Abstract
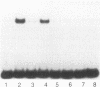
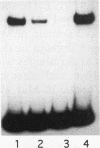
NUC-1, a positive regulatory protein of Neurospora crassa, controls the expression of several unlinked target genes involved in phosphorus acquisition. The carboxy-terminal end of the NUC-1 protein has sequence similarity to the helix-loop-helix family of transcription factors. Bacterially expressed and in vitro-synthesized proteins, which consist of the carboxy-terminal portion of NUC-1, bind specifically to upstream sequences of two of its target genes, pho2+ and pho-4+. These upstream sequences contain the core sequence, CACGTG, a target for many helix-loop-helix proteins. A large loop region (47 amino acids) separates the helix I and helix II domains. Mutations and deletion within the loop region did not interfere with the in vitro or in vivo functions of the protein. Immediately carboxy-proximal to the helix II domain, the NUC-1 protein contains an atypical zipper domain which is essential for function. This domain consists of a heptad repeat of alanine and methionine rather than leucine residues. Analysis of mutant NUC-1 proteins suggests that the helix II and the zipper domains are essential for the protein dimerization, whereas the basic and the helix I domains are involved in DNA binding. The helix I domain, even though likely to participate in dimer formation while NUC-1 is bound to DNA, is not essential for in vitro dimerization.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol Cell Biol. 1985 Sep;5(9):2272–2278. doi: 10.1128/mcb.5.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993 Jan 29;72(2):233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993 Jan 29;72(2):211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990 Apr 6;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Brennan T. J., Chakraborty T., Olson E. N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990 May 4;61(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Dang C. V., Dolde C., Gillison M. L., Kato G. J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., McGuire M., Buckmire M., Lee W. M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989 Feb 16;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Davis L. J., Halazonetis T. D. Both the helix-loop-helix and the leucine zipper motifs of c-Myc contribute to its dimerization specificity with Max. Oncogene. 1993 Jan;8(1):125–132. [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Edmondson D. G., Olson E. N. Helix-loop-helix proteins as regulators of muscle-specific transcription. J Biol Chem. 1993 Jan 15;268(2):755–758. [PubMed] [Google Scholar]
- Ellenberger T., Fass D., Arnaud M., Harrison S. C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994 Apr 15;8(8):970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- Ellis H. M., Spann D. R., Posakony J. W. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990 Apr 6;61(1):27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994 Jan 1;13(1):180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Carr C. S., Parent L. A., Sharp P. A. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991 Dec;5(12A):2342–2352. doi: 10.1101/gad.5.12a.2342. [DOI] [PubMed] [Google Scholar]
- Grotelueschen J., Peleg Y., Glass N. L., Metzenberg R. L. Cloning and characterization of the pho-2+ gene encoding a repressible alkaline phosphatase in Neurospora crassa. Gene. 1994 Jun 24;144(1):147–148. doi: 10.1016/0378-1119(94)90223-2. [DOI] [PubMed] [Google Scholar]
- Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991 Feb 1;192(2):262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Herlitze S., Koenen M. A general and rapid mutagenesis method using polymerase chain reaction. Gene. 1990 Jul 2;91(1):143–147. doi: 10.1016/0378-1119(90)90177-s. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kadesch T. Helix-loop-helix proteins in the regulation of immunoglobulin gene transcription. Immunol Today. 1992 Jan;13(1):31–36. doi: 10.1016/0167-5699(92)90201-h. [DOI] [PubMed] [Google Scholar]
- Kang S. Functional domains of the transcriptional activator NUC-1 in Neurospora crassa. Gene. 1993 Aug 25;130(2):259–264. doi: 10.1016/0378-1119(93)90428-6. [DOI] [PubMed] [Google Scholar]
