Abstract
The non-histone chromatin binding protein high mobility group AT-hook 2 (HMGA2) is expressed in stem cells and many cancer cells, including tumor initiating cells, but not translated in normal human somatic cells. The presence of HMGA2 is correlated with advanced neoplastic disease and poor prognosis for patients. We had previously demonstrated a role of HMGA2 in DNA repair pathways. In the present study, we employed different human tumor cell models with endogenous and exogenous expression of HMGA2 and show that upon DNA damage, the presence of HMGA2 caused an increased and sustained phosphorylation of the ataxia telangiectasia and Rad3-related kinase (ATR) and its downstream target checkpoint kinase 1 (CHK1). The presence of activated pCHK1Ser296 coincided with prolonged G2/M block and increased tumor cell survival, which was enhanced further in the presence of HMGA2. Our study, thus, identifies a novel relationship between the ATR-CHK1 DNA damage response pathway and HMGA2, which may support the DNA repair function of HMGA2 in cancer cells. Furthermore, our data provide a rationale for the use of inhibitors to ATR or CHK1 and HMGA2 in the treatment of HMGA2-positive human cancer cells.
Introduction
The small high mobility group AT-hook (HMGA) non-histone chromatin binding proteins HMGA1 and HMGA2 are composed of an acidic C-terminal tail and three separate N-terminal lysine- and arginine-rich AT-hook domains, which facilitate binding to the minor groove of short stretches of AT-rich DNA [1]. HMGA2 is expressed in embryonic stem (ES) cells, during embryogenesis, in some fetal tissues, and in some cancer cells. The protein is usually not detectable in normal adult somatic cells [2]. Phenotypically, HMGA1/2-positive cells display improved resistance to therapies that introduce chemical modifications of DNA bases, such as oxidation and alkylation [3–5]. HMGA2 knockout mice exhibit a pygmy phenotype with greatly reduced fat tissues, and male mice are infertile [6,7]. By contrast, tissue-specific overexpression of full-length or ubiquitous expression of a truncated protein lacking the C-terminal tail results in gigantism, lipomatosis, and mesenchymal tumors [8,9].
We showed recently that HMGA2 remains associated with chromatin throughout the cell cycle in pluripotent human ES cells and that HMGA2 expression levels are further elevated during human embryoid body formation [10]. Furthermore, HMGA2 seems to be involved in the regulation of key human genes linked to mesenchymal cell lineage differentiation, adipogenesis, and human ES cell proliferation control [11]. We also demonstrated that HMGA1 and HMGA2 are linked to DNA base excision repair and this may have important implications for genome stability in ES cells and during early development and carcinogenesis [5].
Unique among DNA architectural chromatin binding factors, the HMGA genes are considered proto-oncogenes. HMGA1/2 proteins are consistently overexpressed in nearly all types of naturally occurring cancers and are important for multiple cellular processes including oncogenic transformation [12–15]. It has been recognized that high HMGA1/2 protein levels are associated with increased malignancy, metastatic potential, and poor clinical outcome [13,16–18]. HMGA2 expression is primarily regulated by the miRNAs let-7 and miRNA-98 during oncogenic transformation [19,20], but the molecular mechanisms linking let-7 and HMGA2 with chemoresistance in cancer cells and cancer stem/initiating cells remain elusive [21].
Exposure of cells to DNA-damaging agents results in the activation of a signaling cascade aimed at arresting the cell cycle to repair the DNA damage or trigger apoptosis. The ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related kinase (ATR) are related phosphatidylinositol 3-kinase-related kinases with important functions in the DNA damage response (DDR) pathways. ATM and its downstream target checkpoint kinase 2 (CHK2) constitute the main response to double-stranded DNA breakage [22]. The activation of the ATR and its downstream target CHK1 generally occurs in response to UV and agents that inhibit DNA replication forks [23–25]. ATR and CHK1 participate in the stabilization of forks, repair of DNA damage, and the inhibition of late origin firing [26–29]. The interaction between ATR and the ATR-interacting protein is essential for the phosphorylation of CHK1 and cells depleted of CHK1 accumulate multiple DNA breaks and undergo P53-independent apoptosis [30,31]. Recent evidence shows that the activated ATR-CHK1 pathway in response to fork inhibition preferentially inhibits the activation of new replication fork factories, defined as clusters of one or more adjacent replication origins [32–34]. This strategy conserves replication capacity for already active replicon clusters where forks are inhibited rather than engaging new replication factories, and this minimizes the risk of apoptosis [35]. Although different DNA-damaging agents can preferentially activate one of the two DDR signaling pathways [36], both ATM-CHK2- and ATR-CHK1-mediated DDRs are required for cell survival [31].
ATM was recently shown to interact with and phosphorylate HMGA2, and phosphorylated HMGA2 activated a positive feedback loop by upregulating ATM expression [37]. In the present study, we demonstrate a novel interaction between ATR-CHK1 and HMGA2 and provide evidence for a new cytoprotective role of HMGA2 by sustaining ATR-CHK1 phosphorylation. In four different cancer cell models used here, we show that the antiapoptotic activity of HMGA2 is mediated by activated pCHK1. Depletion of HMGA2, CHK1, or both factors resulted in mitotic cell cycle arrest, increased number of nuclear γ-H2AX foci, and caspase 3/7-mediated apoptosis with decreased resistance to the genotoxic agents methyl methane sulfonate (MMS) and hydroxyurea (HU). Thus, our data provide first evidence for an active role of HMGA2 in the ATR-CHK1 DDR pathway.
Materials and Methods
Cell Lines and Culture Conditions
We used established lung cancer A549-HMGA2 transfectants [5] and generated stable transfectants of the undifferentiated thyroid cancer cell line UTC8505 expressing human HMGA2 as described previously [38]. Transfectants were cultured in DME-F12 medium (Thermo Scientific, Ottawa, Ontario) plus 10% fetal calf serum (FCS; Sigma, Oakville, Ontario) and 500 µg/ml geneticin (Life Technologies, Burlington, Ontario). The fibrosarcoma cell line HT1080 (C1) harboring a doxycycline-inducible shHMGA2 construct was generated by standard lentiviral transduction using a Tet-on shHMGA2 construct, pTRIPz-shHMGA2 (Origene, Rockville, MD). Cells were grown in DME-F12 supplemented with 10% FCS and 3 µg/ml puromycin (Sigma). Induction of shHMGA2 was achieved with 4 µg/ml doxycycline (Sigma) for 48 hours with repeated doses every 24 hours and resulted in significant down-regulation of endogenous HMGA2. Human rhabdomyosarcoma (RD) cells were propagated in DME-F12/10% FCS. All cell lines were maintained in a humidified incubator under 5% CO2 at 37°C.
Comet Assay
UTC8505 mock and HMGA2 transfectants (106 cells/ml) were cultured in serum-free DME-F12 medium and comet assays were performed as described earlier [5]. Images were obtained at 400-fold magnification using a chroma filter at 385-nm excitation/450-nm emission with a Z1 microscope (Zeiss, Jena, Germany). We used the (http://www.autocomet.com/products_cometscore.php) Comet Score Version 1.5 analysis software. DNA damage was quantified by the Olive tail moment [39].
