Abstract
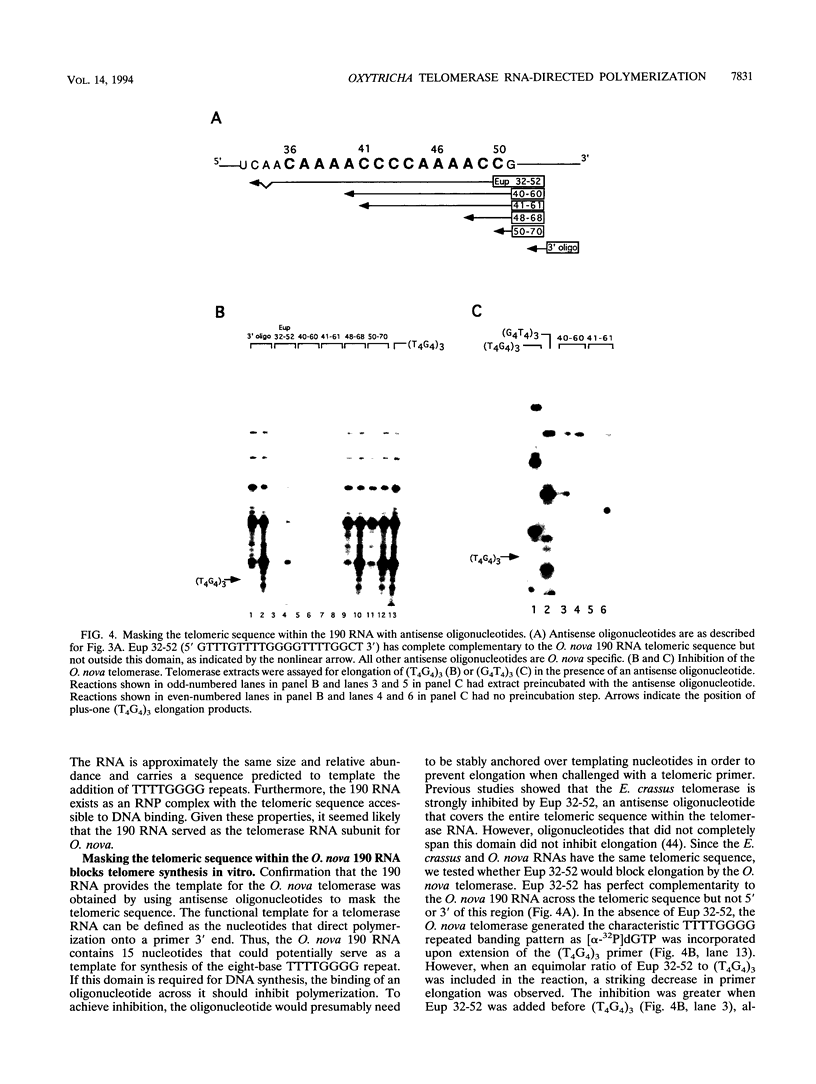
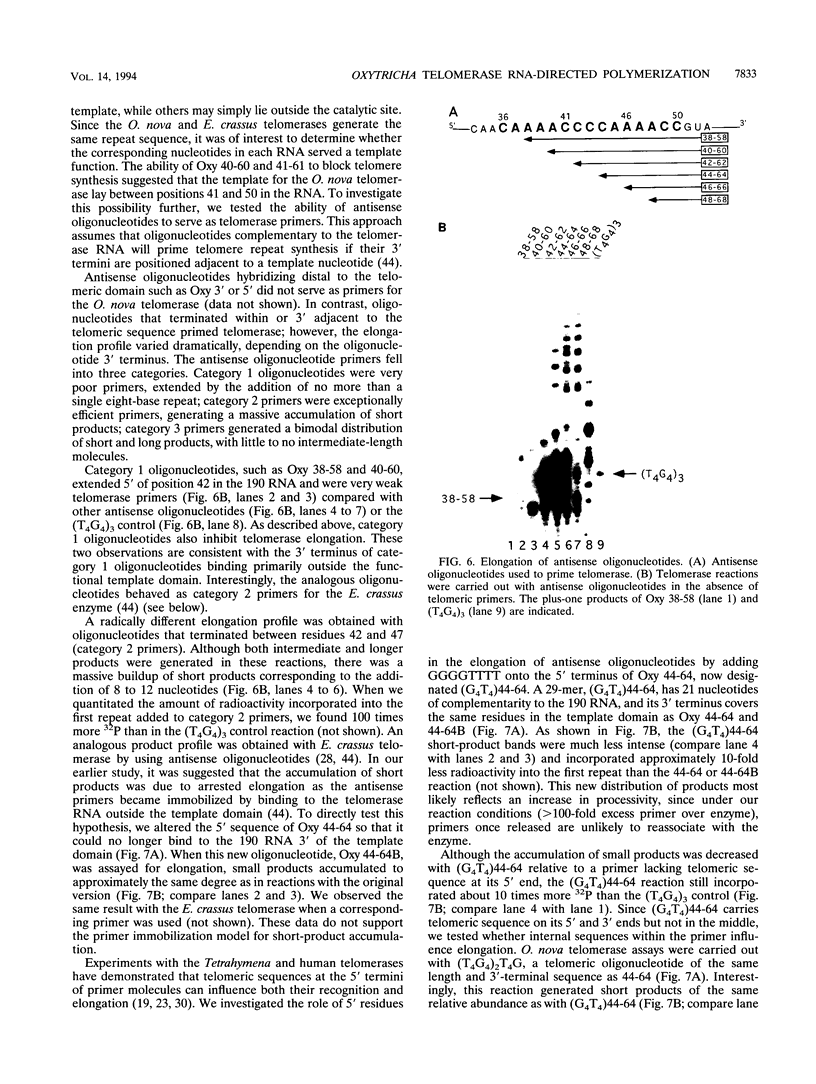
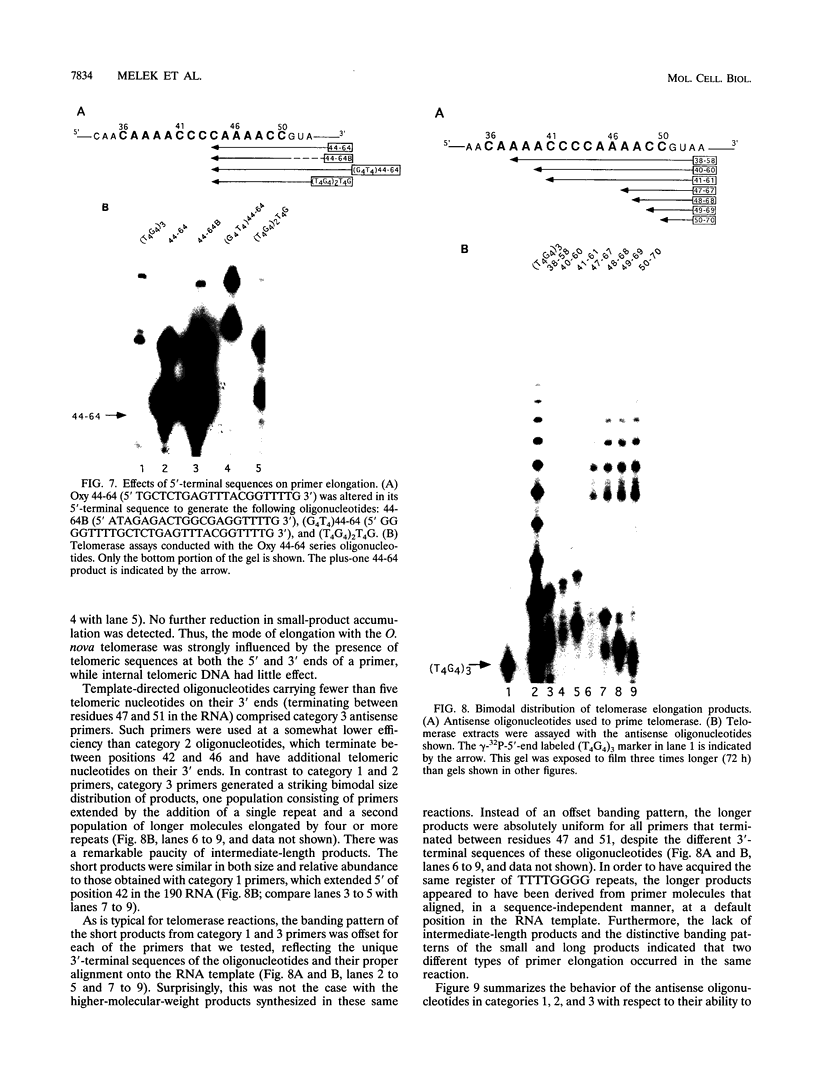
The telomerase reverse transcriptase uses an essential RNA subunit as a template to direct telomeric DNA synthesis. The 190-nucleotide Oxytricha nova telomerase RNA was identified by using an oligonucleotide probe complementary to the predicted CCCCAAAA template. This RNA displays extensive sequence similarity to the Euplotes crassus telomerase RNA and carries the same 5' CAAAACCCCAAAACC 3' telomeric domain. Antisense oligonucleotides were used to map the boundaries of the functional template and to investigate the mechanism of primer recognition and elongation. On the basis of their ability to inhibit or to prime telomerase, oligonucleotides were classified into three categories. Category 1 oligonucleotides, which extended 5' of residue 42 in the RNA, abolished elongation of (T4G4)3 and (G4T4)3 primers in vitro. In contrast, oligonucleotides terminating between residues 42 and 50 (categories 2 and 3), served as efficient telomerase primers. We conclude that the O. nova template comprises residues 42 to 50 in the 190-nucleotide RNA, a different set of nucleotides than are used by the E. crassus enzyme. Category 2 primer reactions amassed short products, and their abundance could be decreased by altering the 5' sequence of the primer, consistent with the two-primer-binding-site model for telomerase. Category 3 primers generated a bimodal distribution of short and long products, each having a unique elongation profile. The long-product profile is inconsistent with sequence-specific primer alignment. Rather, each primer was extended by the same register of TTTTGGGG repeats, suggesting shuttling to a default position within the template. The parallels between telomerase and RNA polymerase elongation mechanisms are discussed.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Autexier C., Greider C. W. Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev. 1994 Mar 1;8(5):563–575. doi: 10.1101/gad.8.5.563. [DOI] [PubMed] [Google Scholar]