- Kang S., Metzenberg R. L. Insertional mutagenesis in Neurospora crassa: cloning and molecular analysis of the preg+ gene controlling the activity of the transcriptional activator NUC-1. Genetics. 1993 Feb;133(2):193–202. doi: 10.1093/genetics/133.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Metzenberg R. L. Molecular analysis of nuc-1+, a gene controlling phosphorus acquisition in Neurospora crassa. Mol Cell Biol. 1990 Nov;10(11):5839–5848. doi: 10.1128/mcb.10.11.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Klein E. S., Simmons D. M., Swanson L. W., Rosenfeld M. G. Tissue-specific RNA splicing generates an ankyrin-like domain that affects the dimerization and DNA-binding properties of a bHLH protein. Genes Dev. 1993 Jan;7(1):55–71. doi: 10.1101/gad.7.1.55. [DOI] [PubMed] [Google Scholar]
- Lamb P., McKnight S. L. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991 Nov;16(11):417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Davis R. L., Wright W. E., Kadesch T., Murre C., Voronova A., Baltimore D., Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991 Jul 26;66(2):305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Legrain M., De Wilde M., Hilger F. Isolation, physical characterization and expression analysis of the Saccharomyces cerevisiae positive regulatory gene PHO4. Nucleic Acids Res. 1986 Apr 11;14(7):3059–3073. doi: 10.1093/nar/14.7.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczynski J. F., Rose G. D. Loops in globular proteins: a novel category of secondary structure. Science. 1986 Nov 14;234(4778):849–855. doi: 10.1126/science.3775366. [DOI] [PubMed] [Google Scholar]
- Ma P. C., Rould M. A., Weintraub H., Pabo C. O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994 May 6;77(3):451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Mann B. J., Akins R. A., Lambowitz A. M., Metzenberg R. L. The structural gene for a phosphorus-repressible phosphate permease in Neurospora crassa can complement a mutation in positive regulatory gene nuc-1. Mol Cell Biol. 1988 Mar;8(3):1376–1379. doi: 10.1128/mcb.8.3.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B. J., Bowman B. J., Grotelueschen J., Metzenberg R. L. Nucleotide sequence of pho-4+, encoding a phosphate-repressible phosphate permease of Neurospora crassa. Gene. 1989 Nov 30;83(2):281–289. doi: 10.1016/0378-1119(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg R. L., Chia W. Genetic control of phosphorus assimilation in Neurospora crassa: dose-dependent dominance and recessiveness in constitutive mutants. Genetics. 1979 Nov;93(3):625–643. doi: 10.1093/genetics/93.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Ogawa N., Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce S., Benezra R. The loop region of the helix-loop-helix protein Id1 is critical for its dominant negative activity. Mol Cell Biol. 1993 Dec;13(12):7874–7880. doi: 10.1128/mcb.13.12.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. A new bind for Myc. Trends Genet. 1992 Mar;8(3):91–96. doi: 10.1016/0168-9525(92)90196-b. [DOI] [PubMed] [Google Scholar]
- Rabindran S. K., Haroun R. I., Clos J., Wisniewski J., Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993 Jan 8;259(5092):230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Starovasnik M. A., Blackwell T. K., Laue T. M., Weintraub H., Klevit R. E. Folding topology of the disulfide-bonded dimeric DNA-binding domain of the myogenic determination factor MyoD. Biochemistry. 1992 Oct 20;31(41):9891–9903. doi: 10.1021/bi00156a006. [DOI] [PubMed] [Google Scholar]
- Sun X. H., Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991 Jan 25;64(2):459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- Torres R., Schreiber-Agus N., Morgenbesser S. D., DePinho R. A. Myc and Max: a putative transcriptional complex in search of a cellular target. Curr Opin Cell Biol. 1992 Jun;4(3):468–474. doi: 10.1016/0955-0674(92)90013-3. [DOI] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronova A., Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W. E., Sassoon D. A., Lin V. K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989 Feb 24;56(4):607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Kuromitsu Z., Ogawa N., Oshima Y. Mode of expression of the positive regulatory genes PHO2 and PHO4 of the phosphatase regulon in Saccharomyces cerevisiae. Mol Gen Genet. 1989 May;217(1):31–39. doi: 10.1007/BF00330939. [DOI] [PubMed] [Google Scholar]