Induction and Recovery Time Kinetic Assays
Cells (105 cells/well) in six-well plates were grown overnight. For induction time kinetics, DNA damage was induced by MMS treatment for 2, 5, 10, 15, 20, and 30 minutes and protein extracts were collected. For recovery time kinetics, cells were treated with MMS for 30 minutes and washed thoroughly twice with 1x phosphate-buffered saline (PBS) to remove MMS. Cells received fresh medium and were allowed to recover from MMS damage for 0, 1, 2, 4, 6, and 24 hours before total protein extraction.
Immunoblot Analysis
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto nitrocellulose membrane (VWR, Mississauga, Ontario) followed by blocking with 5% skimmed milk in 1x Tris-buffered saline and 0.1% Tween 20 (pH 7.6). Membranes were incubated with the primary antibody overnight at 4°C in 5% BSA containing 1x Tris-buffered saline and 0.1% Tween 20 followed by the secondary antibody in blocking buffer for 1 hour at room temperature and detection of bands with the ECL Kit (Pierce, Nepean, Ontario). Primary rabbit polyclonal antibodies used were against phospho-ATR (pATRSer428), phospho-CHK1 (pCHK1Ser296), phospho-CHK2 (pCHK2Thr68), total ATR, total CHK1, and total CHK2 (all from Cell Signaling Technology, Pickering, Ontario) and goat HMGA2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Anti-rabbit IgG (Cell Signaling Technology) and rabbit anti-goat IgG (Santa Cruz Biotechnology) conjugated with HRP were used as secondary antibodies.
Immunofluorescence
The C1 human fibrosarcoma cells were cultured on glass slides plus/minus doxycycline (4 µg/ml) for 48 hours to attain HMGA2+ and HMGA2low cells, respectively. Cells were treated with MMS (Sigma) for 30 minutes and then washed twice with 1x PBS. After fixation with 3.7% formaldehyde, cells were blocked with 1% BSA containing 0.1% Triton X-100 plus 5% serum. Incubation with the goat anti-HMGA2 (Santa Cruz Biotechnology) and mouse anti-γ-H2AX (Millipore, Billerica, MA) in blocking solution was done at 4°C overnight before incubation with fluorescein isothiocyanate-conjugated swine anti-goat (DAKO, Burlington, Ontario) and Alexa Fluor 594-conjugated rabbit anti-mouse (Life Technologies) for 1 hour at room temperature. Slides were counterstained with 0.1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma) and cover-slipped (Vector Laboratories, Burlington, Ontario), and imaging of cell nuclei was done using a Z1 microscope (Zeiss).
Cell Cycle Analysis by Fluorescent-Activated Cell Sorting
For cell cycle fluorescent-activated cell sorting (FACS) analysis, 2 x 106 cells were exposed to 2 mM MMS for 30 minutes at 37°C and allowed to recover for 0, 1, 2, 4, 6, and 24 hours before being harvested and fixed in 70% ethanol at 4°C overnight. Cells were washed with ice-cold PBS followed by treatment with 1 µg/ml RNAse for 2 hours at 37°C. One hour before FACS, cells were stained with 10 µg/ml propidium iodide (Sigma). Cell cycle profiles were obtained with a MoFloXDP flow cytometer (Beckman Coulter, Mississauga, Ontario) using Summit 5.2 software.
RNA Silencing
Cells (105 cells/well) in six-well plates were cultured overnight and transfected with 100 nM HMGA2 siRNA (Sigma), 50 nM CHK1 siRNA, and 50 nM scrambled control siRNA (Cell Signaling Technology) using siLentFect lipid reagent (Bio-Rad, Ontario, Canada). Protein lysates were collected at 24, 48, and 72 hours and protein levels of HMGA2 and CHK1 were assessed by Western blot. Immunodetection of human CHK2 was performed on lysates of siCHK1-treated cells to demonstrate the specificity of the CHK1 knockdown.
Caspase 3/7 and Metabolic Activity Assays
Cells were seeded at 7 x 103 cells/well in 96-well plates and grown overnight before siRNA knockdown of HMGA2 and/or CHK1 for 24 hours. Thereafter, cells were subjected to MMS treatment for 30 minutes followed by 24- and 48-hour recovery periods. Caspase-Glo 3/7 (Promega, Madison, WI) and WST assays (Roche Diagnostics, Laval, Quebec) were performed according to the manufacturer's protocol to determine the percentage of apoptotic and metabolically active cells, respectively, using a 96-well plate reader (Perkin Elmer, Woodbridge, Ontario).
Fluorescent Detection of Active Caspase 3/7
Qualitative detection of apoptosis through active caspase 3/7 on live cells was performed using Fluorescent Labeled Inhibitor of Caspases (FLICA) Apoptosis Detection Kit (Immunocytochemistry Technologies, Bloomington, MN). Untreated and doxycycline-treated C1 fibrosarcoma cells were grown on poly-d-lysine-coated eight-chamber culture slides (BD Biosciences, Mississauga, Ontario) and transfected with CHK1 siRNA followed by MMS treatment and 24-hour recovery. The carboxyfluorescein-labeled (FAM-DEVD-FMK) FLICA probe that covalently binds to active caspases 3 and 7 was added to the cells and incubated for 1 hour at 37°C under 5% CO2. Cells were then counterstained with Hoechst 33342 and fluorescence was analyzed with a Z1 microscope (Zeiss).
Co-immunoprecipitation
Protein extraction and co-immunoprecipitation (Co-IP) were performed as described previously [40]. Briefly, cells (treated and control) were washed with 1x PBS, scraped, and collected by centrifugation. PBS was discarded and the cell pellet was resuspended in two volumes of lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM sodium chloride, 25 mM sodium fluoride, 0.1 mM sodium orthovanadate, 0.2% Triton X-100, 0.3% NP-40, and protease inhibitors. Protein extracts were collected by incubation on ice for 30 minutes followed by centrifugation at 20,800g for 30 minutes. For IP, 1 mg of protein was mixed with 2 µg of anti-ATR goat polyclonal antibody (Santa Cruz Biotechnology) and left rotating at 4°C for 4 hours. The antigen-antibody mixture was then incubated with a 1:1 mixture of protein A-sepharose slurry (GE Healthcare Life Sciences, Baie-D'Urfé, Quebec) and protein G-agarose (Roche Diagnostics) beads for 16 hours at 4°C. The complexes were washed four times with lysis buffer and the immunoprecipitates were eluted using sodium dodecyl sulfate elution buffer.
Statistical Analysis
All the experiments were performed at least in triplicate. Repeated measures two-way analysis of variance (mixed model) and Bonferroni posttests were performed to determine significance of the grouped analyses. For all tests, P < .05 was considered as statistically significant. Error bars represent the standard deviation of the mean.
Results
Characterization of UTC8505-HMGA2 Transfectants
We generated stable HMGA2 transfectants of UTC8505 (Figure 1A) and used previously established A549-HMGA2 human lung carcinoma cell transfectants [5]. UTC8505-HMGA2 clones 4 and 12 expressed HMGA2 and showed exclusive nuclear expression of immunoreactive HMGA2 as determined by Western blot analysis and immunofluorescence (Figure 1, A and B). In all cell models studied, we confirmed MMS-mediated induction of DNA strand breaks by detection of γ-H2AX foci at DNA-damaged sites as demonstrated for UTC8505 transfectants as a representative cell model in Figure 1C. Employing comet assays, we had previously shown a reduced tail moment upon MMS treatment in A549 clones expressing HMGA2 [5]. MMS treatment and subsequent comet assays also resulted in a significantly smaller tail moment with UTC8505-HMGA2 transfectants compared to mock cells (Figure 1D), thus confirming the protective function of HMGA2 against induced DNA damage.