- Avilion A. A., Harrington L. A., Greider C. W. Tetrahymena telomerase RNA levels increase during macronuclear development. Dev Genet. 1992;13(1):80–86. doi: 10.1002/dvg.1020130113. [DOI] [PubMed] [Google Scholar]
- Balakrishnan R., Frohlich M., Rahaim P. T., Backman K., Yocum R. R. Appendix. Cloning and sequence of the gene encoding enzyme E-1 from the methionine salvage pathway of Klebsiella oxytoca. J Biol Chem. 1993 Nov 25;268(33):24792–24795. [PubMed] [Google Scholar]
- Blackburn E. H. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- Borukhov S., Sagitov V., Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993 Feb 12;72(3):459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Collins K., Greider C. W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993 Jul;7(7B):1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979 Sep 11;7(1):179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Gumport R. I., Uhlenbeck O. C. Dinucleoside pyrophosphate are substrates for T4-induced RNA ligase. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4839–4842. doi: 10.1073/pnas.74.11.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Gray J. T., Cech T. R. Oxytricha telomere-binding protein: separable DNA-binding and dimerization domains of the alpha-subunit. Genes Dev. 1993 May;7(5):870–882. doi: 10.1101/gad.7.5.870. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Zakian V. A. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986 Oct 24;47(2):195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989 Jan 26;337(6205):331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985 Dec;43(2 Pt 1):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Greider C. W. Telomerase is processive. Mol Cell Biol. 1991 Sep;11(9):4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L. A., Greider C. W. Telomerase primer specificity and chromosome healing. Nature. 1991 Oct 3;353(6343):451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- Johnson T. L., Chamberlin M. J. Complexes of yeast RNA polymerase II and RNA are substrates for TFIIS-induced RNA cleavage. Cell. 1994 Apr 22;77(2):217–224. doi: 10.1016/0092-8674(94)90314-x. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Swanton M. T., Donini P., Prescott D. M. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3' terminus. Proc Natl Acad Sci U S A. 1981 May;78(5):3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J Mol Biol. 1992 May 20;225(2):221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Blackburn E. H. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993 Oct;13(10):6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hendrick L. L., Cech T. R. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994 Aug 15;8(16):1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lundblad V., Szostak J. W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989 May 19;57(4):633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Mantell L. L., Greider C. W. Telomerase activity in germline and embryonic cells of Xenopus. EMBO J. 1994 Jul 1;13(13):3211–3217. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin G. B. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991 Oct 3;353(6343):454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- Morin G. B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989 Nov 3;59(3):521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Orum H., Nielsen H., Engberg J. Spliceosomal small nuclear RNAs of Tetrahymena thermophila and some possible snRNA-snRNA base-pairing interactions. J Mol Biol. 1991 Nov 20;222(2):219–232. doi: 10.1016/0022-2836(91)90208-n. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. M. Telomere structure in Euplotes crassus: characterization of DNA-protein interactions and isolation of a telomere-binding protein. Mol Cell Biol. 1990 Jul;10(7):3421–3431. doi: 10.1128/mcb.10.7.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowse K. R., Avilion A. A., Greider C. W. Identification of a nonprocessive telomerase activity from mouse cells. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G. A., Chamberlin M. J., Kane C. M. Contacts between mammalian RNA polymerase II and the template DNA in a ternary elongation complex. Nucleic Acids Res. 1993 Jan 11;21(1):113–118. doi: 10.1093/nar/21.