies present in serum from infected individuals and the amount of captured antibody can be quantified using fluorescent secondary antibody. In this way, a comprehensive profile of antibodies resulting after infection or exposure can be determined that is characteristic of the type of infection and the stage of disease [13] [20] [21]. The goals of this research are to develop a more detailed understanding of how the immune system responds to infection and to identify serodiagnostic and subunit vaccine antigens.
Another application for this empirical data is to train an algorithm to predict reactive antigens in silico, and several articles from our group apply enrichment analyses to identify proteomic features that tend to be seen more frequently in the immunodominant and serodiagnostic antigen sets [7] [14]. The antigens were classified according to annotated functional features (e.g., clusters of orthologous groups of proteins (COGs)), computationally predicted features (e.g., subcellular localization, physical properties), and protein expression estimated by mass spectrometry (MS); we found that membrane antigens and virulence factors and proteins expressed at high levels tend to be recognized more frequently by the immune system than the rest of the proteome, and this type of analysis predicted ca. 37% of the protein containing as much as 91% of the serodiagnostic antigens. More recently, an article from our group described a sequence-based approach called AntigenPro for predicting protective antigens that have ca. 75% prediction accuracy [23].
In this study, a naïve Bayes classification approach was applied to assign a relative numerical score to each antigen in the Brucella melitensis proteome. This score reflects the relative likelihood that a protein will be reactive based on its functionally annotated or computationally predicted features. Our analyses indicate that i) over 90% of serodiagnostic antigens are predictable from the top 20% of genome ranked by this naïve Bayes classification approach and ii) the antigens with enriched features in the top 20% of the genome account for 100% of serodiagnostic antigens with these features. This approach greatly enhances the predictive efficiency, compared to previous studies, provides a basis for targeted screens of entire proteomes based on likelihood of seroreactivity, and helps determine trends in the humoral immune response to Gram-negative bacteria.
Results
A data set from empirically determined proteome-wide serological analysis of B. melitensis was examined in the context of naturally acquired human infections, using protein microarrays for B. melitensis [5] [24]. The microarray contained 3046 proteins, corresponding to 95% of the proteome. The immunoproteome comprised 1464 antigens that reacted with at least one culture-positive individual, accounting for 48% of the proteome. Within this immunoproteome, 122 antigens were classified ‘seroreactive’ with mean reactivity greater than 2.5 standard deviations above the mean of the negative controls. Within this collection of 122 seroreactive antigens, there were 33 serodiagnostic antigens that distinguished naturally infected Peruvian brucellosis patients and healthy subjects from the same region with sensitivity and specificity >95%. The remaining 89 seroreactive antigens were defined as cross-reactive, because they reacted similarly in both infected and healthy individuals.
We classified the seroreactive and serodiagnostic antigens according to annotated functional features (COGs), computationally predicted features, and protein expression estimated by mass spectrometry (MS) [24]. Enrichment analyses indicated that ten features were significantly enriched in seroreactive and serodiagnostic antigen sets (Table 1) [24]. These enriching features are i) functionally annotated COGs U, M, N, and O, ii) computationally predicted features (TMHMM = 1, SignalP >0.7, pSort outer membrane, pSort periplasmic, and pI < 5), and iii) MS evidence of expression. All together, these ten features accounted for 37% of the proteome and included 91% of the serodiagnostic antigens.
Table 1.
List of Ten Enriching Features Previously Identified in Seroreactive, Serodiagnostic, and Cross-Reactive Antigen Setsa)
| Enriching features | Proteins on chip | Seroreactive antigens | Serodiagnostic antigens | Cross-reactive antigens | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hits | Enrichmentb) | p-Value | Hits | Enrichment | p-Value | Hits | Enrichment | p-Value | ||
| Functionally annotated COGs | ||||||||||
| U: Intracellular trafficking and secretion | 48 | 8 | 4.3 | 4.05E − 04 | 3 | 6.2 | 1.17E − 02 | 5 | 3.6 | 1.11E − 02 |
| M: Cell envelope biogenesis, outer membrane | 166 | 13 | 2.0 | 1.18E − 02 | 3 | 1.8 | 2.31E − 01 | 10 | 2.1 | 2.58E − 02 |
| N: Cell motility and secretion | 30 | 4 | 3.5 | 2.66E − 02 | 1 | 3.3 | 2.62E − 01 | 3 | 3.5 | 5.25E − 02 |
| O: Posttranslational modification, protein turnover, chaperones | 114 | 13 | 3.0 | 3.54E − 04 | 3 | 2.6 | 1.04E − 01 | 10 | 3.1 | 1.30E − 03 |
| Computationally predicted features | ||||||||||
| TMHMM = 1 | 204 | 35 | 4.3 | 1.44E − 14 | 11 | 5.0 | 4.87E − 06 | 24 | 4.0 | 1.21E − 09 |
| SignalP > 0.7 | 337 | 41 | 3.0 | 8.46E − 12 | 13 | 3.6 | 2.16E − 05 | 28 | 2.8 | 1.09E − 07 |
| pSort outer membrane | 32 | 5 | 3.9 | 8.02E − 03 | 4 | 11.5 | 3.32E − 04 | 1 | 1.1 | 6.15E − 01 |
| pSort periplasmic | 88 | 12 | 3.4 | 1.53E − 04 | 4 | 4.2 | 1.40E − 02 | 8 | 3.1 | 3.67E − 03 |
| pI < 5 | 242 | 16 | 1.7 | 3.92E − 02 | 2 | 0.8 | 1.00E + 00 | 14 | 2.0 | 1.44E − 02 |
| Protein expression evidenced by MS | 356 | 41 | 2.9 | 5.28E − 11 | 20 | 5.2 | 1.87E − 11 | 21 | 2.0 | 1.19E − 03 |
| Total proteins on chip | 3046 | 122 | 33 | 89 | ||||||
Data from recently published results [24].