Figure 1.
Characterization of stable HMGA2 transfectants. (A) Representative immunoblot and (B) immunofluorescent images showing nuclear presence of HMGA2 in two UTC8505-HMGA2 stable transfectants. (C) Immunofluorescence detection of the DNA damage response marker γ-H2AX indicating MMS-induced DNA damage in UTC8505 cells. Green stain, HMGA2; red stain, γ-H2AX; blue stain, nucleus. (D) Quantification of three independent comet assays of UTC8505-HMGA2 (clone 4) and mock (clone 2) transfectants upon MMS-induced DNA damage is shown. MMS was effective in causing DNA damage, as shown by a markedly longer tail moment compared to untreated cells. There was a significant reduction in tail moment upon MMS treatment in the presence of HMGA2, suggesting increased DNA repair capability in UTC-HMGA2 transfectants compared to mock. (*P < .05).
HMGA2 Causes Sustained Phosphorylation of ATR and Downstream Target CHK1
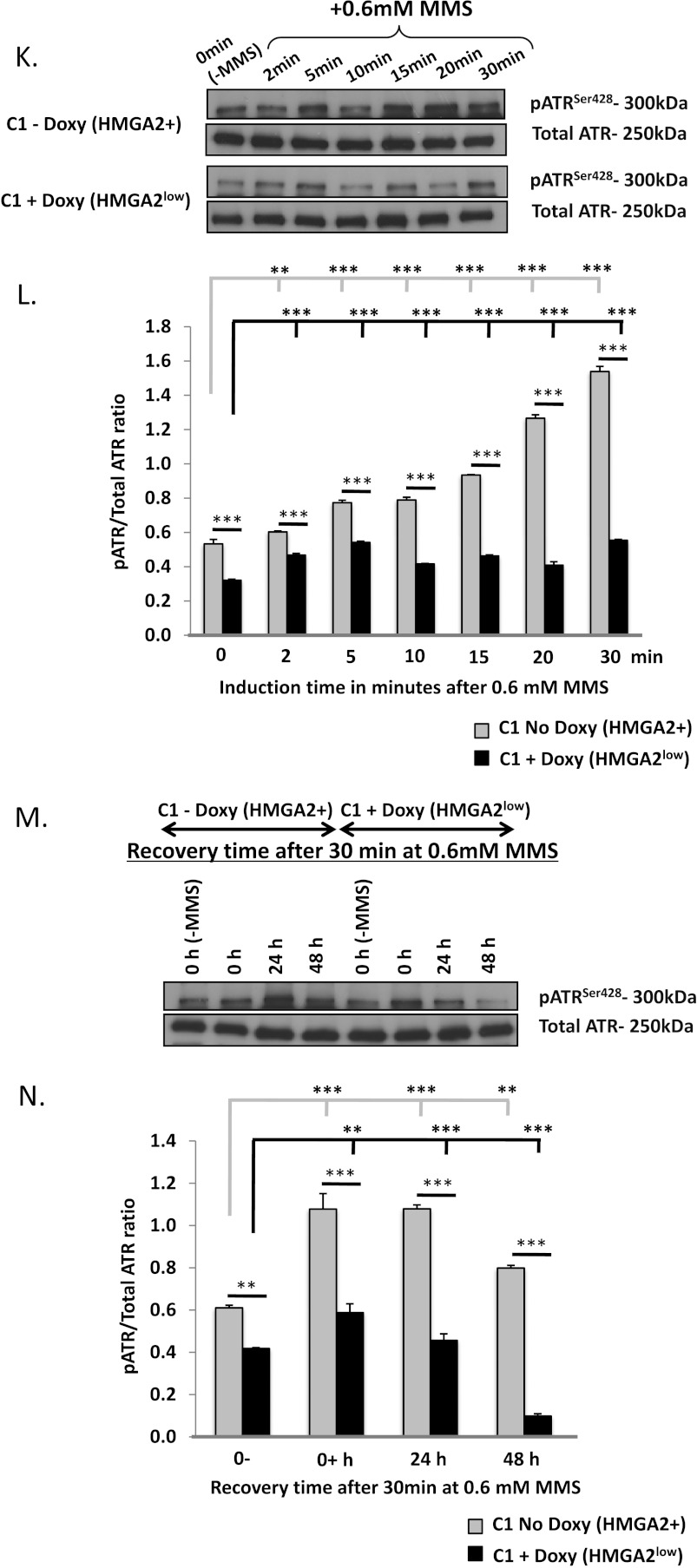
We wanted to identify if the DNA protective role of HMGA2 extended to the ATR-CHK1 DNA damage signaling pathway. In induction time kinetics, UTC8505-HMGA2 (Figure 2, A and B), A549-HMGA2 (Figure W1A), and corresponding mock were exposed to 2 mM and 5 mM MMS, respectively, for 0, 2, 5, 10, 15, 20, and 30 minutes. Immunoblot analysis of the protein extracts collected at these time points showed a gradual increase in the level of pCHK1S296 over time with a maximal phosphorylation at 30 minutes. The level of pCHK1S296 in response to MMS treatment was significantly increased in the presence of HMGA2 (Figures 2, A and B, and W1A). Next, we performed recovery experiments by inducing DNA damage in HMGA2 and mock transfectants of UTC8505 and A549 for 30 minutes, followed by a recovery period in fresh culture medium without MMS. Western blot analysis of pCHK1S296 in protein lysates collected at 0, 1, 2, 4, 6, and 24 hours of recovery time revealed a gradual dephosphorylation of activated CHK1 to baseline values after 4 hours in mock (Figure 2, C and D). By contrast, we observed sustained high-level presence of pCHK1S296 even at 24 hours of recovery in the presence of HMGA2 (Figure 2, C and D). We confirmed the presence of activated upstream CHK1 regulator pATRSer428 upon MMS treatment in UTC8505 cells (Figure 2, E–H). Similar recovery results for pCHK1S296 were obtained for A549-HMGA2 and mock transfectants, but recovery data were less pronounced in A549 cells (Figure W1B).
Figure 2.
(A and E) Representative immunoblots and (B and F) densitometric analysis of induction time kinetics for pCHK1Ser296 and pATRSer428 in UTC8505-HMGA2 and mock plus/minus MMS treatment. For recovery time kinetics, HMGA2 and mock transfectants were initially exposed to MMS for 30 minutes, and then cells were washed and cultured in normal culture medium without MMS. At the indicated time points, immunoreactive pCHK1Ser296 and pATRSer428 were determined. (C and G) Representative Western blots and (D and H) densitometric analysis done from three independent sets of experiments are shown. (I) Schematic and Western blot showing the down-regulation of HMGA2 upon doxycycline (doxy) treatment in human fibrosarcoma (C1) cells with doxy-inducible shHMGA2 construct. (J) Recovery time kinetics for pCHK1Ser296 performed in C1 cells in the presence (HMGA2low) and absence of doxy (HMGA2+). (K and M) Representative immunoblots and (L and N) densitometric analysis of induction time kinetics and recovery time kinetics for pATRSer428 in C1 cells in the presence (HMGA2low) and absence of doxy (HMGA2+) plus/minus MMS treatment. The presence of HMGA2 coincided with increased and prolonged pATRSer428 and pCHK1Ser296 levels. *P < .05, **P < .01, and ***P < .001. 0-, no MMS; 0+, MMS-treated but no recovery time.
