Abstract
Lipids serve essential functions in cells as signaling molecules, membrane components, and sources of energy. Defects in lipid metabolism are implicated in a number of pandemic human diseases, including diabetes, obesity, and hypercholesterolemia. approaches for disease prevention and treatment. Numerous studies have shown that the zebrafish is an excellent model for vertebrate lipid metabolism. In this chapter, we review studies that employ zebrafish to better understand lipid signaling and metabolism.
I. Introduction
Lipids play essential roles in cells as signaling molecules, membrane components, and sources of fuel. Given their necessity for proper cellular function, it is not surprising that defects in lipid metabolism underlie a number of human diseases, including obesity, diabetes, and atherosclerosis (Joffe et al., 2001; McNeely et al., 2001; Watanabe et al., 2008). In 2007–08, one-third of US adults and 18% of children were classified as obese (Flegal et al., 2010; Ogden et al., 2010), with obesity and type 2 diabetes on the rise worldwide (Misra and Khurana, 2008). The globalization of the high-fat western diet and the concurrent rise in the incidence of lipid disorders has provided an impetus to better understand lipid metabolism in the context of metabolic dysfunction. This need to investigate the role of lipids in metabolic disease has brought into focus unanswered questions in the field. For instance, although the genes involved in cholesterol and fatty acid (FA) uptake in intestinal cells have been identified, their exact mechanisms of action are highly debated or largely unknown (Klett and Patel, 2004; Nassir et al., 2007; Nickerson et al., 2009; Shim et al., 2009; Stahl et al., 1999). Such gaps in our understanding of these genes and how they function hinder the development of effective therapeutics for lipid disorders and reveal a need to create better approaches to address them.
In this chapter, we will review novel approaches undertaken to study lipid signaling and metabolism using the zebrafish model organism, with an emphasis on presenting a diverse array of techniques employed to visualize lipid metabolism during various stages of zebrafish development.
A. The Need for Whole Animal Studies of Lipid Metabolism
In vitro studies have laid much of the groundwork for our biochemical understanding of vertebrate lipid metabolism and continue to comprise many of the studies done in the field today; however, a number of caveats arise when attempting to study lipid metabolism in vitro. Such studies are often performed in transformed cultured cells, such as liver HepG2, intestinal Caco2, and adipocyte 3LT3 cells. These cell lines, comprised of a single cell type, cannot duplicate the cellular heterogeneity of an entire organ, such as the intestine, which is composed of stem, enteroendocrine, immune, and goblet cells. These multiple cell types are known to influence each other through paracrine signaling that can have global effects on lipid uptake and processing. Furthermore, bile, intestinal mucus, and the gut microbiota are all known to greatly influence dietary lipid processing and absorption in the intestine (Backhed et al., 2004; Field et al., 2003; Kruit et al., 2006; Martin et al., 2008; Moschetta et al., 2005; Pack et al., 1996; Titus and Ahearn, 1992; Turnbaugh et al., 2008), and are absent in cultured cell models. For these reasons, employing whole animal in vivo strategies, in addition to cultured cell work, is vital to better understand how metabolic dysfunction arises and manifests itself in an organism.
The importance of utilizing whole animal models to identify drugs to ameliorate metabolic dysfunction is exemplified by how the cholesterol absorption inhibitor ezetimibe (Zetia, Vitorin; Merck-Schering Plough) was developed (Van Heek et al., 1997), and how its mechanism of action was defined (Van Heek et al., 2000). Studies of bile duct-cannulated rats treated with ezetimibe revealed that the bioactive compound responsible for the diminished effect on cholesterol absorption was a glucur-onidated form of ezetimibe. Since in vitro studies cannot recreate the complex interplay of neural, chemical, and hormonal cues known to regulate metabolic processes in vivo, there is a need for whole animal approaches to study lipid metabolism as it plays out in a multicellular context.
B. Larval Zebrafish as a Model of Vertebrate Lipid Metabolism
The larval zebrafish is well suited for whole animal studies of lipid metabolism. Larval zebrafish possess many of the same gastrointestinal organs present in humans (e.g., the liver, intestine, exocrine and endocrine pancreas, and gallbladder) (Lieschke and Currie, 2007; Pack et al., 1996; Schlegel and Stainier, 2006; Wallace and Pack, 2003; Wallace et al., 2005) as well as the specialized cell types involved in lipid absorption and processing (e.g., intestinal enterocytes, fat-storing adipocytes, hepatocytes in the liver, and acinar cells of the pancreas) (Wallace et al., 2005). These digestive organs and the cell types present in them are formed using similar developmental programs as in mammals with hhex (Wallace et al., 2001), pdx1 (Yee et al., 2005), and shha (Wallace and Pack, 2003) genes playing critical roles in liver and pancreas organogenesis in zebrafish. Notch and its ligands, Delta and Jagged, play a role in the developing pancreas, both by maintaining undifferentiated precursors and regulating acinar, exocrine, and endocrine differentiation (Apelqvist et al., 1999; Esni et al., 2004; Hald et al., 2003; Jensen et al., 2000; Murtaugh et al., 2003; Zecchin et al., 2004). Due to the optical transparency of larvae, these organs and their multiple cell types can all be directly observed through the body wall without the need for invasive surgical manipulations.
Work from our lab and others has shown that zebrafish express many of the genes needed to transport and metabolize lipids, such as the lipoprotein gene microsomal triglyceride transfer protein (MTP) (Marza et al., 2005) and the FA transport protein (slc27a) and acyl-CoA synthetase (acsl) gene families (Thisse et al., 2005; Thisse et al., 2004; Miyares, unpublished). Zebrafish express the putative cholesterol transporter Niemann-Pick C1-Like 1 (npc1l1) (Farber, unpublished) and treatment of larvae with the human drug ezetimibe, which works through an NPC1L1-dependent pathway and is used to treat hypercholesterolemia, blocks intestinal cholesterol absorption (Clifton et al., 2010). Apolipoprotein C2, a gene needed for lipoprotein assembly in humans, is expressed and required during zebrafish larval development; larvae injected with an apoc2 morpholino exhibit an unabsorbed yolk phenotype (Pickart et al., 2006). Additionally, the enzymes needed to synthesize lipid-signaling molecules, such as prostaglandins and thromboxanes, are highly conserved in zebrafish and these enzymes can be inhibited by commonly used nonsteroidal anti-inflammatory drugs (Grosser et al., 2002). We have presented here only a small sampling of the numerous studies that document the homologies between human and zebrafish lipid metabolism and validate the zebrafish as a model for investigating vertebrate lipid metabolism.
II. Lipid Metabolism in Developing Zebrafish
During the first four days of development, a zebrafish embryo relies entirely on its yolk sac for the nutrients needed to sustain its growth and survival. Yolk lipids are the source of essential fat-soluble vitamins and triacylglycerol (TAG), as well as cholesterol, a required component of cell membranes and a precursor for bile acids (Babin etal., 1997; Bownes, 1992; Munoz et al., 1990). Lipids enter the developing embryo at the yolk and embryo interface, an area termed the yolk syncytial layer (YSL). In the YSL, lipoproteins (e.g., apoE, apoA1, apocII, and vitellogenin) and a host of lipid-modifying enzymes (e.g., Mtp) transport lipids from the yolk ball to the embryo (Babin et al., 1997; Marza et al., 2005). Once the circulatory system forms, yolk, hepatic, and intestinal lipids are transported by lipoproteins to specific target tissues throughout the organism via the bloodstream.
By 5–6 days postfertilization (dpf), the yolk is depleted and larvae must eat to acquire lipids. Both in the wild and the laboratory, zebrafish consume a lipid-rich diet (≥ 10% by weight) high in TAG, phospholipids, and sterols (Enzler et al., 1974; Spence et al., 2008). Prior to absorption by the intestine, these yolk lipids must be processed and solubilized by the digestive enzymes and bile that make up the intraluminal intestinal milieu. As in humans, zebrafish bile is produced by hepatocytes and secreted into an extensive network of intrahepatic ducts, which drains into the gall bladder. In response to hormonal stimulation triggered by food consumption, bile is released into the intestinal lumen to emulsify dietary fat and facilitate its absorption by intestinal enterocytes. The main components of bile are bile acids, phospholipids, and salts, which together promote the formation of micelles by inserting themselves between lipid bilayers and reducing the surface tension to allow for membrane curvature (Tso and Fujimoto, 1991; Verkade and Tso, 2001). After dietary fat is emulsified, TAG and phospholipids must be broken down by luminal lipases to release free FA or mono- and di-acylglycerols, which can then enter the specialized absorptive cells (enterocytes) that line the gut (Thomson et al., 1993). The exocrine pancreas is the main source of these fat-splitting enzymes in larval zebrafish (Hama et al., 2009) and known in mammals to secrete lipase- and protease-rich pancreatic juice into the gall bladder (Layer and Keller, 1999). After being emulsified and broken down, TAG can be absorbed by enterocytes, the main absorptive cell type of the zebrafish intestine. These cells are highly reminiscent of mammalian enterocytes (Buhman et al., 2002), with the characteristic microvilli and basal nuclei (Fig. 1).
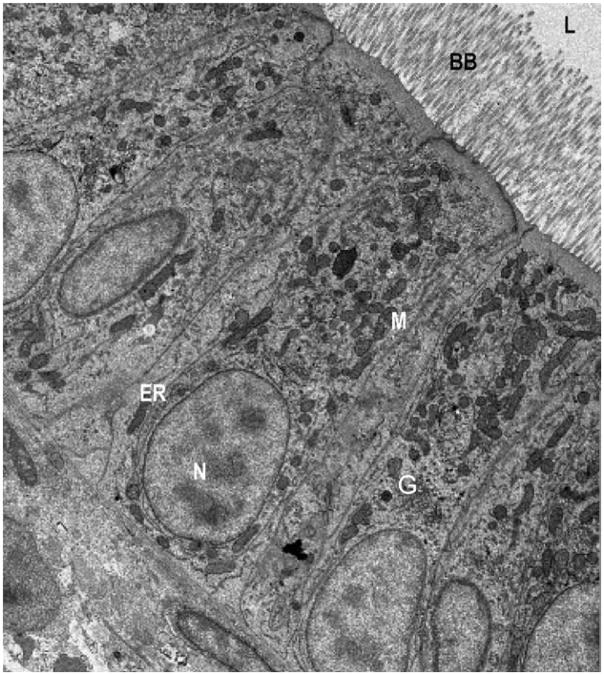
Fig. 1.
The intestinal enterocytes of zebrafish and mammals exhibit a high degree of morphological similarity. Electron micrograph of enterocytes from a larval zebrafish (6 dpf). Zebrafish enterocytes exhibit the typical characteristics of mammalian polarized intestinal cells including apical microvilli, which extend into the intestinal lumen (L) and form the brush border, as well as basal nuclei (N). Organelles and subcellular details including mitochondria (M), Golgi bodies (G), and endoplasmic reticulum (ER) are apparent throughout the enterocytes. For comparison, see mouse EMs in Buhman et al. (2002).
