Abstract
PIK3CA encodes the p110α subunit of the mitogenic signaling protein phosphatidylinositol 3-kinase (PI3K). PIK3CA mutations in the helical binding domain and the catalytic subunit of the protein have been associated with tumorigenesis and treatment resistance in various malignancies. Characteristics of patients with PIK3CA-mutant lung adenocarcinomas have not been reported.
We examined EGFR, KRAS, BRAF, HER2, PIK3CA, AKT1, NRAS, MEK1, and ALK in patients with adenocarcinoma of the lung to identify driver mutations. Clinical data were obtained from the medical records of individuals with mutations in PIK3CA.
Twenty-three of 1125 (2%, 95% confidence interval (CI) 1–3%) patients had a mutation in PIK3CA, 12 in Exon 9 (10 E545K, 2 E542K) and 11 in Exon 20 (3 H1047L, 8 H1047R). The patients (57% women) had a median age of 66 at diagnosis (range 34–78). Eight patients (35%) were never smokers. Sixteen of 23 (70%, 95% CI 49 – 86%) had coexisting mutations in other oncogenes - 10 KRAS, 1 MEK1, 1 BRAF, 1 ALK rearrangement, and 3 EGFR exon 19 deletions.
We conclude that PIK3CA mutations occur in lung adenocarcinomas, usually concurrently with EGFR, KRAS, and ALK. The impact of PIK3CA mutations on the efficacy of targeted therapies such as erlotinib and crizotinib is unknown. Given the high frequency of overlapping mutations, comprehensive genotyping should be performed on tumor specimens from patients enrolling on clinical trials of PI3K and other targeted therapies.
Keywords: lung adenocarcinoma, oncogene, PIK3CA
Introduction
The identification and targeting of specific oncogenic driver-mutations has revolutionized the treatment of lung adenocarcinoma. While mutations in KRAS were identified decades ago and remain a target of investigation, the first therapeutic advance in lung cancer was the discovery of mutations in the epidermal growth factor receptor (EGFR) gene in patients who had experienced dramatic benefit from treatment with EGFR tyrosine kinase inhibitors(1–3). Since then, a number of other oncogenic driver-mutations (missense mutations, insertions and deletions) have been identified in BRAF, PIK3CA, and HER2. In addition, the discovery of the ALK fusion protein in lung cancer(4, 5) led to the rapid identification of the ALK inhibitor crizotinib revealing a 61% overall response rate in patients with ALK rearrangements(6, 7) and leading to expedited approval by the US Food and Drug Administration. This early experience has bolstered the growing enthusiasm for our ability to target therapy for an individual based on the presence of a specific driver mutation in their tumor specimen.
The characteristics of EGFR-mutant and ALK-rearranged lung cancer have been well described(4, 8). Recent reports have summarized the characteristics of BRAF(9) and HER2-mutant populations(10). However, the patients with tumors harboring mutations in PIK3CA have not been characterized.
PIK3CA encodes the p110α subunit of phosphatidylinositol 3-kinase (PI3K), an integral signaling molecule in the pathways driven by growth factor receptors such as HER/ERBB2. Activated PI3K phosphorylates AKT and leads to downstream activation of mTOR (mammalian target of rapamycin) which is essential for cell survival and proliferation. Since activating mutations and over-expression of PI3K are known to be oncogenic, the PI3K pathway has been intensively studied(11).
Multiple mutations have been identified in PIK3CA, the oncogene that encodes the p110α subunit of PI3K(12). The mutations that occur with regularity and in highly conserved regions of the gene lead to amino acid substitutions in the helical binding domain encoded by Exon 9 (E542K, E545K) and in the catalytic subunit of p110α encoded by Exon 20 (H1047R or L). Mutations in the helical binding domain interfere with p85 binding and allow activation of PI3K. The mutations in the catalytic subunit are thought to increase kinase activity(13). These specific mutations have been shown to be sufficient for tumorigenesis both in vivo and in vitro(14, 15).