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. P., Blackburn E. H. A conserved secondary structure for telomerase RNA. Cell. 1991 Oct 18;67(2):343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Sandell L. L., Zakian V. A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993 Nov 19;75(4):729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Shippen-Lentz D., Blackburn E. H. Functional evidence for an RNA template in telomerase. Science. 1990 Feb 2;247(4942):546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- Shippen-Lentz D., Blackburn E. H. Telomere terminal transferase activity from Euplotes crassus adds large numbers of TTTTGGGG repeats onto telomeric primers. Mol Cell Biol. 1989 Jun;9(6):2761–2764. doi: 10.1128/mcb.9.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippen D. E., Blackburn E. H., Price C. M. DNA bound by the Oxytricha telomere protein is accessible to telomerase and other DNA polymerases. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):405–409. doi: 10.1073/pnas.91.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippen D. E. Telomeres and telomerases. Curr Opin Genet Dev. 1993 Oct;3(5):759–763. doi: 10.1016/s0959-437x(05)80095-4. [DOI] [PubMed] [Google Scholar]
- Watson J. D. Origin of concatemeric T7 DNA. Nat New Biol. 1972 Oct 18;239(94):197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Bradley J. D., Attardi L. D., Blackburn E. H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990 Mar 8;344(6262):126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Prescott D. M. Telomere terminal transferase activity in the hypotrichous ciliate Oxytricha nova and a model for replication of the ends of linear DNA molecules. Nucleic Acids Res. 1988 Jul 25;16(14B):6953–6972. doi: 10.1093/nar/16.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A. Structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- de Lange T. Activation of telomerase in a human tumor. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):2882–2885. doi: 10.1073/pnas.91.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]