Values indicate the fold enrichment.
As is important for diagnostic antigen discovery, we have been developing a better classifier of protein-antigenicity prediction to increase the likelihood of identifying serodiagnostic antigens and to apply it on a high-throughput scale to existing or new proteomes. To better quantify the relationship between all of these features and the seroreactivity of the proteins, here, we used a naïve Bayes classification scheme to rank all expression-confirmed proteins in the B. melitensis proteome according to the probability of antigenicity [7] [25].
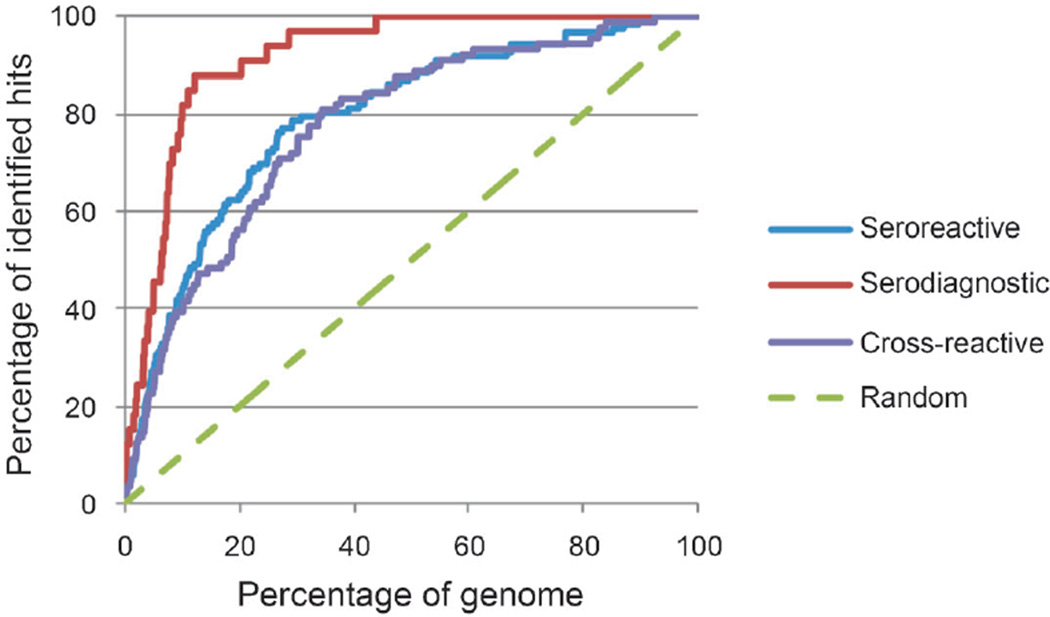
The naïve Bayes model makes the strong assumption that each feature is conditionally independent of all other features. While this assumption is not always accurate, in practice, this method often out-performs more sophisticated models. The likelihood ratio was calculated using all the proteomic features for all proteins in B. melitensis, and the proteins were ranked according to this calculation for seroreactive, serodiagnostic, or cross-reactive antigens. To assess the ranking scheme, we examined where the seroreactive or serodiagnostic hits were ranked. For example, by doing so, we would need to screen only 20% of the genome to be able to identify 63% of all seroreactive antigens, or 56% of cross-reactive antigens, or even more importantly, 91% of serodiagnostic antigens (Tables 2 – 4). As the list of proteins increased, more seroreactive and serodiagnostic antigens were identified (Fig. 1). Precisely, 100% of serodiagnostic hits were among the top 44% of the entire list, but 92% of the proteome had to be sampled to identify all 100% of cross-reactive hits. We take this as evidence that antibody responses against the cross-reactive antigens are derived from previous exposure to unrelated infections and not associated with the active infection in the infected group.
Table 2.
Naïve Bayes Ranking of Seroreactive Antigens
| Top ranked antigens [%] | Antigens on chip | Seroreactive antigens | |||
|---|---|---|---|---|---|
| Number | Proportion [%] | Enrichmenta) | p-Value | ||
| 1 | 30 | 7 | 6 | 5.83 | 1.31E − 04 |
| 2 | 60 | 13 | 11 | 5.41 | < 1.00E − 04 |
| 5 | 152 | 33 | 27 | 5.42 | < 1.00E − 04 |
| 10 | 304 | 54 | 44 | 4.43 | < 1.00E − 04 |
| 20b) | 617 | 77 | 63 | 3.12 | < 1.00E − 04 |
| 25 | 761 | 88 | 72 | 2.89 | < 1.00E − 04 |
| 30 | 913 | 96 | 79 | 2.63 | < 1.00E − 04 |
| 40 | 1218 | 99 | 81 | 2.03 | < 1.00E − 04 |
| 50 | 1523 | 107 | 88 | 1.75 | < 1.00E − 04 |
| 60 | 1827 | 112 | 92 | 1.53 | < 1.00E − 04 |
| 70 | 2132 | 115 | 94 | 1.35 | < 1.00E − 04 |
| 75 | 2284 | 115 | 94 | 1.26 | < 1.00E − 04 |
| 80 | 2436 | 118 | 97 | 1.21 | < 1.00E − 04 |
| 90c) | 2732 | 122 | 100 | 1.11 | < 1.00E − 04 |
| 100 | 3046 | 122 | 100 | 1.00 | 1.00E + 00 |
Values indicate the fold enrichment.