To determine if the presence of endogenous HMGA2 was required for sustained phosphorylation of CHK1, we employed the HMGA2+ human fibrosarcoma cell line C1 with doxycycline-inducible short hairpin (sh) HMGA2 expression for the suppression of HMGA2 (Figure 2I). Similar to the other two human cancer cell models tested, C1 cells showed sustained presence of pCHK1S296 at 24-hour recovery after 0.6 mM MMS treatment. HMGA2 suppression resulted in a gradual decrease of pCHK1S296 levels, suggesting that the prolonged CHK1 phosphorylation was at least in part dependent on the presence of HMGA2 (Figure 2J). We confirmed the presence of activated upstream CHK1 regulator pATRSer428 upon MMS treatment in C1 cells (Figure 2, K–N). Total CHK1 levels remained unchanged throughout the recovery period in both HMGA2+ and HMGA2low conditions (data not shown).
Sustained CHK1 Phosphorylation in the Presence of HMGA2 Prolongs G2/M Arrest
DNA insult created by genotoxicants activates DNA damage signaling pathways, which arrest the cell cycle to facilitate damage repair and/or trigger apoptotic death of irreparably damaged cells [41]. CHK1 phosphorylation prevents mitotic entry by inducing G2/M arrest and eliminates cells with impaired DNA [41–43]. We performed FACS analysis to determine the effect of sustained phosphorylation of CHK1 on cell cycle distribution. After exposure to 2 mM MMS for 30 minutes, UTC8505-HMGA2 and mock were cultured in normal medium for 1, 2, 4, 6, and 24 hours before FACS analysis. Cells not exposed to MMS were used as controls. FACS cell cycle profiles revealed a gradual transition from G1/S to G2/M phase in both mock and HMGA2 transfectants during the recovery period from 1 to 24 hours. At all recovery time points investigated, HMGA2+ cells displayed increased accumulation of cells in G2/M phase in comparison with mock controls (Figure W2, A and B). At 24-hour recovery, only 17% of the HMGA2+ cells were in G1 and 55% were arrested in G2/M, while in mock 38% of cells were in G1 and 43% were found in G2/M (Figure 3, A and B). An additional subpopulation of HMGA2+ cells with double the normal DNA content and comprising 11% of total HMGA2 transfectants was detected at 24-hour recovery (Figure 3A). Prolonged G2/M arrest in HMGA2 transfectants coincided with the prolonged presence of pCHK1S296 even after 24 hours of recovery (Figure 2, C and D). By contrast, in mock the G2/M transition and the presence of 38% of cells in G1 (Figure 3B) coincided with low levels of pCHK1S296 observed at 24 hours of recovery (Figure 2, C and D).
Figure 3.

Sustained presence of pCHK1Ser296 prolonged G2/M arrest in HMGA2+ cancer cells. (A and B) Cell cycle profiling by FACS done at 0- and 24-hour recovery periods from MMS treatment in UTC8505-HMGA2 (A) and mock (B) cells. See Figure W2, A and B, for cell cycle distribution at 1-, 2-, 4-, and 6-hour recovery periods from MMS. Untreated cells were used as control.
HMGA2-Mediated Enhanced CHK1 Signaling Inhibits Apoptosis
To test the hypothesis that HMGA2 facilitates the DNA repair process by increasing activated pCHK1S296 and arresting the cell cycle as well as preventing apoptosis, we performed siCHK1-specific knockdown experiments. Upon siCHK1 treatment, Western blot confirmed a significant, specific, and sustained down-regulation of CHK1, whereas cellular CHK2 levels remained unchanged in the HMGA2 or mock transfectants of UTC8505 (Figure 4A) and A549 (Figure W4A). Caspase 3/7 activity increased upon siCHK1 knockdown and was enhanced further upon MMS treatment at 48-hour recovery. By contrast, in the presence of CHK1, caspase 3/7 activity was markedly reduced in both the UTC8505 and the A549 cell models studied at 24 hours (Figures W3A and W4B) and 48 hours (Figures 4B and W4C). Metabolic activity decreases in cells undergoing apoptosis [44]. CHK1 knockdown followed by 30 minutes of MMS treatment and consecutive recovery for 24 hours (Figures W3B and W4D) and 48 hours (Figures 4C and W4E) revealed a decrease in the metabolic activity in UTC8505 and A549 HMGA2 and mock transfectants, supporting a role for CHK1 in cell survival. The combined CHK1 knockdown and MMS treatment caused an even higher reduction in metabolic activity at 48-hour recovery. Thus, sustained activation of the DDR signaling factor pCHK1S296 on an HMGA2+ background caused increased chemoresistance of these cancer cells to the DNA-damaging agent MMS.
Figure 4.
(A) Immunoblot detection upon silencing of CHK1 by CHK1 siRNA in UTC8505-HMGA2 and mock. CHK1 silencing was specific and did not affect expression of CHK2 protein. A non-silencing randomized sequence was used as a control in the siRNA experiments and equal loading of protein lysates was confirmed with β-actin. (B) Higher activation of caspase 3/7 and (C) suppression of metabolic activity upon CHK1 knockdown at 48 hours of recovery in HMGA2 and mock transfectants. There was no significant difference between the no siRNA and control siRNA-treated cells with respect to both caspase 3/7 activity and metabolic activity. *P < .05, **P < .01, and ***P < .001.