Following food consumption, zebrafish accumulate cytoplasmic lipid drops (LD) in their enterocytes (Walters, unpublished). From there, fats are likely burned via oxidative pathways in the mitochondria or peroxisomes or packaged into chylomicrons, which are secreted from the basolateral surface of enterocytes into lymphatic or blood vessels (Field, 2001; Levy et al., 2007). In chickens, chylomicron production and secretion is highly conserved with the exception that lipoproteins are secreted from the intestine directly into the portal vein and thus are termed portomicrons (Bensadoun and Rothfeld 1972; Griffin et al., 1982). This difference has led some to propose that a similar portomicron process occurs in fish (Robinson and Mead 1973); however, careful ultrastructural studies and isotopic lipid labeling experiments suggest that fish and mammals produce chylomicrons containing largely similar lipoproteins (Sire et al., 1981; Skinner and Rogie, 1978). While it remains to be seen how closely the zebrafish system will model human intestinal lipoprotein metabolism, it is likely that many of the mechanisms of lipoprotein production are conserved.
III. Yolk Metabolism During Early Vertebrate Development
To mobilize lipids stockpiled in the yolk sac, rodents express the same genes required for lipoprotein production as those expressed in fully differentiated intestinal enterocytes and liver hepatocytes (e.g., MTP and the apolipoproteins apoE, apoB, and apo-A-IV and apo-A-I (Elshourbagy et al., 1985; Farese et al., 1996; Plonne et al., 1996)). Without these genes, the rapid transport of essential nutrients from the yolk to developing embryonic tissues is impaired and development cannot proceed normally. Not surprisingly, mice null for MTP (Raabe et al., 1998) and apoB (Farese et al., 1996) are embryonic lethal, and DGAT-2 null mice, which lack the enzyme needed to synthesize diacylglycerol, exhibit stunted embryonic growth and die perinatally (Stone et al., 2004).
The zebrafish YSL expresses a number of lipoprotein-encoding mRNAs that are also expressed later in the larval intestine and liver, including MTP (Marza et al., 2005), apoC2 (Farber Lab, unpublished), intestinal FA binding protein (Sharma et al., 2004), and apoE and apoA-I (Babin et al., 1997). Due to the parallel gene expression patterns observed between the embryonic YSL and larval digestive organs, we hypothesize that yolk utilization during early zebrafish embryogenesis is a regulated process highly analogous to intestinal and hepatic lipoprotein-mediated lipid transport.
To study the role of lipid metabolism genes in the YSL during early zebrafish development, we utilize synthetic antisense morpholinos (MO) to attenuate gene expression and assay subsequent yolk metabolism. MOs are widely used in the zebrafish community to knock down mRNA levels (Heasman, 2002; Nasevicius and Ekker, 2000), and numerous studies have phenocopied mutants by targeting particular mRNA transcripts using this method (Dutton et al., 2001; Karlen and Rebagliati, 2001; Urtishak et al., 2003). By injecting MOs targeting lipid-specific genes into the yolk of 1–4 cell stage embryos, followed by yolk injection of fluorescent lipid analogs, such as BODIPY-labeled FA (Fig. 2A), we can directly assess the necessity of a given gene for yolk metabolism during early larval development. Following sequential injections of the morpholino and BODIPY-labeled FA, embryos are allowed to develop and total larval lipids are extracted at 2–3 dpf. We assay the incorporation of the fluorescent FA into metabolites using thin layer chromatography (TLC). This approach allows one to identify metabolic abnormalities in morphants that may not exhibit obvious morphological phenotypes. Furthermore, we can assay the effects of essential genes at earlier stages of larval development prior to lethality caused by insufficient transcript amounts.
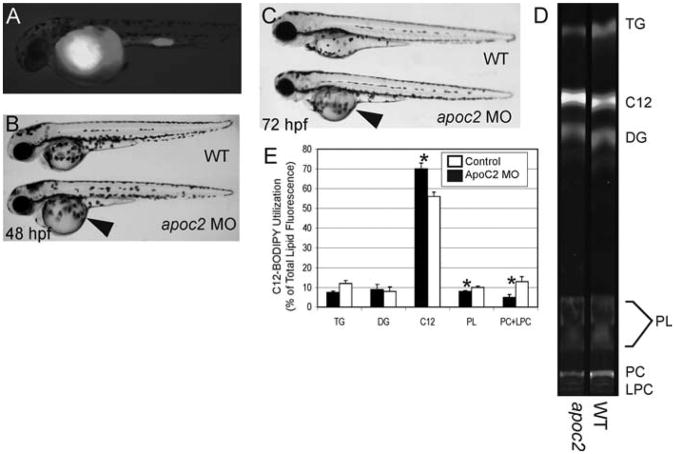
Fig. 2.
BODIPY lipid analogs enable studies of yolk metabolism during early zebrafish development. To assay the function of apoc2 during zebrafish development, embryos were injected with an apoc2 morpholino at the 1–4 cell stage followed by injection of a fluorescent fatty acid (BODIPY-C12) at 24 h postfertilization (hpf). (A) At 48 hpf, embryos injected with BODIPY-C12 retain the fluorescent analog primarily in their yolk. (B, C) apoc2 morphants exhibit an enlarged yolk phenotype (arrowhead) at 48 and 72 hpf, indicating that apoc2 is necessary for yolk utilization during larval development. (D, E) To determine the metabolic consequences of apoc2 deficiency, 1 dpf wild-type and apoc2 morphant larvae were injected with BODIPY-C12 and assayed using fluorescent thin layer chromatography (TLC) 2 days later. (D) TLC analysis shows that BODIPY-C12 is incorporated primarily into triacylglycerol (TAG), diacylglycerol (DG), and phosphatidylcholine (PC) in both wild-type and apoc2 morphant larvae. (E) apoc2 morphants exhibit defects in PC and lysophosphatidylcholine (LPC) metabolism. Total lipid fluorescence was quantified from TLC plates run with total lipids extracted from wild-type and apoc2 morphants. Triacylglycerol (TAG), diacylglycerol (DG), BODIPY C12:0 (C12), phospholipids (PL), PC, and LPC.* p < 0.05.
We are currently using this technique to better understand how the apolipoprotein apoc2 functions in yolk utilization during early zebrafish development. This gene was first identified in a MO screen targeting secreted proteins of the unknown function. In a screen of approximately 100 MOs, we identified one with an unab-sorbed yolk phenotype (Fig. 2B and 2C). TLC analysis revealed that these larvae exhibit metabolic defects in phospholipid production, as morphants incorporated less BODIPY-C12 into phosphatidylcholine (PC) and lysophosphatidylcholine (LPC) (Fig. 2D and 2E). Unabsorbed yolk and phospholipid metabolic deficiency in apoc2 morphants suggest that this gene has a function in the yolk or YSL during zebrafish development that is unrelated to its known role in the activation of lipo-protein lipase in peripheral tissues (Jong et al., 1999).
IV. Lipid Signaling During Early Zebrafish Development
It is not surprising that many lipid-signaling molecules are derived from membrane phospholipids. Such a system allows for rapid transmission of extracellular signals via membrane, and constituents to the intracellular environment to activate appropriate signaling cascades and cellular responses. The synthesis of membrane-derived signaling lipids is often dependent on phospholipases, enzymes that cleave phospholipids in response to specific cellular signals. Phospholipase A2 (PLA2) is one such enzyme that catalyzes the hydrolysis of the second fatty acyl bond of glycerophospholipids to liberate lysophospholipid and free FA, both lipid-signaling precursors. In the last decade, PLA2 activity and its products have been implicated in a wide range of cellular phenomena including inflammation, membrane remodeling, and cancer.
Lipid signaling events during early development are not well elucidated and have only been recently explored in zebrafish. Studies examining PLA2 enzymatic activity throughout larval development using whole embryo lysates revealed varying activity levels during different stages of development, with larvae exhibiting a significant PLA2 activity during somitogenesis (Fig. 3A). Pharmacological inhibition of PLA2 activity causes developmental arrest at epiboly (Farber et al., 1999). Further experiments identified the primary source of phospholipase activity to be the Ca2+-dependent cytosolic PLA2 (cPLA2) type, a crucial mediator of stimulusinduced eicosanoid release. cPLA2 activity releases the polyunsaturated FA arachidonic acid from membranes, allowing it to participate in the synthesis of eicosanoids, a potent class of lipid-signaling molecules that exhibit both paracrine and autocrine influences on cells and tissues (Burke and Dennis, 2009; Clark et al., 1991) and are commonly associated with provoking inflammatory and immune responses. The developmental arrest observed in cPLA2 morphants suggests that cPLA2 activity and eicosanoid signaling are essential for early embryonic patterning, pointing to a novel role for lipid signaling during embryogenesis.
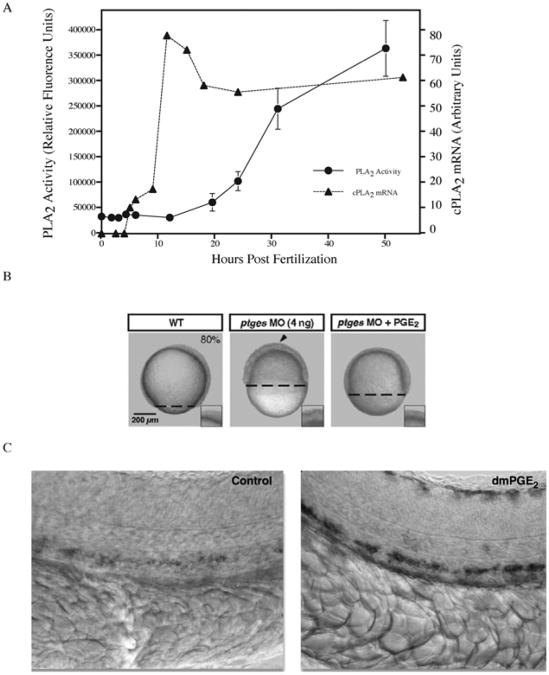
Fig. 3.
Phospholipase activity is required for proper zebrafish development. (A) Cytosolic phospholipase A2 (cPLA2) mRNA levels and PLA2 enzymatic activity were quantified from whole embryo lysates during early stages of zebrafish development. cPLA2 expression peaks during somitogenesis (10 h) while enzymatic activity steadily increases as development proceeds. PLA2 activity is required to generate prostaglandins. (B) Blocking prostaglandin production during early zebrafish development through inhibition of prostaglandin–endoperoxide synthase (Ptgs1) via morpholino knockdown results in developmental arrest at epiboly. Developmental arrest can be rescued by adding back the exogenous enzyme product (PGE2). Reproduced with permission by Development (Speirs et al., 2010). (C) Embryos exposed to the PGE2 analog (16,16-dimethyl-PGE2; dmPGE2) exhibit increased expression of runx11 and cmyb1 as evidenced by in situ hybridization. These genes are expressed in the ventral wall of the dorsal aorta in a region analogous to the mammalian aorta–gonad–mesonephros and are required for mammalian hematopoietic stem cell development. Reprinted by permission from Macmillan Publishers Ltd.: Nature (North et al., 2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–11, copyright 2007.