The top 20% of the genome contain 77 (63%) seroreactive antigens.
The top 90% of the genome contain all 122 (100%) seroreactive antigens.
Table 4.
Naïve Bayes Ranking of Cross-Reactive Antigens
| Top ranked antigens [%] | Antigens on chip | Cross-reactive antigens | |||
|---|---|---|---|---|---|
| Number | Proportion [%] | Enrichmenta) | p-Value | ||
| 1 | 30 | 6 | 7 | 18.46 | < 0.05 |
| 2 | 60 | 11 | 12 | 16.92 | < 0.05 |
| 5 | 152 | 22 | 25 | 13.36 | < 0.05 |
| 10 | 304 | 35 | 39 | 10.63 | < 0.05 |
| 20b) | 617 | 50 | 56 | 7.48 | < 0.05 |
| 25 | 761 | 31 | 35 | 3.76 | < 0.05 |
| 30 | 913 | 64 | 72 | 6.47 | < 0.05 |
| 40 | 1218 | 74 | 83 | 5.61 | < 0.05 |
| 50 | 1523 | 78 | 88 | 4.73 | < 0.05 |
| 60 | 1827 | 82 | 92 | 4.14 | < 0.05 |
| 70 | 2132 | 83 | 93 | 3.59 | < 0.05 |
| 75 | 2284 | 84 | 94 | 3.39 | < 0.05 |
| 80 | 2436 | 84 | 94 | 3.18 | < 0.05 |
| 90 | 2741 | 88 | 99 | 2.96 | < 0.05 |
| 92c) | 2816 | 89 | 100 | 2.92 | < 0.05 |
| 100 | 3046 | 89 | 100 | 2.70 | < 0.05 |
Values indicate the fold enrichment.
The top 20% of the genome contain 50 (56%) cross-reactive antigens.
The top 92% of the genome contain all 122 (100%) cross-reactive antigens.
Fig. 1. Combined naïve Bayes classifier on ranking of seroreactive, serodiagnostic, or cross-reactive antigens.
Genome was ranked by naïve Bayes classifier based on seroreactive, serodiagnostic, or cross-reactive antigens features. As the size of the genome increases, the prediction rate also increases.
To access the probability of prediction using these ten enriched proteomic features, we looked at individual features among the ranked proteins (Table 5). Most features were significantly more enriched in seroreactive antigens in the top 1% of the genome, and the fold enrichment drops as the percentage of genome size increases. Of all these features, COG U was most enriched in the top 1% of the genome. COG N, however, was not identified at all in the top 2% of ranked proteins. Except pI < 5, all enriching features remained at least twofold enriched as 100% of the genome size was reached.
Table 5.
Enrichment of Each Individual Feature in Seroreactive Antigens
| Top ranked antigens [%] |
Antigens on chip |
Seroreactive antigens |
Enrichment of featurea) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COG U | COG M | COG N | COG O | pS om | pS p | SigP | TMHMM | pI | MS | |||
| 1 | 30 | 7 | 25.91 | 3.24 | 0.00 | 17.28 | 8.32 | 15.60 | 5.83 | 6.47 | 8.32 | 4.62 |
| 2 | 60 | 13 | 5.18 | 5.18 | 0.00 | 15.55 | 5.76 | 9.20 | 5.35 | 5.81 | 5.55 | 4.88 |
| 5 | 152 | 33 | 2.59 | 6.05 | 6.48 | 11.11 | 4.99 | 8.61 | 5.51 | 5.71 | 3.12 | 5.26 |
| 10 | 304 | 54 | 5.46 | 6.03 | 2.59 | 6.86 | 4.99 | 5.09 | 4.32 | 5.90 | 2.90 | 4.84 |
| 20 | 617 | 77 | 4.86 | 4.39 | 2.36 | 6.17 | 4.62 | 4.65 | 3.63 | 4.65 | 2.38 | 3.11 |
| 25 | 761 | 88 | 5.32 | 4.44 | 2.99 | 5.10 | 4.30 | 3.71 | 3.45 | 4.39 | 2.15 | 2.93 |
| 30 | 913 | 96 | 5.18 | 3.46 | 3.57 | 3.44 | 4.16 | 3.70 | 3.42 | 4.35 | 2.32 | 2.98 |
| 40 | 1218 | 99 | 4.94 | 2.29 | 3.57 | 3.40 | 4.16 | 3.40 | 3.23 | 4.35 | 2.18 | 2.93 |
| 50 | 1523 | 107 | 4.71 | 2.14 | 3.46 | 3.27 | 4.16 | 3.40 | 3.16 | 4.28 | 1.80 | 2.90 |
| 60 | 1827 | 112 | 4.71 | 2.14 | 3.46 | 3.21 | 4.16 | 3.40 | 3.05 | 4.28 | 1.84 | 2.88 |
| 70 | 2132 | 115 | 4.71 | 1.96 | 3.46 | 3.21 | 4.16 | 3.40 | 3.05 | 4.28 | 1.82 | 2.88 |
| 75 | 2284 | 115 | 4.51 | 1.96 | 3.46 | 3.21 | 4.16 | 3.40 | 3.05 | 4.28 | 1.82 | 2.88 |
| 80 | 2436 | 118 | 4.51 | 1.94 | 3.46 | 3.21 | 4.16 | 3.40 | 3.05 | 4.28 | 1.70 | 2.88 |
| 90 | 2741 | 122 | 4.51 | 2.04 | 3.46 | 3.01 | 4.03 | 3.40 | 3.04 | 4.28 | 1.68 | 2.88 |
| 100 | 3046 | 122 | 4.32 | 2.03 | 3.46 | 2.96 | 3.90 | 3.40 | 3.04 | 4.28 | 1.65 | 2.88 |
Values indicate the fold enrichment. The abbreviations of the enriching features are as follows: pS om, pSort outer membrane; pS p, pSort perplasmic; SigP, SignalP > 0.7; TMHMM, TMHMM = 1; pI, pI < 5; MS, protein expression evidenced by MS. For further details on the enriching features, cf. Table 1.