The rate of PIK3CA mutations reported in NSCLC is estimated from 1–4%(16–19). In the United States alone, this represents 9,000 patients per year who may benefit from a therapy targeting the PI3K pathway. Interestingly, unlike other oncogenic driver mutations in lung adenocarcinoma which are rarely found in squamous cell carcinoma, PIK3CA has been reported to be amplified and mutated in squamous cell carcinoma as well as adenocarcinoma of the lung(20). Preliminary data in the genotyping of squamous cell carcinoma performed at our institution confirms the occurrence of PIK3CA mutations in 2% of squamous lung cancer(21).
As part of the ongoing Memorial Sloan-Kettering Lung Cancer Mutation Analysis Project, we have routinely tested for PIK3CA mutations in patients with adenocarcinoma of the lung since 2009(22). Given the prevalence of PIK3CA mutations in other diseases, multiple drugs targeting PI3K and AKT/m-TOR are in development, including trials targeting PIK3CA mutations in lung cancers, we evaluated their clinical and molecular characteristics to learn more about this patient population.
Methods
Between January 2009 and June 2010, all patients evaluated by the thoracic medical oncology and surgery services were offered participation in an institutional tissue analysis program entitled the Lung Cancer Molecular Analysis Project. In patients with sufficient tissue, assessment for driver mutations was performed in 9 genes: EGFR, KRAS, BRAF, HER2, PIK3CA, AKT1, NRAS, MEK1, and ALK. EGFR exon 19 deletions were identified through a PCR-based assay(23). EGFR exon 20 and 21 mutations, as well as activating mutations in KRAS, BRAF, HER2, PIK3CA, AKT1, NRAS, and MEK1, were assessed using a Mass spectrometry-based nucleic acid assay using the Sequenom™ platform. The platform was designed to include mutations in these genes that have been reported as activating. All detected mutations were confirmed by direct sequencing. These methods have been previously described(24). Rearrangements involving ALK were determined by the ALK breakpoint fluorescence in situ hybridization assay (Vysis LSI ALK Dual Color).
Clinical characteristics were obtained from the medical record. Smoking definitions are as follows: never (<100 cigarettes lifetime), current (active smoker within the past year), former (>100 cigarettes lifetime and no tobacco use within the last year). Pathological stage was determined at the time of surgery according to the AJCC, 7th edition TNM staging system(25). Binary correlative variables were evaluated with the Fisher exact test, continuous variables were evaluated with the Wilcoxon signed-rank test. Overall Survival (OS) was calculated among patients diagnosed with stage IIIB/IV lung adenocarcinoma using the Kaplan-Meier method. Disease free survival is calculated from the date of surgery. Patients were followed from the date of diagnosis of Stage IIIB/IV disease until death or the last available follow-up. Group comparison was performed with the log-rank test. All research was performed under appropriate institutional review board/privacy board protocols and waivers.