Arachidonic acid can act as a substrate for a number of other lipid modifying enzymes (e.g., cyclooxygenase) that are critical for cell movements required to pattern the early zebrafish embryo. Cha et al. (2006) found that PGE2, an eicosanoid downstream of cyclooxygenase, is essential for the morphogenic movements of convergence and extension (Cha et al., 2006; Cha et al., 2005). Moreover, the arrest of epiboly that results from the inhibition of cyclooxygenase 1 (also known as Prostaglandin–endoperoxide synthases, Ptgs1) using an antisense morpholino oligonucleotide can be completely rescued by the addition of PGE2 (Speirs et al., 2010) (Fig. 3B).
The importance of PGE2 during zebrafish development was further demonstrated in a small molecule screen performed in zebrafish larvae (36 hpf) by North et al. (2007). Using this approach PGE2 was found to impact hematopoietic stem cell proliferation as evidenced by a dramatic increase in the expression of hematopoietic markers in larvae treated with a PGE2 analog (North et al., 2007) (Fig. 3C). Taken together, these data provide evidence for the importance of PLA2-derived lipid mediators during zebrafish development.
A. Lipid Modifications Influence Primordial Germ Cell Migration
In addition to uncovering a role for lipid signaling during the early movements of gastrulation, work on the zebrafish has shown that lipid modifications influence another critical cellular movement during early embryonic development: primordial germ cell (PGC) migration. PGC migration in zebrafish embryos can be visualized in embryos as early as 80% epiboly by injecting in vitro transcribed, capped GFP-nanos mRNA (which consists of the coding sequence of GFP fused to the 3′UTR of the nanos gene) into zebrafish embryos at the one-cell stage. GFP-nanos message and protein are stabilized preferentially in PGCs such that they maintain their fluorescence throughout early development, facilitating the detailed study of PGC migratory behavior (Doitsidou et al., 2002).
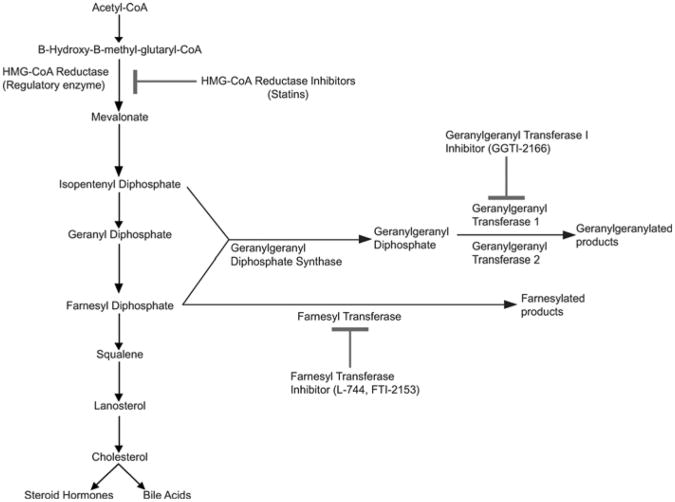
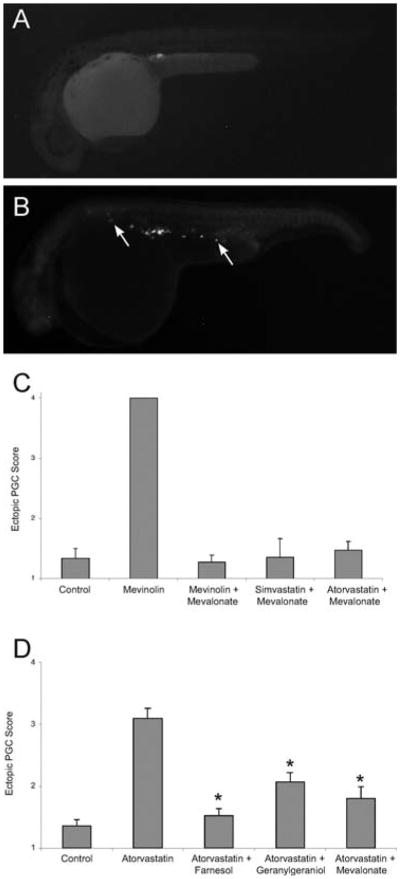
Because lipid metabolism is highly conserved across vertebrates, human drugs can be used to block particular steps in metabolic pathways to determine what pathway or metabolites are necessary for a developmental process, such as PGC migration. In Drosophila, loss of 3-hydroxyl-3-methylglutaryl-CoA reductase (HMGCoAR) results in PGC migration defects (Van Doren et al., 1998). Similarly, studies in zebrafish have shown that pharmacologic inhibition of HMGCoAR by atorvastatin (Lipitor) results in abnormal development and PGC migration (Thorpe et al., 2004). HMGCoAR is a rate-limiting step in the synthesis of cholesterol and is the target of statins (Fig. 4). Zebrafish embryos soaked in statins, either mevinolin (Lovastatin) or simvastatin (Zocor), exhibit developmental arrest, blunted axis elongation, misshapen somites, and head and axial necrosis (Fig. 5). Additionally, PGC migration is profoundly perturbed. Embryos treated with a more hydrophobic statin (Lipitor) exhibit PGC migration defects and only mild morphologic abnormalities.
Fig. 4.
Cholesterol biosynthesis and protein lipidation are highly conserved in vertebrates. Due to the high genetic conservation of these pathways, human drugs can be used to block specific biochemical steps to determine the roles that cholesterol and protein lipidation play in zebrafish development. Reprinted from Development Cell with permission from Elsevier (Thorpe et al., 2004).
Fig. 5.
Embryos treated with statins exhibit developmental defects. In comparison to untreated embryos (A), embryos soaked in a low dose of mevinolin (0.06 μM). (B) exhibit mild developmental defects, as evidenced by tail kinks. (C) Exposure to higher doses of mevinolin (1.2 μM) results in blunted axis elongation, necrosis and developmental arrest. (D) Simvastatin treated embryos (2.0 μM) exhibit similar developmental defects. (E) Dose response of mevinolin on developmental arrest (mean + SEM, n = 3). (F) Dose response of simvastatin on developmental arrest (mean + SEM, n = 3). Reprinted from Development Cell with permission from Elsevier (Thorpe et al., 2004).
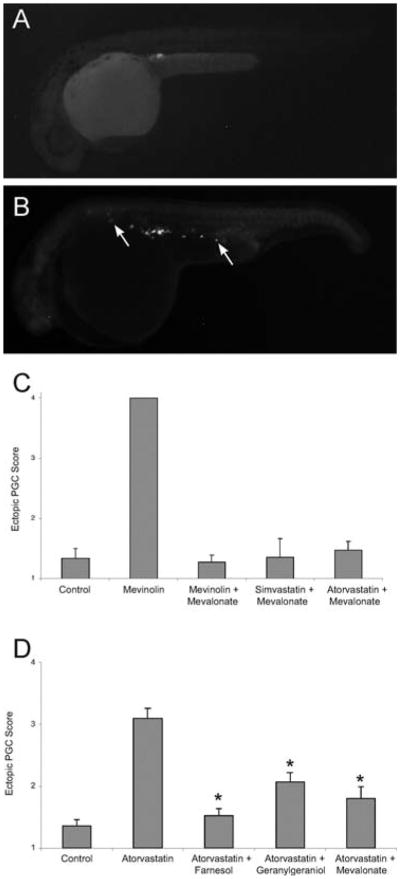
To determine which downstream products of HMGCoAR mediate these developmental defects, a “block and rescue” approach was taken through injection of putative biochemical pathway intermediates downstream of HMGCoAR following statin-mediated inhibition. Injection of mevalonate, the product of HMGCoAR's reduction of ß-hydroxy-ß-methylglutaryl-CoA, completely rescued all the phenotypes associated with the statin treatment (Fig. 6). Similar experiments using intermediates downstream of mevalonate (Fig. 7) indicated that the prenylation pathway, responsible for adding polyunsaturated lipids to proteins, was likely mediating the effect of statins on PGC migration. High doses of the selective farnesyl transferase inhibitors L-744 or FTI-2153 (Crespo et al., 2001; Sun et al., 1999) had no effect on PGC migration. However, embryos treated with geranylgeranyl transferase I (GGTI) inhibitor (GGTI-2166) exhibited a strong PGC migration phenotype with only a slight disruption of the notochord. These data suggest that protein prenylation, specifically by GGTI, is required for correct PGC migration (Thorpe et al., 2004).
Fig. 6.
Isoprenoid intermediates rescue the developmental defects caused by statin treatment. Embryos at early cell stages were injected with mevalonate and then soaked overnight in mevinolin, simvastatin, or atorvastatin. (A) Embryos treated with mevinolin show severe developmental defects at 24 hpf. (B) Embryos injected with the isoprenoid intermediate mevalonate (1–16 cell stages) and then treated with mevinolin exhibit normal morphology. (C) Mevalonate injection rescues the somatic defects observed in embryos treated with different statins. Embryo morphology was scored at 24 hpf. Data represent the MEAN ± SEM from 3–4 experiments. Reprinted from Development Cell with permission from Elsevier (Thorpe et al., 2004).
Fig. 7.
Statin treatment causes abnormal primordial germ cell (PGC) migration in zebrafish embryos. Compared to wild-type (A), embryos treated with statins (B) display ectopic PGCs. The arrows indicate ectopic PGCs that have failed to migrate to the developing gonad. (C) The PGC migratory defect observed following statin treatment is prevented by injections of isoprenoid intermediates. Embryos at early stages (1–16 cell) were injected with gfp-nos mRNA and mevalonate and then soaked overnight in mevinolin, simvastatin or atorvastatin. At 24 hpf, embryos were scored for ectopic PGCs, with a score of 1 indicating a wild-type single gonadal cluster and score of 4 indicating no discernable PGC cluster. Data represent the mean ± SEM from 3–4 experiments. (D) The PGC migratory defect observed following statin treatment is prevented by increasing the levels of isoprenoid synthesis intermediates. Embryos injected at the 1–16 cell stage with farnesol, geranylgeraniol or mevalonate and then soaked overnight with atorvastatin (10 μM) show normal PGC migration. Data represents MEAN ± SEM, *p < 0.01 difference from atorvastatin alone. Reprinted from Development Cell with permission from Elsevier (Thorpe et al., 2004).