Abrogation of HMGA2 along with CHK1 Leads to a Robust Increase in Apoptosis
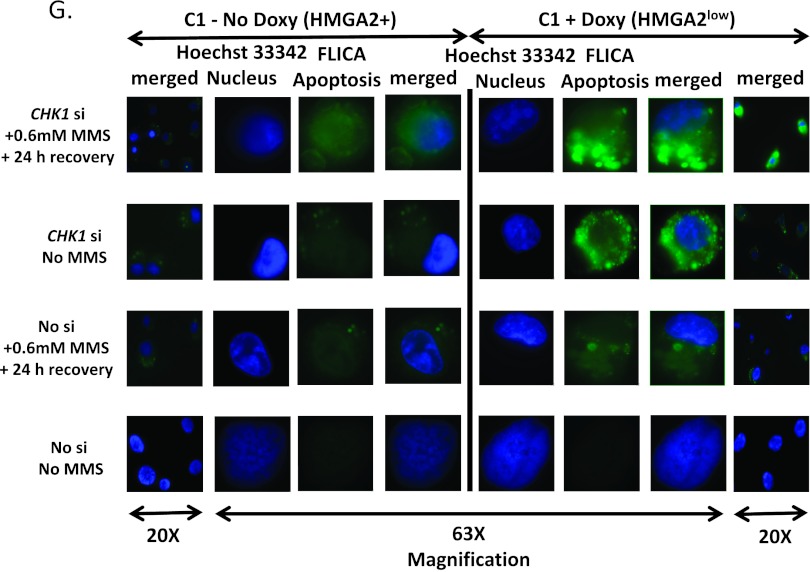
Depletion of HMGA2 enhances the sensitivity of cancer cells to certain DNA-damaging agents due to an impairment in DNA repair capacity [37]. In light of this, we decided to examine the effects of the combined down-regulation of cellular HMGA2 and CHK1. We employed the endogenous HMGA2-producing RD cells and C1 fibrosarcoma stable transfectants with doxycycline-inducible shHMGA2-mediated HMGA2 silencing. RD cells were subjected to siHMGA2 or siCHK1 single knockdown and combined siHMGA2/siCHK1 double knockdown (Figure 5A). Upon siRNA treatment, cells were treated with 1.5 mM MMS and caspase 3/7 and metabolic activity were measured at 24 hours (Figure W5, A and B) and 48 hours of recovery time (Figure 5, B and C). A significant increase in caspase 3/7-mediated apoptosis was observed upon siHMGA2 and siCHK1 single knockdown compared to cells treated with control siRNA (P < .001). A further robust increase in apoptosis of RD cells was observed in siHMGA2/siCHK1 double knockdown combined with MMS treatment at 48-hour recovery (Figure 5B). In agreement with these findings, suppression of HMGA2 in C1 cells increased the caspase 3/7-mediated apoptosis and this was enhanced dramatically with the additional siCHK1 knockdown (Figures 5, D and E, and W5C). Our caspase 3/7 fluorescent detection assays in living cells confirmed the luminometric caspase 3/7 quantitation and revealed changes in cell and nuclear morphology indicative of apoptosis that included membrane blebbing and DNA condensation at 24-hour recovery in cells with reduced HMGA2 levels (Figure 5G). Similar findings were observed upon MMS treatment of doxycycline-treated C1 cells (HMGA2low) and CHK1-depleted cells. The marked decrease in cell vitality observed upon single and double HMGA2/CHK1 knockdown corroborated with the observed increase in apoptosis in both the RD and C1 fibrosarcoma cell models (Figure 5, C and F; for 24-hour recovery, see Figure W5, B and D). Altogether, these results implied that the antiapoptotic action of HMGA2+ cells is augmented by activated CHK1 and, together with the induction of a mitotic block, may support the DNA repair function of HMGA2 in cancer cells.
Figure 5.
(A) Immunoblots showing successful single knockdown of HMGA2 or CHK1 and double knockdown of HMGA2 and CHK1 in RD cells as well as (D) specific siRNA-mediated CHK1 silencing in C1 cells. (B and E) Luminescent detection of caspase 3/7 activity after HMGA2 or CHK1 single knockdown and HMGA2/CHK1 double knockdown at 48 hours of recovery in RD (B) and C1 cells (E). (G) Fluorescent detection of caspase 3/7 activity in CHK1-depleted C1 cells in the presence (HMGA2low) and absence of doxy (HMGA2+) followed by MMS treatment and 24-hour recovery. Cells were labeled with caspase 3/7 substrate FLICA (FAM-DEVD-VMK) for fluorescent detection of apoptotic cells (green). Nuclear staining by Hoechst 33342 revealed nuclear morphology (blue). (C and F) Corresponding WST assays showed a reduction in metabolic activity upon HMGA2 or CHK1 single knockdown and HMGA2/CHK1 double knockdown in RD (C) and C1 cells (F). *P < .05, **P < .01, and ***P < .001.
Enhanced Apoptosis upon HMGA2 and CHK1 Knockdown Extends to Other DNA-Damaging Agents
We also investigated the effect of the genotoxicant and clinical chemotherapeutic agent HU on the protective role of HMGA2 in our cancer cell models. HMGA2 and CHK1-deficient C1 fibrosarcoma cells grown in 96-well plates were treated with 5 mM HU for 24 hours and left to recover for 4 and 6 hours. The level of active caspase 3/7 was significantly higher in HU-treated CHK1-deficient C1 cells (Figure 6A; for 4-hour recovery, see Figure W6A). The combined HMGA2 and CHK1 knockdown further enhanced cell death and an additional HU treatment resulted in a robust apoptotic response (more than two-fold increase) at the 6-hour recovery time point (Figure 6A). Similar to MMS treatment and coinciding with the increase in apoptosis, we observed markedly decreased metabolic cell activity (more than two-fold decrease) at the 6-hour recovery time point in HU-treated HMGA2/CHK1-depleted cells (Figure 6B; for 4-hour recovery, see Figure W6B). Thus, both HMGA2 and CHK1 acted in concert to protect tumor cells from genotoxicants by preventing these cells from entering the apoptotic program.
Figure 6.
HU treatment caused (A) caspase 3/7 activation and (B) reduced metabolic activity at 6 hours of recovery in CHK1-deficient C1 cells. All experiments were done in the presence (HMGA2low) and absence of doxy (HMGA2+). *P < .05, **P < .01, and ***P < .001.
HMGA2 Interacts with ATR and CHK1
We performed Co-IP studies to investigate possible mechanistic interactions between HMGA2 and the ATR-CHK1 pathway. We found that HMGA2 and CHK1 co-immunoprecipitated with ATR in both endogenous HMGA2-producing C1 human fibrosarcoma (Figure 7A) and in HMGA2 overexpressing UTC8505 transfectants (Figure 7C). This ATR interaction with HMGA2 involved the participation of pATRSer428 and pCHK1S296 and, upon MMS exposure of UTC8505-HMGA2 transfectants, resulted in a marked increase in the recruitment of total ATR and pATRSer428. Intriguingly, DNAse digest before IP did not affect the interaction between HMGA2 and ATR-CHK1, indicating that chromatin was not required for this protein complex formation (Figure 7, A and B). However, HMGA2 was not essential for the interaction of ATR with CHK1 as demonstrated for HMGA2-negative UTC8505 mock cells (Figure 7C). Thus, in an HMGA2+ cellular context, the presence of HMGA2 in the ATR-CHK1 complex aided in the survival of stem cells and cancer cells.
Figure 7.
Co-IP revealing the interaction of HMGA2 with ATR and CHK1 in (A) C1 human fibrosarcoma cells and (C) UTC8505 cells. HMGA2 and CHK1 were co-immunoprecipitated with affinity-purified anti-ATR goat polyclonal antibody. This protein-protein interaction was found to be DNA independent and observed (B) after cleavage of DNA with 5 µg/ml of DNAse. Normal goat IgG was used as negative control. (D) Proposed model for the protective mechanism of HMGA2 against genotoxicants in cancer cells. Upon genotoxic insult, HMGA2 engages in a complex with pATR-pCHK1 and increases and prolongs ATR-CHK1 phosphorylation status. This prolongs G2/M arrest, facilitates DNA damage repair, and increases survival and chemoresistance of cancer cells.
Discussion
Here, we have demonstrated that HMGA2 exerts a marked cell protective function against DNA insult through the activation of the ATR-CHK1 pathway. HMGA2 overexpression is known to be associated with many malignant tumors [13] and linked to poor prognosis and metastasis [45,46]. Gene expression profiling of 30 cancer cell lines revealed that HMGA2 contributes to resistance against 4 of the 11 drugs tested [47]. However, this phenotype may be even more widespread because HMGA2 is found to be associated with rare tumor-inducing cells [21].