Results
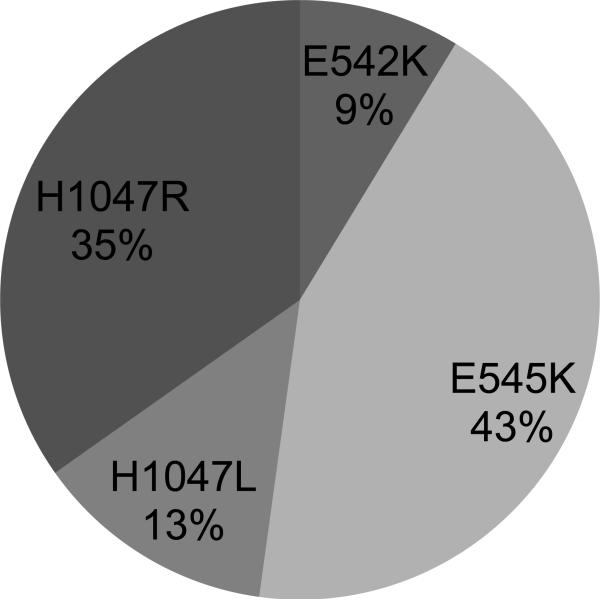
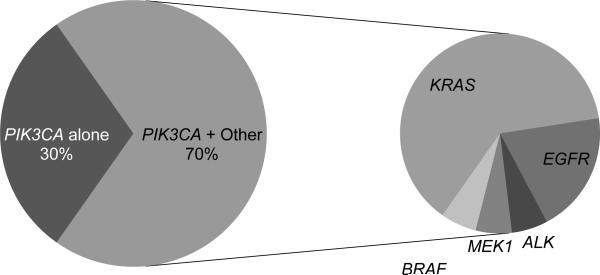
Twenty-three of 1125 (2%, 95% CI 1–3%) patients had a mutation in PIK3CA (10 E545K, 2 E542K, 3 H1047L, 8 H1047R) (Figure 1). There were no mutations identified in R88, N345, C420, and M1043. The clinical characteristics of the PIK3CA-mutant patients are presented in Table 1. Sixteen of 23 (70%, 95% CI 49–86%) had coexisting mutations in other oncogenes - 10 KRAS, 1 MEK1, 1 BRAF, 1 ALK rearrangement, and 3 EGFR exon 19 deletions (Figure 2). This is 1% (95% CI, <1–4%) of the 260 EGFR-mutant cases, 3% (95% CI, <1–16%) of the 34 ALK-rearranged cases, and 3% (95% CI, 1–5%) of the 355 KRAS-mutant cases.
Figure 1.
PIK3CA mutations in lung adenocarcinoma
Table 1.
Clinical Characteristics
| Characteristic (N=23) | PIK3CA-positive N=23 | PIK3CA-negative N=1102 | p-value |
|---|---|---|---|
|
| |||
| Age – Median (Range) | 66 (34–78) | 66 (24–96) | 0.99 |
|
| |||
| Sex – Female (%) | 13 (57%) | 602 | 0.83 |
|
| |||
| Smoking History | |||
| Never | 8 (35%) | 280 (25%) | 0.34 |
| Ever-smoker | |||
| --Former | 10 (43%) | 668 (61%) | |
| --Current | 5 (22%) | 154 (14%) | |
|
| |||
| Stage | |||
| Early (IA–IIIA) | 9 (39%) | 644 (58%) | 0.2 |
| Advanced (IIIB/IV) | 14 (61%) | 458 (42%) | |
Figure 2.
Coexisting mutations in patients with PIK3CA-mutant lung adenocarcinoma
Patients had a median follow-up of 13 months (range 3 – 60 months). Of the 9 patients with early stage disease, 5 received neoadjuvant chemotherapy and all underwent a complete resection. Four of the 5 patients treated with neoadjuvant chemotherapy received a cisplatin or carboplatin-based doublet. The other patient had the co-mutation BRAF V600E and had a marked treatment response to neoadjuvant gefitinib on a clinical trial(26). Five patients have had disease recurrence and 3 have died of disease. The mutation data, treatment course, pathological stage and survival of the patients are presented in Table 2.
Table 2.