V. Visualizing Lipid Metabolism in Larval and Adult Zebrafish
A. Lipophilic Dyes
Lipophilic dyes, known as lysochromes, are one of the first tools used to visualize lipids in cells. These dyes are capable of labeling a variety of lipids and lipidcontaining structures including TAG, FA, and lipoproteins. Dyes, such as oil red O (ORO), sudan black B, and nile red, were initially used to label LDs in tissue sections and cultured cells and continue to be used today. Marza and colleagues (Marza et al., 2005) utilized sudan black B to identify LDs in histological sections of fed adult zebrafish. The authors found that feeding a high-fat meal increased the expression of MTP in intestinal epithelial cells. This protein is required for proper assembly and secretion of hepatic and intestinal ApoB-containing lipoproteins, chylomicrons, and very low-density lipoproteins (VLDL) (Gordon et al., 1995). Their observation that LDs are coincident with an upregulation of MTP expression is consistent with MTP's known function in humans (Marza et al., 2005).
Lysochromes can also be used to visualize endogenous lipid stores in whole fixed zebrafish to generate an overall picture of neutral lipid localization during development. Schlegel and Stainier used ORO to assess the consequence of MTP knockdown (via MO) on lipid absorption in whole zebrafish larvae (Schlegel and Stainier, 2006). MTP morphants exhibited decreased yolk consumption and an inability to absorb dietary neutral lipids, resulting in death by 6 dpf Although lysochromes such as sudan black B and ORO consistently label neutral lipids in tissue sections and fixed larvae, fixation techniques are laborious and staining procedures have been shown to cause artificial fusion of adjacent LDs and mislocalization of the LD marker, adipose differentiation-related protein (Adrp/Perilipin2) (Fukumoto and Fujimoto, 2002). More recent techniques to visualize LDs have focused on staining these drops in vivo.
Greenspan et al. (1985) first showed the utility of nile red (9-diethylamino-5H-benzo[a]phenoxazine-5-one) to label intracellular LD in live cultured peritoneal macrophages and smooth muscle cells. Nile red is an uncharged heterocyclic molecule that only fluoresces in a hydrophobic environment. Labeled neutral lipids fluoresce a yellow-gold to red color, depending on their relative hydrophobicity, with no detectable damage or deformation of dye-infused tissues (Fowler and Greenspan, 1985). More recently, Jones et al. (2008) used nile red to visualize neutral lipid deposits in live zebrafish larvae. They initially demonstrated that daily exposure of larvae to nile red-containing embryo media for 4 days (from 3 to 7 dpf) consistently labeled lipid-rich tissues. The authors then sought to test the effects of known pharmacological inhibitors of triglyceride metabolism on total larval lipid content. Treatment with nicotinic acid, a potent pharmacological inhibitor of adipocyte lipolysis (Carlson, 1963), resulted in an increase in total triglyceride content and decreased cholesterol levels. Treatment with resveratrol, a compound known to inhibit FA synthase (Tian, 2006), resulted in a decrease in total triglyceride content as detected by nile red staining. Total triglyceride content was further decreased when resveratrol was supplemented with norepinephrine (Jones et al., 2008).
While fluorescent dyes and stains are useful for identifying lipid deposits in cells and tissues, issues arise regarding the distribution and affinity properties of these compounds. Nonspecific labeling of tissues devoid of lipid deposits may be observed and staining and washing procedures must then be carefully optimized to minimize this effect. Additionally, nile red staining does not distinguish between FA and cholesterol in vivo, although some discrimination based on staining intensity of tissue sections is possible (Fowler and Greenspan, 1985).
B. BODIPY Fatty Acid Analogs
The wide variety of fluorescent lipid analogs commercially available allows one to fully exploit the optical clarity of zebrafish larvae to study lipid metabolism. One type of analog widely used in cultured cells to visualize lipid dynamics is BODIPY-labeled FA. These analogs consist of an acyl chain of variable length attached to the BODIPY (4,4-difluoro-4-bora-3a, 4a-diaza-S-indacene) fluorescent moiety. First synthesized by Treibs and Kreuzer in 1968 (Treibs and Kreuzer, 1969), the BODIPY fluorophore possesses a number of advantageous qualities including high photostability, strong and narrow wavelength emission in the visible spectrum, and an overall uncharged state (Monsma et al., 1989; Pagano et al., 1991).
To administer BODIPY FA analogs to live zebrafish larvae, we developed a feeding assay that utilizes liposomes to create a suspension of relatively hydrophobic FA analogs in embryo media (Carten, unpublished). Following a short liposome feed, digestive organ structure and metabolic function can be assessed, as the fluorescent FAs accumulate readily throughout numerous larval organs and tissues. With this assay, we have observed that different chain length FAs (short, medium, and long) accumulate in distinct patterns throughout digestive organs and tissues, with each chain length suited to visualize particular larval organs and cellular structures. Long chain fatty acids (LCFA) appear in cytoplasmic LD in enterocytes and hepatocytes. Short chain fatty acids (SCFA) accumulate primarily in the hepatic and pancreatic ducts and are particularly suited to illuminate ductal networks. Medium chain fatty acids (MCFA) accumulate in LDs throughout a wide range of cell types, as well as ductal and arterial networks, and reveal the subcellular structures of multiple cell types. Because this feeding assay enables the rapid assessment of digestive function and FA metabolism in live zebrafish larvae, it has the potential for use in genetic and pharmacologic screens to identify genes involved in FA metabolism and potential drug targets.
C. BODIPY Cholesterol
In addition to FA analogs, a number of sterol analogs are available for use with cultured cells and zebrafish larvae (e.g., NBD-cholesterol, BODIPY-cholesterol). Initially created to visualize cholesterol partitioning into membranes, these analogs were found to preferentially enter into liquid-disordered domains, making them less useful for studies of sterol trafficking in cells (Li et al., 2006). To address these limitations a new BODIPY-tagged cholesterol analog was synthesized with a modified fluorophore linker (Li and Bittman, 2007). Recent studies utilizing the improved BODIPY-cholesterol analog found it to partition into the cholesterol-rich liquid-ordered membrane domain (Ariola et al., 2009) and interact with membranes in ways similar to native sterols, making it a powerful new tool for imaging sterol trafficking in live cells (Marks et al., 2008).
Studies done in the zebrafish have found that BODIPY-cholesterol localizes to the yolk of developing zebrafish larvae (Holtta-Vuori et al., 2008). Ongoing work in the Farber lab is examining the localization of BODIPY-cholesterol in zebrafish larval intestinal enterocytes after a high-fat meal and comparing its localization to that of LD (revealed by BODIPY-labeled FA). Recent data suggest that the initial uptake of sterol and of FA segregate into nonoverlapping compartments (Walters, unpublished). These types of experiments will ultimately enable the development of a clearer model for the uptake and trafficking of dietary lipid in intestinal enterocytes.
D. Fluorescent Reporters of Lipid Metabolism
The ability to perform forward genetic studies in zebrafish by mutagenizing the entire genome and screening for particular phenotypes has made this vertebrate model popular (Driever et al., 1996; Haffter et al., 1996). Mutagenesis methods commonly utilized by the zebrafish community include gamma-ray irradiation to generate large deletions and translocations (Fisher et al., 1997), soaking founder fish in mutagenic chemicals such as ethylnitrosourea (ENU) to generate point mutations, (Driever et al., 1996; Haffter et al., 1996), and retrovirus- or transposon-mediated gene insertions (Chen and Farese, 2002; Ivics et al., 2004). We perform an ongoing ENU-mutagenesis screen to search for mutations that perturb lipid processing. To screen for mutants defective in lipid processing, we soak zebrafish larvae in fluorescent lipid reporters that are swallowed and allow lipid processing to be visualized in vivo (Farber et al., 2001).
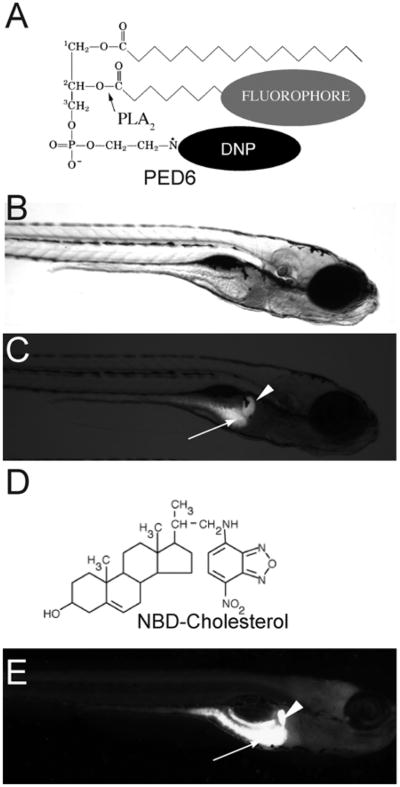
One fluorescent reporter we routinely use in our screen is the phosphoethanolamine analog PED6, [N-((6-(2,4-dinitro-phenyl)amino)hexanoyl)-1-palmitoyl-2-BODIPY-FL-pentanoyl-sn-glycerol-3-phosphoethanolamine] (Fig. 8A). Following ingestion, PED6 is cleaved by phospholipase A2 (PLA2), resulting in the release of the fluorescent labeled acyl chain (Farber et al., 1999) (Fig. 8C). When zebrafish larvae (5 dpf) are immersed in media containing PED6, bright green fluorescence is observed in the intestine, gall bladder, and liver (Fig. 8E). We also utilize the sterol analog 22-NBD-cholesterol (22-[N-(7-nitronbenz-2-oxa-1,3-diazol-4-yl) amino]-23,24-bisnor-5-cholen-3-ol) (Fig. 8B) to visualize cholesterol absorption in live larvae and screen for mutants (Fig. 8F). This reagent is different from PED6 in that it continuously fluoresces and due to its hydrophobicity it is slightly more difficult to administer via feeding.
Fig. 8.
PED6 and NBD-cholesterol visualize lipid uptake in larval zebrafish. (A) The chemical structures of PED6. The BODIPY-labeled acyl chain of PED6 is normally quenched by the dinitrophenyl group at the sn-3 position. Upon PLA2 cleavage at the sn-2 position, the BODIPY-labeled acyl chain is unquenched and can fluoresce. Bright field (B) and fluorescent (C) images of 5 dpf larva following soaking in PED6 for 6 h. PED6 labeling reveals lipid processing in the gall bladder (arrowhead) and intestine (arrow). (D) The chemical structure of NBD-cholesterol. The NBD-cholesterol analog contains a NDB fluorophore where the alkyl tail at the terminal end of cholesterol would normally reside. (E) Soaking zebrafish larvae (5 dpf) in NBD-cholesterol (3 mg/ml, solubilized with fish bile) for 2 h visualizes cholesterol uptake in the gall bladder (arrowhead) and intestine (arrow).