We recently demonstrated that HMGA1 and HMGA2 can be linked to the base excision repair machinery and significantly protect tumor cells against damaging chemical modifications to single DNA bases due to chemotherapeutic insult [5]. In the present study, we extended these findings to the protective function of HMGA2 in human UTC cells and demonstrated the superior ability of HMGA2-overexpressing UTC to repair MMS-induced DNA damage, suggesting that the protective role of HMGA2 extends to a broad range of different cancer cells. Further, the presence of HMGA2 resulted in significant and sustained hyperphosphorylation of ATR and CHK1 at residues Ser428 and Ser296, respectively, upon induction of DNA strand breaks by MMS. This finding is supporting the previous observation that HMGA2 is responsible for the hyperphosphorylation and prolonged phosphorylation of the DNA-dependent protein kinase catalytic subunit (DNA-PKc, another phosphatidylinositol 3-kinase-related kinase member) at Thr2609 and Ser2056 upon induction of DNA insult by doxorubicin [48]. The ability of HMGA2 to facilitate hyperphosphorylation of DNA-PKc and ATR-CHK1 kinases may reflect yet another strategy employed by HMGA2+ tumor cells to resist DNA-damaging agents, with HMGA2 capitalizing on the known cooperation of DNA-PKc with ATM and ATR pathways to phosphorylate the checkpoint-related proteins [49]. In fission yeast, sustained phosphorylation of CHK1 is maintained by the interaction between pCHK1S345 and either 14-3-3 [50,51] or Crb2 [52] proteins, which protect the phosphorylation status of CHK1 [53]. While HMGA2 was not essential for ATR-CHK1 interaction in our human cancer cell models, increased and sustained ATR and CHK1 phosphorylation was dependent on the presence of HMGA2, suggesting a novel role for HMGA2 in modulating the activity status of the ATR-CHK1 signaling complex.
CHK1 is a potential target for anticancer therapy and many CHK1-selective inhibitors are in various stages of clinical trials [54–56]. CHK1 plays important roles in cell cycle arrest, non-homologous end joining [57], and homologous recombination repair mechanisms [30]. Phosphorylation-dependent activation of CHK1 is essential for regulating cell cycle delay and cell survival [58]. We hypothesized that the MMS-induced sustained accumulation of pCHK1S296 in the presence of HMGA2 may prolong the cell cycle arrest to facilitate effective DNA repair and prevent premature entry into mitosis resulting in apoptosis. Mitotic arrest at the G2/M checkpoint remains active until completion of DNA repair and is controlled and regulated by CHK1 [42,59–62]. In Saccharomyces pombe, sustained CHK1 phosphorylation is essential in maintaining the G2 DNA damage checkpoint to prevent the entry of damaged cells into mitosis [53]. We identified the persistent presence of pCHK1S296 in the presence of HMGA2 to be accompanied by a prolonged G2/M arrest. Thus, HMGA2 appeared to confer in tumor cells the capacity for timely repair of DNA lesions and this was facilitated by a CHK1-mediated cell cycle delay.
We asked if the increased activation of CHK1 signaling and mitotic arrest observed in HMGA2 clones can promote cell survival by exerting an antiapoptotic effect. The siRNA-mediated knockdown of CHK1 in HMGA2+ cancer cells resulted in caspase 3/7-mediated apoptotic cell death and this apoptotic response was further amplified in the presence of DNA-damaging agents MMS and HU in all HMGA2+ cell models studied. These results are consistent with the previous findings that CHK1 inhibitors enhance the cytotoxicity induced by DNA-damaging agents [42,62] and the role of the ATR-CHK1 pathway activation in preventing caspase 3-mediated apoptosis in response to ionizing radiation (IR)- or UV-induced DNA damage [63,64]. Furthermore, the combined knockdown of both HMGA2 and CHK1, in our fibrosarcoma model of shHMGA2-regulated suppression of HMGA2, significantly increased caspase 3/7 activity and cytotoxicity. Again, this effect was further augmented by the addition of DNA-damaging agents. This increase in apoptosis coincided with a marked decrease in cell viability, a finding consistent with recent reports demonstrating increased sensitivity of cancer cells to both radiotherapeutics and chemotherapeutics upon depletion of HMGA2 [5,37,65]. HMGA2 gene silencing had been reported to upregulate apoptosis in well-differentiated liposarcomas overexpressing HMGA2 and in serous ovarian carcinomas [66,67]. Here, we identified HMGA2 as a novel interaction partner of ATR-CHK1 complex in cancer cells. This interaction involved phosphorylated ATR/CHK1 but was independent of the presence of DNA. It is tempting to speculate that the increased and extended phosphorylation status of ATR and CHK1 in the presence of HMGA2 reflects a new function of HMGA2 aimed at extending the activity of this pathway. This involved a prolonged cell cycle arrest to provide sufficient time for DNA damage repair and prevent cancer cell apoptosis. Importantly, formation of a functional ATR-CHK1 complex was readily observed in cancer cells devoid of HMGA2. However, cancer cells low in or devoid of HMGA2 failed to respond with prolonged phosphorylation of ATR and CHK1 and more easily succumbed to cell death when exposed to genotoxicants. We summarized these findings in a proposed model (Figure 7D). In the presence of the genotoxic agents MMS and HU, HMGA2 through interaction with the ATR-CHK1 complex, facilitated sustained activation of this DDR pathway and this contributed to an increase in chemoresistance in HMGA2-positive cancer cells. What could be the possible underlying mechanisms? CHK1 phosphorylation requires ATR phosphorylation and this is a highly regulated process [68]. At ssDNA-damaged sites, ATR interacts with the 9-1-1 complex (Rad9-Rad1-Hus1) and pTOPBP1 and the latter is necessary for ATR activation [69–71]. ATM-mediated phosphorylation of TOPBP1 can also enhance ATR phosphorylation [72]. Acting as a linker protein, HMGA2 at damaged DNA sites may increase the local concentration of 9-1-1 complex, TOPBP1, and ATR, resulting in sustained ATR phosphorylation through physical proximity between activated TOPBP1 and its ATR substrate. HMGA2 may also facilitate ATM-dependent ATR activation [72]. HMGA2 and ATM are known to interact and this results in increased pATM levels [37]. ATM kinase activity enhances phosphorylation of TOPBP1, which, in turn, promotes ATR phosphorylation. Thus, HMGA2 may act as a novel link between ATM activation and prolonged phosphorylation of ATR in HMGA2+ cells. Finally, we cannot exclude that the possible recruitment of HMGA2 to a preformed pATR-CHK1 complex can block ATR and CHK1 phosphorylated sites from phosphatase attack, thus preventing pATR and/or pCHK1 from reverting into their inactive non-phosphorylated forms.
Conventional chemotherapies often fail primarily due to the development of multidrug chemoresistance and this appears to be in part a function of cancer-initiating cells (CICs) [73]. The re-expression of HMGA2 in cancer cells as opposed to their non-malignant counterparts promotes both chemoresistance and genomic stability through survival-promoting ATR-CHK1 activation and enhanced DDR. It is tempting to suggest a protective role for HMGA2 in cancer cell subsets with stem-like properties [21], and this has important clinical relevance, as tumor recurrence may be attributed to the presence of an HMGA2-positive CIC pool. Our data identify HMGA2 as an important contributor to chemoresistance through ATR-CHK1 signaling in HMGA2-expressing cancers and warrant the development of small inhibitors that target both the chromatin-binding property of HMGA2 and the ATR-CHK1 pathway to effectively bypass the antiapoptotic actions of HMGA2 and ATR/CHK1. The ability to block these mechanisms of chemoresistance in HMGA2-expressing cancer cells will likely also target and possibly incapacitate the CIC pool, thereby providing a marked advance in the treatment of some chemoresistant and aggressive HMGA2+ cancer types.