As we looked into enriching features in each combined category, we also observed the highest probability of predicting seroreactive and serodiagnostic antigens in the top 1% of ranked genome (Tables 6 and 7). Of 15 antigens predicted to have COGs U, M, N, and O, 6 were identified to be seroreactive, leading to a prediction rate of 40% for seroreactive antigens (Table 6). The probability of prediction on seroreactive antigens is 23% for computationally predicted features, 19% for MS evidence of expression, and 23% for proteins with all ten features (Table 6). In the top 1% of the ranked genome, serodiagnostic antigens were found in 29% of proteins with COGs U, M, N, O features, 17% of proteins with computationally predicted features, 17% of proteins with MS evidence of expression, and 17% of proteins with all these ten features (Table 7). These prediction rates were significantly higher compared to an expected prediction with no applied selection criteria. Indeed, we would expect the prediction rates on seroreactive antigens by chance to be 11, 10, 12, and 9% and the prediction rates on serodiagnostic antigens by chance to be 3, 3, 6, and 3% for these four categories of proteins, respectively. With the attempt to identify a larger percentage of the seroreactive or serodiagnostic proteins, the overall prediction rate decreased.
Table 6.
Probability of Predicting Seroreactive Antigens by Enriching Featuresa) in Proteins Ranked by Naïve Bayes Classification
| Top ranked antigens [%] |
Antigens on chip |
Seroreactive antigens |
COGs | Computational prediction | MS Protein expression | Sum of all ten features | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted | Hits | Rate | Predicted | Hits | Rate | Predicted | Hits | Rate | Predicted | Hits | Rate | |||
| 1 | 30 | 7 | 15 | 6 | 0.40 | 30 | 7 | 0.23 | 27 | 5 | 0.19 | 30 | 7 | 0.23 |
| 2 | 60 | 13 | 30 | 10 | 0.33 | 60 | 13 | 0.22 | 46 | 9 | 0.20 | 60 | 13 | 0.22 |
| 5 | 152 | 33 | 53 | 15 | 0.28 | 151 | 33 | 0.22 | 76 | 16 | 0.21 | 152 | 33 | 0.22 |
| 10 | 304 | 54 | 97 | 24 | 0.25 | 270 | 47 | 0.17 | 129 | 25 | 0.19 | 304 | 54 | 0.18 |
| 20 | 617 | 77 | 136 | 27 | 0.20 | 440 | 59 | 0.13 | 289 | 36 | 0.12 | 617 | 77 | 0.12 |
| 25 | 761 | 88 | 176 | 34 | 0.19 | 488 | 59 | 0.12 | 341 | 40 | 0.12 | 753 | 88 | 0.12 |
| 30 | 913 | 96 | 237 | 36 | 0.15 | 544 | 64 | 0.12 | 344 | 41 | 0.12 | 866 | 95 | 0.11 |
| 40 | 1218 | 99 | 286 | 36 | 0.13 | 578 | 64 | 0.11 | 349 | 41 | 0.12 | 952 | 95 | 0.10 |
| 50 | 1523 | 107 | 302 | 36 | 0.12 | 633 | 65 | 0.10 | 353 | 41 | 0.12 | 1027 | 96 | 0.09 |
| 60 | 1827 | 112 | 304 | 36 | 0.12 | 668 | 67 | 0.10 | 355 | 41 | 0.12 | 1066 | 98 | 0.09 |
| 70 | 2132 | 115 | 318 | 36 | 0.11 | 670 | 67 | 0.10 | 356 | 41 | 0.12 | 1083 | 98 | 0.09 |
| 75 | 2284 | 115 | 320 | 36 | 0.11 | 670 | 67 | 0.10 | 356 | 41 | 0.12 | 1085 | 98 | 0.09 |
| 80 | 2436 | 118 | 321 | 36 | 0.11 | 686 | 67 | 0.10 | 356 | 41 | 0.12 | 1102 | 98 | 0.09 |
| 90 | 2732 | 122 | 329 | 37 | 0.11 | 691 | 67 | 0.10 | 356 | 41 | 0.12 | 1115 | 99 | 0.09 |
| 100 | 3046 | 122 | 338 | 37 | 0.11 | 696 | 67 | 0.10 | 356 | 41 | 0.12 | 1128 | 99 | 0.09 |
The enriching features are classified into three categories: i) COGs (COG U, M, N, and O) for proteins assigned to one or more of these four COGs, ii) computational predictions with enriching features for proteins predicted to have one or more of the following features, i.e., TMHMM = 1, SignalP > 0.7, pSort outer membrane, pSort periplasmic, or pI < 5, and iii) protein expression as detected by mass spectrometry. For further details on the enriching features, cf. Table 1.
Table 7.