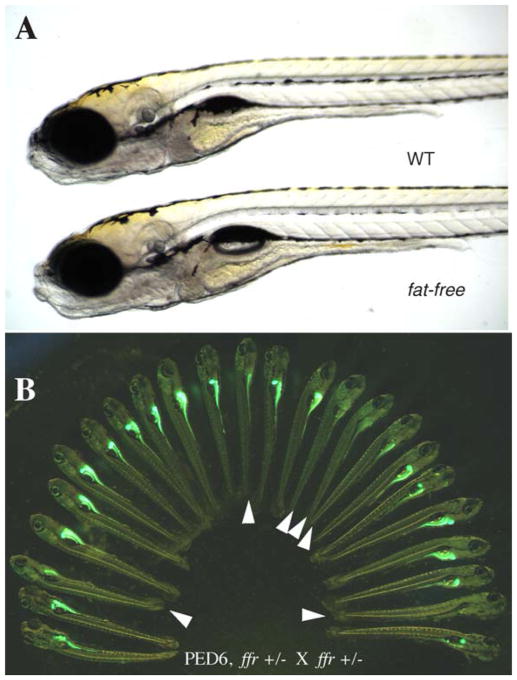
Because PED6 and NBD-cholesterol provide rapid readouts for digestive organ morphology and lipid processing, we used them to perform the first forward genetic physiologic screen in zebrafish to identify new genes that regulate lipid metabolism. One mutation identified with these lipid reporters, fat-free (ffr), was a recessive lethal mutation. Although ffr mutants appeared morphologically normal, they exhibited significantly diminished fluorescence in their intestine and the gall bladder following PED6 treatment (Fig. 9) (Farber et al., 2001). ffr larvae were further characterized using NBD-cholesterol to determine if the uptake and trafficking of sterol-like molecules was also impaired. In contrast to wild-type larvae, which accumulated NBD-cholesterol in their gall bladders within a few hours of feeding, ffr mutants were unable to concentrate NBD-cholesterol in their gall bladders. This observation suggests that ffr larvae have a significant defect in bile secretion and/or transport. When ffr mutants were incubated with BODIPY FL-C5, a medium chain FA analog, they had nearly normal digestive organ fluorescence. Because this medium chain length FA is less hydrophobic and is not dependent on emulsifiers, such as bile, for absorption, this further suggests that the ffr mutation attenuates biliary synthesis or secretion (Ho et al., 2003).
Fig. 9.
PED6 labeling of fat-free(ffr) larvae reveals defective lipid processing. (A) ffr larvae appear morphologically normal in comparison to wild-type siblings (6 dpf). (B). ffr mutants have diminished intestinal and gall bladder fluorescence when labeled with PED6, indicating abnormal lipid processing.
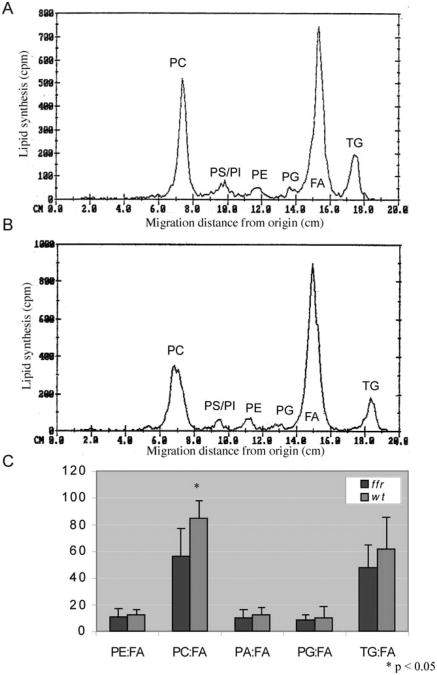
To better characterize the metabolic defects of ffr larvae, we immersed mutants in media containing radioactive oleic acid (250 mCi/mmol, 3 h), followed by whole embryo lipid extraction and TLC analysis. We found that ffr larvae have significantly reduced radioactivity incorporated into phosphatidylcholine (PC) fraction (p<0.05) in comparison to wild-type larvae (Fig. 10). PC is a major component of bile and its overall reduction in ffr mutants is consistent with impaired liver lipid synthesis.
Fig. 10.
ffr larvae have abnormal phospholipid metabolism as shown by thin layer chromatography (TLC). To identify the metabolic defects in ffr larvae, mutants and wild-type larvae (4 dpf) were incubated with radioactive oleic acid (C18:1) for 20 h, followed by total lipid extraction and radioactive TLC analysis. (A, B) Chromatograms generated from scans of TLC plates run with total lipids extracted from wild-type (A) and ffr (B) larvae. They-axis reflects radioactivity counts (cpm; 2–3% of actual activity detected for 3H); the x-axis shows various oleic acid metabolites determined by the migration distance from the start of the TLC plate measured in centimeters (cm). The major metabolites derived from oleic acid (here FA) are PC, phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and TAG. (C) Comparison of relative FA metabolites between ffr and wild-type larvae reveals that ffr mutants have significantly decreased PC production (n = 9, mean ± SD). Reprinted from Methods in Enzymology with permission from Elsevier (Ho et al., 2003).
The positional cloning of ffr indicated that this gene encodes a protein with no known function. Further sequence analysis revealed ffr to be well conserved from invertebrates to mammals. Further studies found ffr to regulate a number of cellular processes including Golgi structure/maintenance, protein sorting, vesicle trafficking, and intestinal lipid absorption and processing (Ho et al., 2006). Recent in vivo and in vitro work suggests that ffr is a novel Arf GTPase family effector that regulates the phospholipase D (PLD) activity. ffr mutants have excess PLD, leading to higher levels of phosphatidic acid (Wang et al., 2006), which is known to increase membrane curvature (Begle et al., 2009). The excessive membrane curvature in ffr mutants may cause abnormal Golgi-vesicle trafficking of nutrient transporters to the cell surface, leading to higher extracellular blood glucose levels and impaired lipid metabolism (Liu et al., 2010).
After identifying the molecular nature of the zebrafish ffr mutation, the human ortholog was located by BLAST searches of public databases. While the human gene was present in these databases, there was no data on its possible function. This example demonstrates the power of forward genetic methods in zebrafish to assign functions to mammalian genes. Other reports of forward genetic screens demonstrate the utility of PED6 and similar synthetic analogs to identify genes involved in digestive processes. For example, Wantanabe et al. used PED6 to identify genes that regulate liver morphogenesis and bile synthesis in Medaka fish (Watanabe et al., 2004).
VI. Triple Screen: Phospholipase, Protease and Swallowing Function Assays
Although PED6 has proven successful in identifying abnormal lipase activity in zebrafish mutants, there can be significant variability in the digestive tract fluorescence observed between wild-type siblings (Fig. 11A). Such variable labeling can make the use of PED6 in genetic screens difficult, as an increase or decrease in fluorescence cannot be attributed solely to mutations and may reflect interindividual differences in reporter ingestion. To address this issue, it was reasoned that PED6 could be used in conjunction with other reporters of digestive function to create a physiologically relevant readout of in vivo digestive processes.
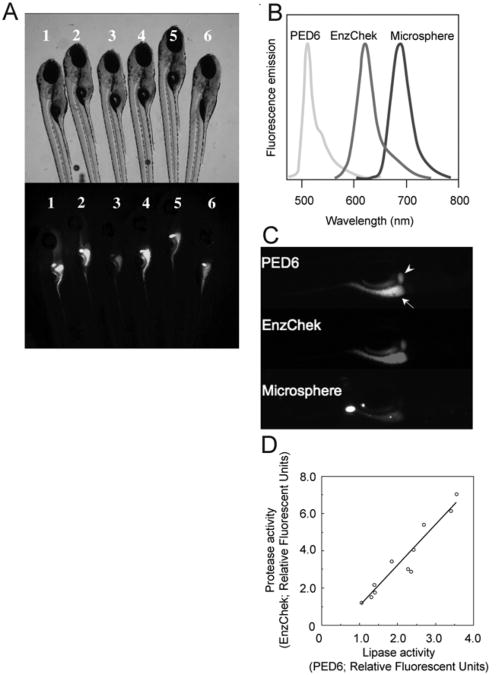
Fig. 11.
Concurrent feeding of PED6, EnzChek, and fluorescent microspheres enables the assessment of larval digestive function. To correct for interindividual variability observed in wild-type siblings (6 dpf) labeled with PED6 (A), multiple reporters of the digestive function can be used. (B) PED6, EnzChek and microspheres each fluoresce at distinct wavelengths, allowing simultaneous screening for lipase and protease activities, and swallowing function, respectively. (C) PED6 and EnzChek signal in the gall bladder (arrowhead) and intestine (arrow) of wild-type zebrafish. Microspheres in the intestine indicate normal ingestion. (D) Intestinal protease and phospholipase activity correlate, validating the use of the ratio of PED6 to EnzChek signal as readout of the digestive function. Figure from Hama et al. (2008), Am. J. Physiol. Gastrointest. Liver Physiol., used with permission from Am. Physiol. Soc.
Hama et al. (Hama et al., 2009) recently demonstrated that concurrent feeding of PED6, EnzChek protease reporter (Invitrogen Inc.), and nonhydrolyzable microspheres allows one to monitor lipase, protease, and swallowing activities, respectively, in larval zebrafish. Since the three reporters fluoresce at distinct wavelengths, simultaneous viewing of all three signals is possible (Figs. 11B and 11C). The EnzChek protease reporter consists of the phosphoprotein casein labeled with multiple red or green BODIPY fluorophores. The proteolytic cleavage of the quenched reporter generates highly fluorescent casein fragments, with total fluorescence proportional to enzyme activity (Jones et al., 1997). EnzChek has been previously used to detect the activity of metallo, serine, acid and thiol proteases in a number of biological systems (Haugland and Kang, 1988; Jones et al., 1997; Menges et al., 1997; Sarment et al., 1999). Unlike PED6 and EnzChek, which report enzymatic function, nonhydrolyzable microspheres assess swallowing and intestinal lumen integrity.
Administering this screening cocktail to larval zebrafish revealed a correlation between the intestinal protease and phospholipase activity, consistent with the hypothesis that the variance observed in PED6 fluorescence was partly due to differing amounts of PED6 consumed by each larva (Fig. 11D). This work demonstrated that the ratio of PED6 to EnzChek fluorescence can serve as a readout of the digestive function, since the ratio in individual larvae is unaffected by differences in reporter ingestion.
After validating the triple screening method, Hama et al. (2009) demonstrated the utility of this assay in evaluating the role of the exocrine pancreas in the digestive function. The exocrine pancreas secretes many of the gastric lipases and proteases needed for the breakdown and subsequent uptake of nutrients (Layer and Keller, 1999). Previous work has shown that morpholino knockdown of the ptf1a transcription factor can selectively prevent exocrine pancreas development (Lin et al., 2004). Analysis of ptf1a mutants (5 dpf) using the triple screen found that these larvae retain normal levels of lipase activity yet have reduced protease activity. Further investigation revealed that there is a marked increase in pancreatic phospholipase A2 (groups IB and III) expression between 5 and 6 dpf, supporting the notion that the exocrine pancreas is not the main source of phospholipase activity in 5 dpf fish. However, 6 dpf ptf1a mutant larvae exhibit decreased amounts of both protease and lipase activity, suggesting the exocrine pancreas begins providing gastric lipases after 5 dpf.