Supplementary Material
Acknowledgments
We thank Y. Liu for the generation of human fibrosarcoma cell line C1 and H. Taubert (University of Nuremburg, Erlangen, Germany) for providing the RD cell line. We thank Dana Henderson and Andreea Nistor for excellent technical support.
Abbreviations
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related kinase
- CHK
checkpoint kinase
- CICs
cancer-initiating cells
- DDR
DNA damage response
- DNA-PKc
DNA-dependent protein kinase catalytic subunit
- FLICA
Fluorescent Labeled Inhibitor of Caspases
- HMG
high mobility group
- HU
hydroxyurea
- MMS
methyl methane sulfonate
- RD
rhabdomyosarcom
Footnotes
This work was financed by the Singapore Biomedical Research Council (BMRC No. 07/1/22/19/532) to P.D. S.H.K. and T.K. are both grateful for financial support from the Natural Sciences and Engineering Council of Canada, the Manitoba Institute of Child Health, and the Department of Surgery Research Fund at the Faculty of Medicine, University of Manitoba. The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 2.Gattas GJF, Quade BJ, Nowak RA, Morton CC. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer. 1999;25:316–322. [PubMed] [Google Scholar]
- 3.Liau S-S, Whang E. HMGA1 is a molecular determinant of chemoresistance to gemcitabine in pancreatic adenocarcinoma. Clin Cancer Res. 2008;14:1470–1477. doi: 10.1158/1078-0432.CCR-07-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park S-M, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 5.Summer H, Li O, Bao Q, Zhan L, Peter S, Sathiyanathan P, Henderson D, Klonisch T, Goodman SD, Dröge P. HMGA2 exhibits dRP/AP site cleavage activity and protects cancer cells from DNA-damage-induced cytotoxicity during chemotherapy. Nucleic Acids Res. 2009;37:4371–4384. doi: 10.1093/nar/gkp375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chieffi P, Battista S, Barchi M, Di Agostino S, Pierantoni GM, Fedele M, Chiariotti L, Tramontano D, Fusco A. HMGA1 andHMGA2 protein expression in mouse spermatogenesis. Oncogene. 2002;21:3644–3650. doi: 10.1038/sj.onc.1205501. [DOI] [PubMed] [Google Scholar]
- 7.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 8.Battista S, Fidanza V, Fedele M, Klein-Szanto AJP, Outwater E, Brunner H, Santoro M, Croce CM, Fusco A. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999;59:4793–4797. [PubMed] [Google Scholar]
- 9.Zaidi MR, Okada Y, Chada KK. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–7459. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 10.Li O, Vasudevan D, Davey CA, Dröge P. High-level expression of DNA architectural factor HMGA2 and its association with nucleosomes in human embryonic stem cells. Genesis. 2006;44:523–529. doi: 10.1002/dvg.20242. [DOI] [PubMed] [Google Scholar]
- 11.Li O, Li J, Dröge P. DNA architectural factor and proto-oncogene HMGA2 regulates key developmental genes in pluripotent human embryonic stem cells. FEBS Lett. 2007;581:3533–3537. doi: 10.1016/j.febslet.2007.06.072. [DOI] [PubMed] [Google Scholar]
- 12.Di Cello F, Hillion J, Hristov A, Wood LJ, Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R, Resar LMS. HMGA2 participates in transformation in human lung cancer. Mol Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 14.Wood LJ, Maher JF, Bunton TE, Resar LMS. The oncogenic properties of the HMG-I gene family. Cancer Res. 2000;60:4256–4261. [PubMed] [Google Scholar]
- 15.Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta (BBA) 2010;1799:48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Rogalla P, Drechsler K, Kazmierczak B, Rippe V, Bonk U, Bullerdiek J. Expression of HMGI-C, a member of the high mobility group protein family, in a subset of breast cancers: relationship to histologic grade. Mol Carcinog. 1997;19:153–156. doi: 10.1002/(sici)1098-2744(199707)19:3<153::aid-mc2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 17.Meyer B, Loeschke S, Schultze A, Weigel T, Sandkamp M, Goldmann T, Vollmer E, Bullerdiek J. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46:503–511. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 18.Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004;64:2024–2029. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 19.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 Regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 21.Dröge P, Davey CA. Do cells let-7 determine stemness? Cell Stem Cell. 2008;2:8–9. doi: 10.1016/j.stem.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Lim D-S, Kim S-T, Xu B, Maser RS, Lin J, Petrini JHJ, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 23.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 24.Byun TS, Pacek M, Yee M-C, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward IM, Minn K, Chen J. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J Biol Chem. 2004;279:9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- 26.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 27.Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 29.Zachos G, Rainey MD, Gillespie DAF. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003;22:713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez R, Meuth M. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Mol Biol Cell. 2006;17:402–412. doi: 10.1091/mbc.E05-07-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadoni N, Cardoso MC, Stelzer EHK, Leonhardt H, Zink D. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci. 2004;117:5353–5365. doi: 10.1242/jcs.01412. [DOI] [PubMed] [Google Scholar]
- 34.Gillespie PJ, Blow J. Clusters, factories and domains: the complex structure of S-phase comes into focus. Cell Cycle. 2010;9:3218–3226. doi: 10.4161/cc.9.16.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galet C, Min L, Narayanan R, Kishi M, Weigel NL, Ascoli M. Identification of a transferable two-amino-acid motif (GT) present in the C-terminal tail of the human lutropin receptor that redirects internalized G protein-coupled receptors from a degradation to a recycling pathway. Mol Endocrinol. 2003;17:411–422. doi: 10.1210/me.2002-0161. [DOI] [PubMed] [Google Scholar]
- 36.Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261:16037–16042. [PubMed] [Google Scholar]
- 37.Palmieri D, Valentino T, D'Angelo D, De Martino I, Postiglione I, Pacelli R, Croce CM, Fedele M, Fusco A. HMGA proteins promote ATM expression and enhance cancer cell resistance to genotoxic agents. Oncogene. 2011;30:3024–3035. doi: 10.1038/onc.2011.21. [DOI] [PubMed] [Google Scholar]
- 38.Hombach-Klonisch S, Bialek J, Trojanowicz B, Weber E, Holzhausen H-JR, Silvertown JD, Summerlee AJ, Dralle H, Hoang-Vu C, Klonisch T. Relaxin enhances the oncogenic potential of human thyroid carcinoma cells. Am J Pathol. 2006;169:617–632. doi: 10.2353/ajpath.2006.050876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olive PL, Banáth JP, Durand RE. Detection of etoposide resistance by measuring DNA damage in individual Chinese hamster cells. J Natl Cancer Inst. 1990;82:779–783. doi: 10.1093/jnci/82.9.779. [DOI] [PubMed] [Google Scholar]