Probability of Predicting Serodiagnostic Antigens by Enriching Featuresa) in Proteins Ranked by Naïve Bayes Classification
| Top ranked antigens [%] |
Antigens on chip |
COGs | Computational prediction | MS Protein expression | Sum of all ten features | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted | Hits | Rate | Predicted | Hits | Rate | Predicted | Hits | Rate | Predicted | Hits | Rate | ||
| 1 | 30 | 14 | 4 | 0.29 | 30 | 5 | 0.17 | 30 | 5 | 0.17 | 30 | 5 | 0.17 |
| 2 | 60 | 26 | 5 | 0.19 | 60 | 7 | 0.12 | 54 | 7 | 0.13 | 60 | 7 | 0.12 |
| 5 | 152 | 51 | 6 | 0.12 | 142 | 13 | 0.09 | 90 | 11 | 0.12 | 152 | 15 | 0.10 |
| 10 | 304 | 86 | 9 | 0.10 | 211 | 18 | 0.09 | 191 | 19 | 0.10 | 304 | 27 | 0.09 |
| 20 | 617 | 148 | 10 | 0.07 | 415 | 20 | 0.05 | 239 | 20 | 0.08 | 575 | 30 | 0.05 |
| 100 | 3046 | 338 | 10 | 0.03 | 696 | 20 | 0.03 | 356 | 20 | 0.06 | 1128 | 30 | 0.03 |
The enriching features are classified into three categories: i) COGs (COG U, M, N, and O) for proteins assigned to one or more of these four COGs, ii) computational predictions with enriching features for proteins predicted to have one or more of the following features, i.e., TMHMM = 1, SignalP > 0.7, pSort outer membrane, pSort periplasmic, or pI < 5, and iii) protein expression as detected by mass spectrometry. For further details on the enriching features, cf. Table 1.
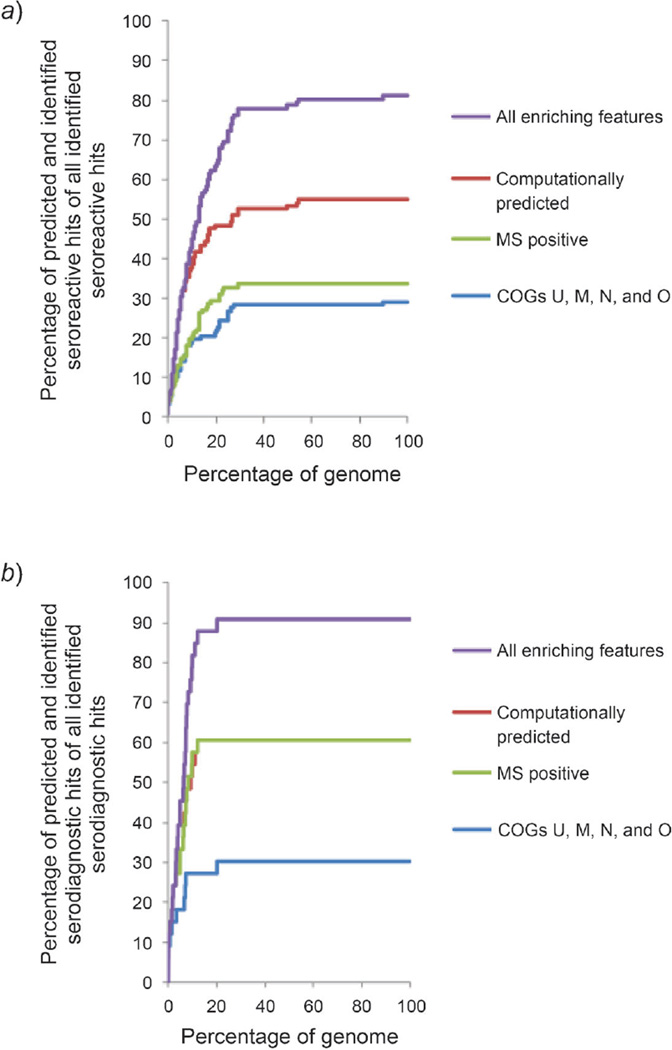
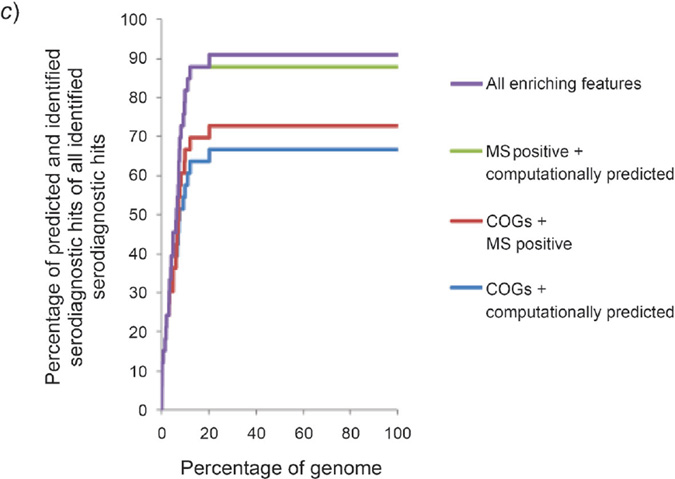
These categorized enriched proteomic features predicted up to 91% of serodiagnostic hits in the top ranked 20% of the proteome. The COGs, the computationally predicted features, and the MS positive proteins accounted for 30, 61, and 61% of the serodiagnostic hits, respectively. Combining the pool of computationally predicted and MS positive proteins predicted 88% of the hits. All together, the three categories accounted for 91% of the serodiagnostic antigens (Table 8). Interestingly, the naïve Bayes classification successfully ranked the antigens in an efficient way, so that 91% of serodiagnostic antigens with enriched features were within the top 20% of the ranked genome (Fig. 2,b and c, and Table 8), and 72% of seroreactive antigens with enriched features were within the top 25% of the ranked genome (Fig. 2, a, and Table 6).
Table 8.