The regulation of phospholipase and protease activity was also examined using the triple screening method. The peptide hormone Cholecystokinin (CCK) facilitates digestion by causing the secretion of gastric enzymes into the small intestine after food consumption (Raybould, 2007). Release of CCK into the circulatory system activates the CCK receptor A (CCK-RA) in the exocrine pancreas. Larvae (5 dpf) treated with CCK-RA antagonist showed a reduction in protease activity but had unaffected lipase activity. Much like the ptf1a mutants, 6 dpf larvae had lower levels of both protease and lipase activity. Not surprisingly, the effect of CCK-RA antagonist was abolished in ptf1a morphants. This work suggests that CCK signaling regulates zebrafish secretion of exocrine pancreas-derived intestinal proteases earlier in development (5 dpf) and phospholipases later (6 dpf).
The utility of employing multiple reagents to screen for the digestive function using larval zebrafish has been demonstrated by others in the field. Recent work from Clifton et al. utilized a multi-tiered approach to identify and describe novel compounds that inhibit dietary lipid absorption (Clifton et al., 2010). In the initial screen, larvae (5 pdf) were incubated in PED6 after compound exposure and assayed for a reduction in gall bladder fluorescence. Results were validated in adult fish and inactive or toxic compounds were removed from the study. Further narrowing was done through the use of fluorescent microspheres to confirm swallowing, as previously described. Absorption of fluorescent cholesterol (NBD-cholesterol) and FA analogs (SCFA BODIPY-C5, LCFA BODIPY-C16) was also assayed to better understand the mechanism of action for seven potentially novel compounds and two known lipid absorption inhibitors, orlistat and ezetimibe. Interestingly, the authors found that ezetimibe was found to reduce metabolism of both PED6 and BODIPY-C16 in addition to NBD-cholesterol. Five of the seven compounds identified in the initial PED6 screen had comparable results. AM1–43, a dye commonly used to visualize endocytosis, and the EnzChk protease assay were used to screen for physiological disruptions, while the lipophilic dye ORO was used to determine whether reduced gall bladder fluorescence was due to reduced lipid processing. AM1–43 staining after ezetimibe treatment was significantly reduced, supporting a disruptive role in endocytosis, perhaps more pronounced than previously described. Those with reduced AM1–43 staining underwent further testing to determine whether cholesterol synthesis was normal: larvae were pretreated with a reagent that extracts membrane cholesterol (methyl-ß-cyclodextran;MßC) and observed for recovery. By using a series of well-established assays, three compounds were identified for testing in mammals and ezetimibe was implicated as playing a larger role in intestinal endocytic dynamics than previously thought. These studies further establish the zebrafish system as a useful and translatable model for testing and prioritizing new compounds for follow-up studies in mammals.
As with any in vivo system, there is inherent variability (developmental timing, intestinal microenvironment), which will result in signal variation. Therefore, caution should be taken to minimize this variability by carefully scoring digestive organ morphology of larvae prior to and after reporter ingestion, discarding sickly larvae that lack swim bladders or exhibit developmental delay, and using properly staged larvae.
VII. Zebrafish Models of Human Dyslipidemias
Recently, an adult zebrafish model for early atheroschlerosis has been developed through administration of a high-cholesterol diet (HCD) (Stoletov et al., 2009). After receiving a 4% cholesterol diet from 5 weeks postfertilization (wpf) to 13–17 wpf, fish developed hypercholesterolemia, as demonstrated by their four-fold plasma total cholesterol levels. No weight gain or increase in TAG was observed. Lipoprotein profiles of HCD-fed fish showed, in addition to the wild-type control HDL fraction, strong fractions for LDL and VLDL. Antibody studies showed that the fish had oxidized plasma lipoproteins, correlating to the development of atherosclerotic lesions. Staining of sections indicated early lesions of atherosclerosis (fatty streaks) while antibody (human L-plastin) staining implicated macrophage infiltration. Live larvae were used to visualize myeloid cell and lipid accumulation, endothelial cell defects, and increased PLA2 activity, signs of early atherosclerosis. This novel model has been used in transplant studies to demonstrate in vivo the requirement of toll-like receptor-4 in macrophage lipid uptake.
The zebrafish has also emerged as a model of glucose metabolism and thus, diabetes, allowing the role lipids play in the onset and pathologies of both subtypes of this disease to be examined. Diabetes was once studied primarily in the context glucose uptake and metabolism; however, the causal effects of dietary lipids on type 2 diabetes are now being examined more closely. Larval zebrafish contain insulin-secreting beta cells that are triggered to release insulin in response to elevated blood glucose and FA levels, much like in humans. Work done by Elo et al. has shown that the exposure of larval and adult zebrafish to high glucose levels results in hyperglycemia (Elo et al., 2007). Furthermore, treatment with the antidiabetic compounds Glipizide and Metformin reduces blood glucose levels in adult zebrafish and lowers the expression of phosphoenolpyruvate carboxykinase, which is regulated by insulin and catalyzes the rate-limiting step of gluconeogenesis, in larval zebrafish (Elo et al., 2007).
VIII. Summary
In this, chapter we have presented novel approaches undertaken using the zebrafish to investigate lipid metabolism and signaling. The optical transparency of zebrafish embryos and larvae can be fully exploited by using transgenic lines, fluorescent reporters, and lipid analogs to visualize metabolic events. Furthermore, the high genetic conservation across lipid signaling and metabolic pathways allows pharmacological regents to be utilized to study the importance of lipids during early development. The high fecundity, small size, and genetic tractability of these organisms make them ideal for high-throughput screening efforts to identify genes involved in lipid metabolism and signaling and thus identify potential therapeutic targets for human diseases.
Despite the immense potential of the zebrafish as a genetic and therapeutic screening tool, these organisms are currently underutilized by the academic and pharmaceutical research communities. While we have discussed some of the exceptions here, many studies use the zebrafish to investigate developmental events such as pattern formation and early neurogenesis, with few taking advantage of the system to study digestive physiology and lipids. We advocate the increased use of zebrafish in metabolic studies and fully expect a continued upward trend in the use of this organism as its contributions to our understanding of human disease become more apparent.
Acknowledgments
The authors would like James Walters and Mike Sepanski of the Carnegie Institution for Science for the EM image of the zebrafish enterocyte. Sections of this chapter contain modified text from our review in Clinical Lipidology: Carten JD, Farber SA: A new model system swims into focus: using the zebrafish to visualize intestinal lipid metabolism in vivo. Clin. Lipidol. 4(4): 501–515 (2009). The article is available at http://www.futuremedicine.com/doi/full/10.2217/clp.09.40.
Abbreviations
- LCFA
long chain fatty acid
- LD
lipid drop
- MCFA
medium chain fatty acid
- MTP
microsomal triglyceride transfer protein
- SCFA
short chain fatty acid
- TAG
triacylglycerol
Footnotes
Financial Disclosure: The authors have no affiliations or financial arrangement with any organization that has a financial interest or stake in the material discussed in this manuscript. The Carnegie Institution does hold a patent together with the University of Penn. on the invention of the author (S.A.F.) that describes the use of fluorescent lipids in zebrafish for high-throughput screening (Pub. No.: US 2009/0136428 A1). The authors have no current consultancies, honoraria, stock ownership or options, expert testimony or royalties regarding the material described. However, the Carnegie Institution and/or U. Penn. can license this technology in the future, potentially providing royalties to the author (S.A.F). Research performed by the authors and described in this manuscript was supported by the Carnegie Institution Endowment and with grants from the US National Institutes of Health (RO1 GM63904 and RO1 DK060369).
References
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Ariola FS, Li Z, Cornejo C, Bittman R, Heikal AA. Membrane fluidity and lipid order in ternary giant unilamellar vesicles using a new bodipy-cholesterol derivative. Biophys J. 2009;96:2696–2708. doi: 10.1016/j.bpj.2008.12.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin PJ, Thisse C, Durliat M, Andre M, Akimenko MA, Thisse B. Both apolipoprotein E and A-I genes are present in a nonmammalian vertebrate and are highly expressed during embryonic development. Proc Natl Acad Sci USA. 1997;94:8622–8627. doi: 10.1073/pnas.94.16.8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begle A, Tryoen-Toth P, de Barry J, Bader MF, Vitale N. ARF6 regulates the synthesis of fusogenic lipids for calcium-regulated exocytosis in neuroendocrine cells. J Biol Chem. 2009;284:4836–4845. doi: 10.1074/jbc.M806894200. [DOI] [PubMed] [Google Scholar]
- Bensadoun A, Rothfeld A. The form of absorption of lipids in the chicken, Gallus domesticus. Proc Soc Exp Biol Med. 1972;141:814–817. doi: 10.3181/00379727-141-36878. [DOI] [PubMed] [Google Scholar]
- Bownes M. Why is there sequence similarity between insect yolk proteins and vertebrate lipases? J Lipid Res. 1992;33:777–790. [PubMed] [Google Scholar]
- Buhman KK, Smith SJ, Stone SJ, Repa JJ, Wong JS, Knapp FF, Jr, Burri BJ, Hamilton RL, Abumrad NA, Farese RV., Jr DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. J Biol Chem. 2002;277:25474–25479. doi: 10.1074/jbc.M202013200. [DOI] [PubMed] [Google Scholar]
- Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res. 2009;50(Suppl):S237–S242. doi: 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA. Studies on the effect of nicotinic acid on catecholamine stimulated lipolysis in adipose tissue in vitro. Acta Med Scand. 1963;173:719–722. doi: 10.1111/j.0954-6820.1963.tb17457.x. [DOI] [PubMed] [Google Scholar]