- 40.Kedar PS, Stefanick DF, Horton JK, Wilson SH. Interaction between PARP-1 and ATR in mouse fibroblasts is blocked by PARP inhibition. DNA Repair (Amst) 2008;7:1787–1798. doi: 10.1016/j.dnarep.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Löffler H, Bochtler T, Fritz B, Tews BR, Ho AD, Lukas J, Bartek J, Krämer A. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle. 2007;6:2541–2548. doi: 10.4161/cc.6.20.4810. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z, Xiao Z, Chen J, Ng S-C, Sowin T, Sham H, Rosenberg S, Fesik S, Zhang H. Human Chk1 expression is dispensable for somatic cell death and critical for sustaining G2 DNA damage checkpoint. Mol Cancer Ther. 2003;2:543–548. [PubMed] [Google Scholar]
- 43.Xiao Z, Xue J, Sowin TJ, Rosenberg SH, Zhang H. A novel mechanism of checkpoint abrogation conferred by Chk1 downregulation. Oncogene. 2004;24:1403–1411. doi: 10.1038/sj.onc.1208309. [DOI] [PubMed] [Google Scholar]
- 44.Yuwen L, Cunxin W, Congyi Z, Haixiang W, Zhiyong W, Songsheng Q. Microcalorimetric study of the metabolism of U-937 cells undergoing apoptosis induced by the combined treatment of hyperthermia and chemotherapy. J Therm Biol. 2002;27:129–135. [Google Scholar]
- 45.Langelotz C, Schmid P, Jakob C, Heider U, Wernecke KD, Possinger K, Sezer O. Expression of high-mobility-group-protein HMGI-C mRNA in the peripheral blood is an independent poor prognostic indicator for survival in metastatic breast cancer. Br J Cancer. 2003;88:1406–1410. doi: 10.1038/sj.bjc.6600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Liu X, Li AY-J, Chen L, Lai L, Lin HH, Hu S, Yao L, Peng J, Loera S, et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin Cancer Res. 2011;17:2570–2580. doi: 10.1158/1078-0432.CCR-10-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Györffy B, Surowiak P, Kiesslich O, Denkert C, Schäfer R, Dietel M, Lage H. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int J Cancer. 2006;118:1699–1712. doi: 10.1002/ijc.21570. [DOI] [PubMed] [Google Scholar]
- 48.Li AYJ, Boo LM, Wang S-Y, Lin HH, Wang CCC, Yen Y, Chen BPC, Chen DJ, Ann DK. Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res. 2009;69:5699–5706. doi: 10.1158/0008-5472.CAN-08-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meek K, Dang V, Lees-Miller SP, Frederick WA. Advances in Immunology. Vol. 99. New York, NY: Academic Press; 2008. pp. 33–58. [DOI] [PubMed] [Google Scholar]
- 50.Chen L, Liu T-H, Walworth NC. Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Genes Dev. 1999;13:675–685. doi: 10.1101/gad.13.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang K, Pereira E, Maxfield M, Russell B, Goudelock DM, Sanchez Y. Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser-345. J Biol Chem. 2003;278:25207–25217. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- 52.Mochida S, Esashi F, Aono N, Tamai K, O'Connell MJ, Yanagida M. Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J. 2004;23:418–428. doi: 10.1038/sj.emboj.7600018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Latif C, den Elzen NR, O'Connell MJ. DNA damage checkpoint maintenance through sustained Chk1 activity. J Cell Sci. 2004;117:3489–3498. doi: 10.1242/jcs.01204. [DOI] [PubMed] [Google Scholar]
- 54.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrett MD, Collins I. Anticancer therapy with checkpoint inhibitors: what, where and when? Trends Pharmacol Sci. 2011;32:308–316. doi: 10.1016/j.tips.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 56.Walton MI, Eve PD, Hayes A, Valenti M, De Haven Brandon A, Box G, Boxall KJ, Aherne GW, Eccles SA, Raynaud FI, et al. The preclinical pharmacology and therapeutic activity of the novel CHK1 inhibitor SAR-020106. Mol Cancer Ther. 2010;9:89–100. doi: 10.1158/1535-7163.MCT-09-0938. [DOI] [PubMed] [Google Scholar]
- 57.Goudelock DM, Jiang K, Pereira E, Russell B, Sanchez Y. Regulatory interactions between the checkpoint kinase Chk1 and the proteins of the DNA-dependent protein kinase complex. J Biol Chem. 2003;278:29940–29947. doi: 10.1074/jbc.M301765200. [DOI] [PubMed] [Google Scholar]
- 58.Capasso H, Palermo C, Wan S, Rao H, John UP, O'Connell MJ, Walworth NC. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J Cell Sci. 2002;115:4555–4564. doi: 10.1242/jcs.00133. [DOI] [PubMed] [Google Scholar]
- 59.Liu Q, Guntuku S, Cui X-S, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 60.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O'Connell MJ. Oncogene. Vol. 20. New York, NY: Nature Publishing Group; 2001. p. 7453. [DOI] [PubMed] [Google Scholar]
- 61.Lossaint G, Besnard E, Fisher D, Piette J, Dulic V. Chk1 is dispensable for G2 arrest in response to sustained DNA damage when the ATM/p53/p21 pathway is functional. Oncogene. 2011;30:4261–4274. doi: 10.1038/onc.2011.135. [DOI] [PubMed] [Google Scholar]
- 62.Chen C, Kennedy R, Sidi S, Look AT, D'Andrea A. CHK1 inhibition as a strategy for targeting fanconi anemia (FA) DNA repair pathway deficient tumors. Mol Cancer. 2009;8:24. doi: 10.1186/1476-4598-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heffernan TP, Kawasumi M, Blasina A, Anderes K, Conney AH, Nghiem P. ATR-Chk1 pathway inhibition promotes apoptosis after UV treatment in primary human keratinocytes: potential basis for the UV protective effects of caffeine. J Invest Dermatol. 2009;129:1805–1815. doi: 10.1038/jid.2008.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D'Angelo D, Borbone E, Palmieri D, Uboldi S, Esposito F, Frapolli R, Pacelli R, D'Incalci M, Fusco A. The impairment of the high mobility group A (HMGA) protein function contributes to the anticancer activity of trabectedin. Eur J Cancer. 2012 doi: 10.1016/j.ejca.2012.10.014. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 66.Pentimalli F, Dentice M, Fedele M, Pierantoni GM, Cito L, Pallante P, Santoro M, Viglietto G, Cin PD, Fusco A. Suppression of HMGA2 protein synthesis could be a tool for the therapy of well differentiated liposarcomas overexpressing HMGA2. Cancer Res. 2003;63:7423–7427. [PubMed] [Google Scholar]
- 67.Malek A, Bakhidze E, Noske A, Sers C, Aigner A, Schäfer R, Tchernitsa O. HMGA2 gene is a promising target for ovarian cancer silencing therapy. Int J Cancer. 2008;123:348–356. doi: 10.1002/ijc.23491. [DOI] [PubMed] [Google Scholar]
- 68.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindsey-Boltz LA, Sancar A. Tethering DNA damage checkpoint mediator proteins topoisomerase IIbeta-binding protein 1 (TopBP1) and Claspin to DNA activates ataxia-telangiectasia mutated and RAD3-related (ATR) phosphorylation of checkpoint kinase 1 (Chk1) J Biol Chem. 2011;286:19229–19236. doi: 10.1074/jbc.M111.237958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- 73.Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, Los M. Cancer stem cell markers in common cancers—therapeutic implications. Trends Mol Med. 2008;14:450–460. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.