Enrichment Summary for Serodiagnostic Antigens with Categorized Enriching Features
| Top ranked antigens [%] |
Antigens on chip |
Percentage of total serodiagnostic antigen hits [%] | ||||||
|---|---|---|---|---|---|---|---|---|
| COGsa) | CPb) | MSc) | COGs + CP | COGs + MS | CP + MS | Sum of features | ||
| 1 | 30 | 12 | 15 | 15 | 15 | 15 | 15 | 15 |
| 2 | 60 | 15 | 21 | 21 | 21 | 21 | 21 | 21 |
| 5 | 152 | 18 | 39 | 33 | 39 | 36 | 45 | 45 |
| 10 | 304 | 27 | 55 | 58 | 58 | 67 | 82 | 82 |
| 20 | 617 | 30 | 61 | 61 | 67 | 73 | 88 | 91 |
| 100 | 3046 | 30 | 61 | 61 | 67 | 73 | 88 | 91 |
COGs: Functionally annotated COGs (COG U, M, N, and O).
CP: Computationally predicted features.
MS: Protein expression as detected by mass spectrometry. For further details on the enriching features, cf. Table 1.
Fig. 2. Categorized enriching features on prediction of seroreactive or serodiagnostic antigens based on naïve Bayes ranking.
a) Prediction of categorized enriching features on seroreactive antigens. Within the top 30% of the ranked genome, COGs (COG U, M, N, and O), computationally predicted features, the MS positive feature, and the ten enriching features altogether could predict 30, 52, 34, and 78% of all seroreactive hits, respectively. b) Prediction of categorized enriching features on serodiagnostic antigens. Within the top 20% of the ranked genome, COGs (COG U, M, N, and O), computationally predicted features, the MS positive feature, and the ten enriching features altogether could predict 30, 61, 61, and 91% of all serodiagnostic hits, respectively. c) Prediction of paired categorized enriching features on serodiagnostic antigens. Pairing two of the categories of the enriching features significantly enhanced the prediction on serodiagnostic antigens compared to prediction from an individual category of enriching features.
Discussion
The humoral immune response is essential for host defense against bacterial pathogens. However, high-throughput tools for understanding the extent and quality of the immune response against pathogens on a genome-wide scale are limited, and understanding of the immune response on a systems biology level has developed very slowly. Emerging efforts include application of reverse vaccinology to discover new subunit vaccine candidates [26– 28], or developing of sequence-based prediction models [23]. This emergence has been driven by the rapid accumulation of whole genome sequencing data from thousands of microorganisms [29]. Here, we have been taking a protein microarray approach to profile the humoral immune response to numerous infectious agents and to identify the complete antibody repertoire associated with each disease, attempting to be as complete and accurate as possible.
Protein microarrays have become a powerful method enabling profiling of pathogen-specific antibody responses generated upon exposure to infectious agents. No other existing approach can provide such a detailed understanding of the humoral immune response to infection.
Protein microarrays can be used to efficiently probe the entire proteome of a given pathogen against large numbers of patient sera samples, which allows for statistically significant identification of the serodiagnostic antigens. In addition to the identification of potential biomarkers for diagnostics and subunit vaccine candidates, protein microarray studies can also provide the basis for a comprehensive and quantitative determination of basic biological characteristics of the entire set of the serodominant and serodiagnostic antigens on a genomic level.
Protein microarrays enable enrichment analyses to identify proteomic features that are enriched in the immunodominant antigen set and predict antigenicity based on annotated and computationally predicted proteomic features. The predictor can be used on a high-throughput scale on existing or new proteomes to identify key antigenic proteins that may have serodiagnostic or protective qualities and may be used in diagnostic tests and in vaccines. We have classified the reactive immunodominant antigens for numerous agents and found features that consistently predict antigenicity.
We have classified reactive antigens according to annotated functional features (COGs), computationally predicted features (e.g., subcellular localization and physical properties), and protein expression estimated by MS. Enrichment analysis identified ten features that were significantly enriched in seroreactive antigens. Combining all proteins with these enriching features would constitute 37% of the B. melitensis genome and reveal 91% of serodiagnostic antigens and 78% of seroreactive antigens. However, the accuracy of this prediction is low, because the immune system develops significant antibody titers against only 10% of the proteins with these enriching features, and 90% of the predicted antigens are false positives. To increase the accuracy of prediction of seroreactive and serodiagnostic antigens over the entire proteome, we used this information in a naïve Bayes classification approach to rank the entire proteome for increased likelihood of seroreactivity. By doing so, we were able to segregate 63% of all seroreactive proteins and 91% of all serodiagnostic proteins within 20% of the proteome. Within the top 30% of the ranked genome, we were able to predict 79% of seroreactive antigens and 97% of serodiagnostic antigens. The prediction is highest among the serodiagnostic antigens, which are the most relevant for diagnostics and subunit vaccines. The naïve Bayes classification successfully ranked the antigens in an efficient way, so that 82% of serodiagnostic antigens with enriched features were within the top 10% of the ranked genome, and up to 72% of seroreactive antigens with enriched features were within the top 25% of ranked genome.
This approach is important and necessary for studies that aim to identify a subset of the proteome that most likely contains seroreactive or serodiagnostic antigens, making the development of vaccines and serodiagnostic tests more effective and efficient. Ultimately, it could be applied toward construction of a universal bacterial proteome array containing the top ranked predicted serodiagnostic antigens from thousands of bacterial genomes. This universal pathogen array could be applied to help identify microorganisms that are the cause of emerging infectious diseases, outbreaks, and bioterrorism attacks.