- Cha YI, Kim SH, Sepich D, Buchanan FG, Solnica-Krezel L, DuBois RN. Cyclooxygenase-1-derived PGE2 promotes cell motility via the G-protein-coupled EP4 receptor during vertebrate gastrulation. Genes Dev. 2006;20:77–86. doi: 10.1101/gad.1374506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha YI, Kim SH, Solnica-Krezel L, Dubois RN. Cyclooxygenase-1 signaling is required for vascular tube formation during development. Dev Biol. 2005;282:274–283. doi: 10.1016/j.ydbio.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Chen HC, Farese RV., Jr Fatty acids, triglycerides, and glucose metabolism: recent insights from knockout mice. Curr Opin Clin Nutr Metab Care. 2002;5:359–363. doi: 10.1097/00075197-200207000-00002. [DOI] [PubMed] [Google Scholar]
- Clark JD, Lin LL, Kriz RW, Ramesha CS, Sultzman LA, Lin AY, Milona N, Knopf JL. A novel arachidonic acid-selective cytosolic PLA2 contains a Ca(2+)-dependent translocation domain with homology to PKC and GAP. Cell. 1991;65:1043–1051. doi: 10.1016/0092-8674(91)90556-e. [DOI] [PubMed] [Google Scholar]
- Clifton JD, Lucumi E, Myers MC, Napper A, Hama K, Farber SA, Smith AB, IIIrd, Huryn DM, Diamond SL, Pack M. Identification of novel inhibitors of dietary lipid absorption using zebrafish. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo NC, Ohkanda J, Yen TJ, Hamilton AD, Sebti SM. The farnesyltransferase inhibitor, FTI-2153, blocks bipolar spindle formation and chromosome alignment and causes prometaphase accumulation during mitosis of human lung cancer cells. J Biol Chem. 2001;276:16161–16167. doi: 10.1074/jbc.M006213200. [DOI] [PubMed] [Google Scholar]
- Doitsidou M, Reichman-Fried M, Stebler J, Koprunner M, Dorries J, Meyer D, Esguerra CV, Leung T, Raz E. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Elo B, Villano CM, Govorko D, White LA. Larval zebrafish as a model for glucose metabolism: expression of phosphoenolpyruvate carboxykinase as a marker for exposure to anti-diabetic compounds. J Mol Endocrinol. 2007;38:433–440. doi: 10.1677/JME-06-0037. [DOI] [PubMed] [Google Scholar]
- Elshourbagy NA, Boguski MS, Liao WS, Jefferson LS, Gordon JI, Taylor JM. Expression of rat apolipoprotein A-IV and A-I genes: mRNA induction during development and in response to glucocorticoids and insulin. Proc Natl Acad Sci USA. 1985;82:8242–8246. doi: 10.1073/pnas.82.23.8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzler L, Smith V, Lin JS, Olcott HS. The lipids of Mono Lake, California, brine shrimp (Artemia salina) J Agric Food Chem. 1974;22:330–331. doi: 10.1021/jf60192a017. [DOI] [PubMed] [Google Scholar]
- Esni F, Ghosh B, Biankin AV, Lin JW, Albert MA, Yu X, MacDonald RJ, Civin CI, Real FX, Pack MA, Ball DW, Leach SD. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- Farber SA, Olson ES, Clark JD, Halpern ME. Characterization of Ca2+-dependent phospholipase A2 activity during zebrafish embryogenesis. J Biol Chem. 1999;274:19338–19346. doi: 10.1074/jbc.274.27.19338. [DOI] [PubMed] [Google Scholar]
- Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, Hendrickson HS, Hendrickson EK, Halpern ME. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- Farese RV, Jr, Veniant MM, Cham CM, Flynn LM, Pierotti V, Loring JF, Traber M, Ruland S, Stokowski RS, Huszar D, Young SG. Phenotypic analysis of mice expressing exclusively apolipoprotein B48 or apolipoprotein B100. Proc Natl Acad Sci USA. 1996;93:6393–6398. doi: 10.1073/pnas.93.13.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field FJ. Regulation of intestinal cholesterol metabolism. In: Mansbach CM, Tso P, Kuksis A, editors. Intestinal Lipid Metabolism. Kluwer Academic; New York: 2001. pp. 235–255. [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II Pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Fisher S, Amacher SL, Halpern ME. Loss of cerebum function ventralizes the zebrafish embryo. Development. 1997;124:1301–1311. doi: 10.1242/dev.124.7.1301. [DOI] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- Fowler SD, Greenspan P. Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem. 1985;33:833–836. doi: 10.1177/33.8.4020099. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Fujimoto T. Deformation of lipid droplets in fixed samples. Histochem Cell Biol. 2002;118:423–428. doi: 10.1007/s00418-002-0462-7. [DOI] [PubMed] [Google Scholar]
- Gordon DA, Wetterau JR, Gregg RE. Microsomal triglyceride transfer protein: a protein complex required for the assembly of lipoprotein particles. Trends Cell Biol. 1995;5:317–321. doi: 10.1016/s0962-8924(00)89054-6. [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin H, Grant G, Perry M. Hydrolysis of plasma triacylglycerol-rich lipoproteins from immature and laying hens (Gallus domesticus) by lipoprotein lipase in vitro. Biochem J. 1982;206:647–654. doi: 10.1042/bj2060647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosser T, Yusuff S, Cheskis E, Pack MA, FitzGerald GA. Developmental expression of functional cyclooxygenases in zebrafish. Proc Natl Acad Sci USA. 2002;99:8418–8423. doi: 10.1073/pnas.112217799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hald J, Hjorth JP, German MS, Madsen OD, Serup P, Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- Hama K, Provost E, Baranowski TC, Rubinstein AL, Anderson JL, Leach SD, Farber SA. In vivo imaging of zebrafish digestive organ function using multiple quenched fluorescent reporters. Am J Physiol Gastrointest Liver Physiol. 2009;296:G445–G453. doi: 10.1152/ajpgi.90513.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland RP, Kang HC. U.S. Patent. 4,774,339 1988
- Heasman J. Morpholino oligos: making sense of antisense? Dev Biol. 2002;243:209–214. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- Ho SY, Lorent K, Pack M, Farber SA. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 2006;3:289–300. doi: 10.1016/j.cmet.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY, Pack M, Farber SA. Analysis of small molecule metabolism in zebrafish. Methods Enzymol. 2003;364:408–426. doi: 10.1016/s0076-6879(03)64023-1. [DOI] [PubMed] [Google Scholar]
- Holtta-Vuori M, Uronen RL, Repakova J, Salonen E, Vattulainen I, Panula P, Li Z, Bittman R, Ikonen E. BODIPY-cholesterol: a new tool to visualize sterol trafficking in living cells and organisms. Traffic. 2008;9:1839–1849. doi: 10.1111/j.1600-0854.2008.00801.x. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Kaufman CD, Zayed H, Miskey C, Walisko O, Izsvak Z. The Sleeping Beauty transposable element: evolution, regulation and genetic applications. Curr Issues Mol Biol. 2004;6:43–55. [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Joffe BI, Panz VR, Raal FJ. From lipodystrophy syndromes to diabetes mellitus. Lancet. 2001;357:1379–1381. doi: 10.1016/S0140-6736(00)04616-X. [DOI] [PubMed] [Google Scholar]
- Jones KS, Alimov AP, Rilo HL, Jandacek RJ, Woollett LA, Penberthy WT. A high throughput live transparent animal bioassay to identify non-toxic small molecules orgenes that regulate vertebrate fat metabolism for obesity drug development. Nutr Metab (Lond) 2008;5:23. doi: 10.1186/1743-7075-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Upson RH, Haugland RP, Panchuk-Voloshina N, Zhou M, Haugland RP. Quenched BODIPY dye-labeled casein substrates for the assay of protease activity by direct fluorescence measurement. Anal Biochem. 1997;251:144–152. doi: 10.1006/abio.1997.2259. [DOI] [PubMed] [Google Scholar]
- Jong MC, Hofker MH, Havekes LM. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol. 1999;19:472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- Karlen S, Rebagliati M. A morpholino phenocopy of the cyclops mutation. Genesis. 2001;30:126–128. doi: 10.1002/gene.1046. [DOI] [PubMed] [Google Scholar]
- Klett EL, Patel SB. Biomedicine. Will the real cholesterol transporter please stand up. Science. 2004;303:1149–1150. doi: 10.1126/science.1095519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruit JK, Groen AK, van Berkel TJ, Kuipers F. Emerging roles of the intestine in control of cholesterol metabolism. World J Gastroenterol. 2006;12:6429–6439. doi: 10.3748/wjg.v12.i40.6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer P, Keller J. Pancreatic enzymes: secretion and luminal nutrient digestion in health and disease. J Clin Gastroenterol. 1999;28:3–10. doi: 10.1097/00004836-199901000-00002. [DOI] [PubMed] [Google Scholar]
- Levy E, Spahis S, Sinnett D, Peretti N, Maupas-Schwalm F, Delvin E, Lambert M, Lavoie MA. Intestinal cholesterol transport proteins: an update and beyond. Curr Opin Lipidol. 2007;18:310–318. doi: 10.1097/MOL.0b013e32813fa2e2. [DOI] [PubMed] [Google Scholar]
- Li Z, Bittman R. Synthesis and spectral properties of cholesterol- and FTY720-containing boron dipyrromethene dyes. J Org Chem. 2007;72:8376–8382. doi: 10.1021/jo701475q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Mintzer E, Bittman R. First synthesis of free cholesterol-BODIPY conjugates. J Org Chem. 2006;71:1718–1721. doi: 10.1021/jo052029x. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, Leach SD. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;270:474–486. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- Liu HY, Lee N, Tsai TY, Ho SY. Zebrafish fat-free, a novel Arf effector, regulates phospholipase D to mediate lipid and glucose metabolism. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbalip.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Marks DL, Bittman R, Pagano RE. Use of Bodipy-labeled sphingolipid and cholesterol analogs to examine membrane microdomains in cells. Histochem Cell Biol. 2008;130:819–832. doi: 10.1007/s00418-008-0509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB, Kochhar S, Lindon JC, Holmes E, Nicholson JK. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marza E, Barthe C, Andre M, Villeneuve L, Helou C, Babin PJ. Developmental expression and nutritional regulation of a zebrafish gene homologous to mammalian microsomal triglyceride transfer protein large subunit. Dev Dyn. 2005;232:506–518. doi: 10.1002/dvdy.20251. [DOI] [PubMed] [Google Scholar]
- McNeely MJ, Edwards KL, Marcovina SM, Brunzell JD, Motulsky AG, Austin MA. Lipoprotein and apolipoprotein abnormalities in familial combined hyperlipidemia: a 20-year prospective study. Atherosclerosis. 2001;159:471–481. doi: 10.1016/s0021-9150(01)00528-7. [DOI] [PubMed] [Google Scholar]
- Menges DA, Ternullo DL, Tan-Wilson AL, Gal S. Continuous assay of proteases using a microtiter plate fluorescence reader. Anal Biochem. 1997;254:144–147. doi: 10.1006/abio.1997.2408. [DOI] [PubMed] [Google Scholar]
- Misra A, Khurana L. Obesity and the metabolic syndrome in developing countries. J Clin Endocrinol Metab. 2008;93:S9–30. doi: 10.1210/jc.2008-1595. [DOI] [PubMed] [Google Scholar]
- Monsma FJ, Jr, Barton AC, Kang HC, Brassard DL, Haugland RP, Sibley DR. Characterization of novel fluorescent ligands with high affinity for D1 and D2 dopaminergic receptors. J Neurochem. 1989;52:1641–1644. doi: 10.1111/j.1471-4159.1989.tb09220.x. [DOI] [PubMed] [Google Scholar]
- Moschetta A, Xu F, Hagey LR, van Berge-Henegouwen GP, van Erpecum KJ, Brouwers JF, Cohen JC, Bierman M, Hobbs HH, Steinbach JH, Hofmann AF. A phylogenetic survey of biliary lipids in vertebrates. J Lipid Res. 2005;46:2221–2232. doi: 10.1194/jlr.M500178-JLR200. [DOI] [PubMed] [Google Scholar]
- Munoz G, Donghi S, Cerisola H. Vitellogenesis in the crayfish Rhynchocinetes typus: role of hepatopancreas in lipid yolk biosynthesis. Cell Mol Biol. 1990;36:531–536. [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nassir F, Wilson B, Han X, Gross RW, Abumrad NA. CD36 is important for fatty acid and cholesterol uptake by the proximal but not distal intestine. J Biol Chem. 2007;282:19493–19501. doi: 10.1074/jbc.M703330200. [DOI] [PubMed] [Google Scholar]
- Nickerson JG, Alkhateeb H, Benton CR, Lally J, Nickerson J, Han XX, Wilson MH, Jain SS, Snook LA, Glatz JF, Chabowski A, Luiken JJ, Bonen A. Greater transport efficiencies of the membrane fatty acid transporters FAT/CD36 and FATP4 compared with FABPpm and FATP1 and differential effects on fatty acid esterification and oxidation in rat skeletal muscle. J Biol Chem. 2009;284:16522–16530. doi: 10.1074/jbc.M109.004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, Orkin SH, Zon LI. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Curtin LR, Lamb MM, Flegal KM. Prevalence of high body mass index in US children and adolescents, 2007–2008. JAMA. 2010;303:242–249. doi: 10.1001/jama.2009.2012. [DOI] [PubMed] [Google Scholar]
- Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, Stemple DL, Driever W, Fishman MC. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- Pagano RE, Martin OC, Kang HC, Haugland RP. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol. 1991;113:1267–1279. doi: 10.1083/jcb.113.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart MA, Klee EW, Nielsen AL, Sivasubbu S, Mendenhall EM, Bill BR, Chen E, Eckfeldt CE, Knowlton M, Robu ME, Larson JD, Deng Y, Schimmenti LA, Ellis LB, Verfaillie CM, Hammerschmidt M, Farber SA, Ekker SC. Genome-wide reverse genetics framework to identify novel functions of the vertebrate secretome. PLoS ONE. 2006;1:e104. doi: 10.1371/journal.pone.0000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plonne D, Stacke A, Weber KU, Endisch U, Dargel R. The pattern of apolipoprotein B100 containing lipoprotein subclasses produced by the isolated visceral rat yolk sac depends on developmental stage and fatty acid availability. Biochim Biophys Acta. 1996;1299:54–66. doi: 10.1016/0005-2760(95)00189-1. [DOI] [PubMed] [Google Scholar]
- Raabe M, Kim E, Veniant M, Nielsen LB, Young SG. Using genetically engineered mice to understand apolipoprotein-B deficiency syndromes in humans. Proc Assoc Am Physicians. 1998;110:521–530. [PubMed] [Google Scholar]
- Raybould HE. Mechanisms of CCK signaling from gut to brain. Curr Opin Pharmacol. 2007;7:570–574. doi: 10.1016/j.coph.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Mead JF. Lipid absorption and deposition in rainbow trout (Salmo gairdnerii) Can J Biochem. 1973;51:1050–1058. doi: 10.1139/o73-137. [DOI] [PubMed] [Google Scholar]
- Sarment DP, Korostoff J, D'Angelo M, Polson AM, Feldman RS, Billings PC. In situ localization and characterization of active proteases in chronically inflamed and healthy human gingival tissues. J Periodontol. 1999;70:1303–1312. doi: 10.1902/jop.1999.70.11.1303. [DOI] [PubMed] [Google Scholar]
- Schlegel A, Stainier DY. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry. 2006;45:15179–15187. doi: 10.1021/bi0619268. [DOI] [PubMed] [Google Scholar]
- Sharma MK, Denovan-Wright EM, Degrave A, Thisse C, Thisse B, Wright JM. Sequence, linkage mapping and early developmental expression of the intestinal-type fatty acid-binding protein gene (fabp2) from zebrafish (Danio rerio) Comp Biochem Physiol B Biochem Mol Biol. 2004;138:391–398. doi: 10.1016/j.cbpc.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Shim J, Moulson CL, Newberry EP, Lin MH, Xie Y, Kennedy SM, Miner JH, Davidson NO. Fatty acid transport protein 4 is dispensable for intestinal lipid absorption in mice. J Lipid Res. 2009;50:491–500. doi: 10.1194/jlr.M800400-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire MF, Lutton C, Vernier JM. New views on intestinal absorption of lipids in teleostean fishes: an ultrastructural and biochemical study in the rainbow trout. J Lipid Res. 1981;22:81–94. [PubMed] [Google Scholar]
- Skinner ER, Rogie A. The isolation and partial characterization of the serum lipoproteins and apolipoproteins of the rainbow trout. Biochem J. 1978;173:507–520. doi: 10.1042/bj1730507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs CK, Jernigan KK, Kim SH, Cha YI, Lin F, Sepich DS, DuBois RN, Lee E, Solnica-Krezel L. Prostaglandin Gbetagamma signaling stimulates gastrulation movements by limiting cell adhesion through Snai1a stabilization. Development. 2010;137:1327–1337. doi: 10.1242/dev.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence R, Gerlach G, Lawrence C, Smith C. The behaviour and ecology of the zebrafish Danio rerio. Biol Rev. 2008;83:13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- Stahl A, Hirsch DJ, Gimeno RE, Punreddy S, Ge P, Watson N, Patel S, Kotler M, Raimondi A, Tartaglia LA, Lodish HF. Identification of the major intestinal fatty acid transport protein. Mol Cell. 1999;4:299–308. doi: 10.1016/s1097-2765(00)80332-9. [DOI] [PubMed] [Google Scholar]
- Stoletov K, Fang L, Choi SH, Hartvigsen K, Hansen LF, Hall C, Pattison J, Juliano J, Miller ER, Almazan F, Crosier P, Witztum JL, Klemke RL, Miller YI. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ Res. 2009;104:952–960. doi: 10.1161/CIRCRESAHA.108.189803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SJ, Myers HM, Watkins SM, Brown BE, Feingold KR, Elias PM, Farese RV., Jr Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J Biol Chem. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- Sun J, Blaskovich MA, Knowles D, Qian Y, Ohkanda J, Bailey RD, Hamilton AD, Sebti SM. Antitumor efficacy of a novel class of non-thiol-containing peptidomimetic inhibitors of farnesyltransferase and geranylgeranyltransferase I: combination therapy with the cytotoxic agents cisplatin, Taxol, and gemcitabine. Cancer Res. 1999;59:4919–4926. [PubMed] [Google Scholar]
- Thisse B, Thisse C. Fast release clones: a high throughput expression analysis. ZFIN Direct Data Submission. 2004 ( http://zfin.org)
- Thisse C, Thisse B. High throughput expression analysis of ZF-Models consortium clones. ZFIN Direct Data Submission. 2005 ( http://zfin.org)
- Thomson AB, Keelan M, Cheng T, Clandinin MT. Delayed effects of early nutrition with cholesterol plus saturated or polyunsaturated fatty acids on intestinal morphology and transport function in the rat. Biochim Biophys Acta. 1993;1170:80–91. doi: 10.1016/0005-2760(93)90178-c. [DOI] [PubMed] [Google Scholar]
- Thorpe JL, Doitsidou M, Ho SY, Raz E, Farber SA. Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation. Dev Cell. 2004;6:295–302. doi: 10.1016/s1534-5807(04)00032-2. [DOI] [PubMed] [Google Scholar]
- Tian WX. Inhibition of fatty acid synthase by polyphenols. Curr Med Chem. 2006;13:967–977. doi: 10.2174/092986706776361012. [DOI] [PubMed] [Google Scholar]
- Titus E, Ahearn GA. Vertebrate gastrointestinal fermentation: transport mechanisms for volatile fatty acids. Am J Physiol. 1992;262:R547–R553. doi: 10.1152/ajpregu.1992.262.4.R547. [DOI] [PubMed] [Google Scholar]
- Treibs A, Kreuzer F. Difluorboryl-Komplexe von Di- und Tripyrrylmethenen. Justus Liebigs Ann Chem. 1969;721:116–120. [Google Scholar]
- Tso P, Fujimoto K. The absorption and transport of lipids by the small intestine. Brain Res Bull. 1991;27:477–482. doi: 10.1016/0361-9230(91)90145-a. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtishak KA, Choob M, Tian X, Sternheim N, Talbot WS, Wickstrom E, Farber SA. Targeted gene knockdown in zebrafish using negatively charged peptide nucleic acid mimics. Dev Dyn. 2003;228:405–413. doi: 10.1002/dvdy.10394. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Broihier HT, Moore LA, Lehmann R. HMG-CoA reductase guides migrating primordial germ cells. Nature. 1998;396:466–469. doi: 10.1038/24871. comment. [DOI] [PubMed] [Google Scholar]
- Van Heek M, Farley C, Compton DS, Hoos L, Alton KB, Sybertz EJ, Davis HR., Jr Comparison of the activity and disposition of the novel cholesterol absorption inhibitor, SCH58235, and its glucuronide, SCH60663. Br J Pharmacol. 2000;129:1748–1754. doi: 10.1038/sj.bjp.0703235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heek M, France CF, Compton DS, McLeod RL, Yumibe NP, Alton KB, Sybertz EJ, Davis HR., Jr Invivo metabolism-based discovery of a potent cholesterol absorption inhibitor, SCH58235, in the rat and rhesus monkey through the identification of the active metabolites of SCH48461. J Pharmacol Exp Ther. 1997;283:157–163. [PubMed] [Google Scholar]
- Verkade HJ, Tso P. Biophysics of Intestinal Luminal Lipids. In: Mansbach CM, Tso P, Kuksis A, editors. Intestinal Lipid Metabolism. Kluwer Academic; New York: 2001. pp. 1–14. [Google Scholar]
- Wallace K, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Yusuff S, Sonntag JM, Chin AJ, Pack M. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis. 2001;30:141–143. doi: 10.1002/gene.1050. [DOI] [PubMed] [Google Scholar]
- Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45:250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Yaginuma R, Ikejima K, Miyazaki A. Liver diseases and metabolic syndrome. J Gastroenterol. 2008;43:509–518. doi: 10.1007/s00535-008-2193-6. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Asaka S, Kitagawa D, Saito K, Kurashige R, Sasado T, Morinaga C, Suwa H, Niwa K, Henrich T, Hirose Y, Yasuoka A, Yoda H, Deguchi T, Iwanami N, Kunimatsu S, Osakada M, Loosli F, Quiring R, Carl M, Grabher C, Winkler S, Del Bene F, Wittbrodt J, Abe K, Takahama Y, Takahashi K, Katada T, Nishina H, Kondoh H, Furutani-Seiki M. Mutations affecting liver development and function in Medaka, Oryzias latipes, screened by multiple criteria. Mech Dev. 2004;121:791–802. doi: 10.1016/j.mod.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Yee NS, Lorent K, Pack M. Exocrine pancreas development in zebrafish. Dev Biol. 2005;284:84–101. doi: 10.1016/j.ydbio.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, Peers B, Bortolussi M, Argenton F. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268:174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]