Conclusions
Naïve Bayes classification has shown to be a convenient and efficient approach to account for a large number of proteomic features and to classify the immune-reactive proteome determined by protein microarray. These results will provide useful insight toward understanding the humoral immune response to B. melitensis infection and provide a systems biology foundation for comparing antigenicity of other Gram-negative bacterial infections.
Experimental Part
Protein microarray data were obtained from our recently published results [24]. The ‘vsn’ package of the Bioconductor suite [30] in the R statistical environment [31] was used to calculate the signal intensity. In addition to the variance correction, this method calculates maximum-likelihood shifting and scaling calibration parameters for each array, such that control probe variance is minimized. This calibration has been shown to minimize experimental effects [32]. Differential analysis of the normalized signals was then performed using a Bayes-regularized t-test adapted from Cyber-T for protein arrays [33] [34]. Benjamini–Hochberg p-value adjustments were applied to account for multiple test conditions [35]. A p value < 0.05 was regarded as significant.
The following software was used: TMHMM v2.0 software [36] for the computational prediction of transmembrane domains, SignalP v3.0 software [37] for the signal-peptide prediction, and PSORTb v2.0.4 software [38] [39] for the cellular-location prediction. The PI/MW tool from the Swiss Institute of Bioinformatics was used to determine isolectric points [40]. The COG information utilized can be found at the National Center for Biotechnology Information (NCBI) [41]. Enrichment statistical analysis was performed in the R statistical environment, using Fisher’s exact test. A combined naïve Bayes classifier approach [25], originally applied by us to classify antigens from the Francisella tularensis proteome [7], was used here to rank all B. melitensis proteins.
Table 3.
Naïve Bayes Ranking of Serodiagnostic Antigens
| Top ranked antigens [%] | Antigens on chip | Serodiagnostic antigens | |||
|---|---|---|---|---|---|
| Number | Proportion [%] | Enrichmenta) | p-Value | ||
| 1 | 30 | 5 | 15 | 15.38 | 1.28E − 05 |
| 2 | 60 | 7 | 21 | 10.77 | < 1.00E − 04 |
| 5 | 152 | 15 | 45 | 9.11 | < 1.00E − 04 |
| 10 | 304 | 27 | 82 | 8.20 | < 1.00E − 04 |
| 20b) | 617 | 30 | 91 | 4.49 | < 1.00E − 04 |
| 25 | 761 | 31 | 94 | 3.76 | < 1.00E − 04 |
| 30 | 913 | 32 | 97 | 3.24 | < 1.00E − 04 |
| 44c) | 1335 | 33 | 100 | 2.28 | < 1.00E − 04 |
| 100 | 3046 | 33 | 100 | 1.00 | 1.00E + 00 |
Values indicate the fold enrichment.
The top 20% of the genome contain 30 (91%) serodiagnostic antigens.
The top 44% of the genome contain all 33 (100%) serodiagnostic antigens.
Acknowledgments
We thank Dr. Arlo Randall (Institute for Genomics and Bioinformatics, School of Information and Computer Sciences, University of California, Irvine) for helpful discussion on naïve Bayes classification.
REFERENCES
- 1.Kunnath-Velayudhan S, Salamon H, Wang HY, Davidow AL, Molina DM, Huynh VT, Cirillo DM, Michel G, Talbot EA, Perkins MD, Felgner PL, Liang X, Gennaro ML. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14703. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, Oloo JA, Blair PL, Aguiar JC, Baldi P, Davies DH, Felgner PL. Proteomics. 2008;8:4680. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundaresh S, Doolan DL, Hirst S, Mu Y, Unal B, Davies DH, Felgner PL, Baldi P. Bioinformatics. 2006;22:1760. doi: 10.1093/bioinformatics/btl162. [DOI] [PubMed] [Google Scholar]
- 4.Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, Burk CR, Waisberg M, Jasinskas A, Tan X, Doumbo S, Doumtabe D, Kone Y, Narum DL, Liang X, Doumbo OK, Miller LH, Doolan DL, Baldi P, Felgner PL, Pierce SK. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6958. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang L, Leng D, Burk C, Nakajima-Sasaki R, Kayala MA, Atluri VL, Pablo J, Unal B, Ficht TA, Gotuzzo E, Saito M, Morrow WJ, Liang X, Baldi P, Gilman RH, Vinetz JM, Tsolis RM, Felgner PL. PLoS Negl. Trop. Dis. 2010;4:e673. doi: 10.1371/journal.pntd.0000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molina DM, Pal S, Kayala MA, Teng A, Kim PJ, Baldi P, Felgner PL, Liang X, de la Maza LM. Vaccine. 2010;28:3014. doi: 10.1016/j.vaccine.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyles JE, Unal B, Hartley MG, Newstead SL, Flick-Smith H, Prior JL, Oyston PC, Randall A, Mu Y, Hirst S, Molina DM, Davies DH, Milne T, Griffin KF, Baldi P, Titball RW, Felgner PL. Proteomics. 2007;7:2172. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 8.Sundaresh S, Randall A, Unal B, Petersen JM, Belisle JT, Gill Hartley M, Duffield M, Titball RW, Davies DH, Felgner PL, Baldi P. Bioinformatics. 2007;23:i508. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 9.Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, Molina DM, Hirst S, Chew JS, Wang D, Tan G, Duffield M, Yang R, Neel J, Chantratita N, Bancroft G, Lertmemongkolchai G, Davies DH, Baldi P, Peacock S, Titball RW. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13499. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tippayawat P, Saenwongsa W, Mahawantung J, Suwannasaen D, Chetchotisakd P, Limmathurotsakul D, Peacock SJ, Felgner PL, Atkins HS, Titball RW, Bancroft GJ, Lertmemongkolchai G. PLoS Negl. Trop. Dis. 2009;3:e407. doi: 10.1371/journal.pntd.0000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beare PA, Chen C, Bouman T, Pablo J, Unal B, Cockrell DC, Brown WC, Barbian KD, Porcella SF, Samuel JE, Felgner PL, Heinzen RA. Clin. Vaccine Immunol. 2008;15:1771. doi: 10.1128/CVI.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Bouman TJ, Beare PA, Mertens K, Zhang GQ, Russell-Lodrigue KE, Hogaboam JP, Peters B, Felgner PL, Brown WC, Heinzen RA, Hendrix LR, Samuel JE. Clin. Microbiol. Infect. 2009;15:156. doi: 10.1111/j.1469-0691.2008.02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. Infect. Immunol. 2008;76:3374. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vigil A, Ortega R, Jain A, Nakajima-Sasaki R, Tan X, Chomel BB, Kasten RW, Koehler JE, Felgner PL. PLoS One. 2010;5:e11447. doi: 10.1371/journal.pone.0011447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doskaya M, Kalantari-Dehaghi M, Walsh CM, Hiszczynska-Sawicka E, Davies DH, Felgner PL, Larsen LS, Lathrop RH, Hatfield GW, Schulz JR, Guruz Y, Jurnak F. Vaccine. 2007;25:1824. doi: 10.1016/j.vaccine.2006.10.060. [DOI] [PubMed] [Google Scholar]
- 16.Mochon AB, Ye J, Kayala MA, Wingard JR, Clancy CJ, Nguyen MH, Felgner PL, Baldi P, Liu H. PLoS Pathog. 2010;6:e1000827. doi: 10.1371/journal.ppat.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driguez P, Doolan DL, Loukas A, Felgner PL, McManus DP. Parasit. Vectors. 2010;3:4. doi: 10.1186/1756-3305-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. Proc. Natl. Acad. Sci. U.S.A. 2005;102:547. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies DH, McCausland MM, Valdez C, Huynh D, Hernandez JE, Mu Y, Hirst S, Villarreal L, Felgner PL, Crotty S. J. Virol. 2005;79:11724. doi: 10.1128/JVI.79.18.11724-11733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, Pablo J, Unal B, Nakajima-Sasaki R, Liang X, Crotty S, Karem KL, Damon IK, Ahmed R, Villarreal L, Felgner PL. Proteomics. 2007;7:1678. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 21.Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, Molina DM, Hirst S, Moss B, Frey SE, Felgner PL. J. Virol. 2008;82:652. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luevano M, Bernard H-U, Barrera-Saldana HA, Trevino V, Garcia-Carranca A, Villa LL, Monk BJ, Tan X, Davies DH, Felgner PL, Kalantari M. Virology. 2010;405:31. doi: 10.1016/j.virol.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnan CN, Zeller M, Kayala MA, Vigil A, Randall A, Felgner PL, Baldi P. Bioinformatics. 2010;26:2936. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang L, Tan X, Juarez S, Villaverde H, Pablo J, Nakajima-Sasaki R, Gotuzzo E, Saito M, Hermanson G, Molina D, Felgner S, Morrow WJ, Liang X, Gilman RH, Davies DH, Tsolis RM, Vinetz JM, Felgner PL. J. Proteome Res. 2011;10:4813. doi: 10.1021/pr200619r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witten IH, Frank E. Data Mining, Practical Machine Learning Tools and Techniques. 2nd edn. Amsterdam, San Francisco: Morgan Kaufman; 2005. [Google Scholar]
- 26.Rappuoli R. Vaccine. 2001;19:2688. doi: 10.1016/s0264-410x(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 27.Vivona S, Gardy JL, Ramachandran S, Brinkman FS, Raghava GP, Flower DR, Filippini F. Trends Biotechnol. 2008;26:190. doi: 10.1016/j.tibtech.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. Science. 2000;287:1816. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 29.Liolios K, Chen I-M, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. Nucleic Acids Res. 2010;38:D346. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. http://Bioconductor.org/. [Google Scholar]
- 31. http://www.R-project.org/. [Google Scholar]
- 32.Kreil DP, Karp NA, Lilley KS. Bioinformatics. 2004;20:2026. doi: 10.1093/bioinformatics/bth193. [DOI] [PubMed] [Google Scholar]
- 33.Baldi P, Brunak SR. Bioinformatics: the Machine Learning Approach. Cambridge, MA: MIT Press; 2001. [Google Scholar]
- 34.Baldi P, Long AD. Bioinformatics. 2001;17:509. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. J. R. Stat. Soc. B. 1995;57:289. [Google Scholar]
- 36.Möller S, Croning MD, Apweiler R. Bioinformatics. 2001;17:646. doi: 10.1093/bioinformatics/17.7.646. http://www.cbs.dtu.dk/services/TMHMM/. [DOI] [PubMed] [Google Scholar]
- 37.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J. Mol. Biol. 2004;340:783. doi: 10.1016/j.jmb.2004.05.028. http://www.cbs.dtu.dk/services/SignalP/. [DOI] [PubMed] [Google Scholar]
- 38.Chitranshi P, Chen CN, Jones PR, Faridi JS, Xue L. Bioinorg. Chem. Appl. 2010:619436. doi: 10.1155/2010/619436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. Bioinformatics. 2005;21:617. doi: 10.1093/bioinformatics/bti057. http://www.psort.org/psortb/. [DOI] [PubMed] [Google Scholar]
- 40. http://ca.expasy.org/tools/pi_tool.html. [Google Scholar]
- 41. http://www.ncbi.nlm.nih.gov.