Molecular and Clinical Characteristics
| PIK3CA | Other | Age | Sex | Tobacco Use | Pack Yrs | Stage | Treatment Neoadjuvant or 1st line | Surv |
|---|---|---|---|---|---|---|---|---|
| E542K | KRAS | 61 | F | Current | 30 | IIB | Cisplatin + Docetaxel | 17+ |
| E542K | BRAF | 78 | F | Former | 30 | IA | Gefitinib | 15 |
| E545K | KRAS | 77 | F | Former | 20 | IB | Carbo + Docetaxel | 3 |
| E545K | - | 70 | F | Never | 0 | IIA | - | 5+ |
| E545K | KRAS | 65 | F | Current | 45 | IIIA | - | 7 |
| E545K | - | 74 | F | Former | 40 | IIIB | Carbo + Pacli + RT | 27 |
| E545K | - | 50 | M | Current | 70 | IV | Carbo + Pacli | 21 |
| E545K | EGFR | 57 | M | Current | 50 | IV | Carbo + Peme | 11 |
| E545K | KRAS | 74 | F | Former | 50 | IIIB | RT | 10 |
| E545K | KRAS | 65 | F | Current | 40 | IV | Carbo + Peme | 6 |
| E545K | KRAS | 71 | M | Former | 55 | IV | Cisplatin + Doce + B | 16 |
| E545K | KRAS | 64 | F | Former | 15 | IV | Unknown | 13 |
| H1047L | KRAS | 66 | M | Former | 30 | IA | - | 3+ |
| H1047L | - | 34 | F | Former | 10 | IV | Cisplatin + Peme | 31 |
| H1047L | EGFR | 61 | M | Never | 0 | IV | Erlotinib | 21* |
| H1047R | - | 71 | M | Former | 45 | IA | Carbo + Pacli + RT | 1+ |
| H1047R | - | 68 | F | Never | 0 | IA | - | 2+ |
| H1047R | KRAS | 38 | M | Never | 0 | IIA | Cisplatin+Doce+B | 4 |
| H1047R | EGFR | 57 | F | Never | 0 | IV | Erlotinib | 12* |
| H1047R | MEK1 | 69 | M | Former | 100 | IV | Peme + Pacli + B | 6* |
| H1047R | KRAS | 76 | M | Never | 0 | IV | Carbo + Peme | 9 |
| H1047R | - | 64 | F | Never | 0 | IV | Peme + Pacli + B | 23* |
| H1047R | ALK | 73 | F | Never | 0 | IV | Erlotinib | 23 |
Abbreviations: Surv (survival – disease free in early stage patients (I–IIIA) and overall in advanced stage patients (IIIB/IV)), Pacli (paclitaxel), Carbo (carboplatin), B (bevacizumab), Peme (pemetrexed), Doce (docetaxel) RT (radiotherapy), + (disease free)
alive with disease
The median survival of the 14 patients with Stage IIIB and IV disease was 21 months. In this small group of patients there was a shorter median survival in patients with a coexisting mutation (EGFR, KRAS, BRAF, ALK) versus those with mutations in PIK3CA alone, median 13 versus 27 months (p=0.03). Three patients had EGFR Exon 19 deletions. One patient treated with 1st-line erlotinib had a prolonged radiographic partial response and then developed T790M-mediated resistance after 15 months on erlotinib. The other patient treated in the 1st-line had a partial radiographic response of 5 months duration and did not undergo a repeat biopsy at the time of progression. The 3rd EGFR-mutant patient did not respond to 2nd line erlotinib; he had no evidence of T790M. The re-biopsy samples were not tested for persistence or loss of the PIK3CA mutation. The patient with an ALK rearrangement was initially treated with erlotinib with upfront progression and subsequently progressed again on docetaxel before being treated with crizotinib with stable disease as the best response. The remainder of patients were treated with standard first-line treatments (Table 2).
There was no difference in the stage or the frequency of coexisting mutations between patients with mutations in the PIK3CA kinase versus helical domain. Mutations in the kinase domain of PIK3CA occurred with higher frequency in patients who were never smokers (p=0.009) (Table 3).
Table 3.
Comparison of PIK3CA Helical (Exon 9) and Kinase (Exon 20) Domain Mutations.
| Characteristic | Helical N (%) | Kinase N (%) | p-value |
|---|---|---|---|
|
| |||
| Stage | |||
| Early (IA–IIIA) | 5 (22) | 4 (18) | 1.0 |
| Advanced (IIIB/IV) | 7 (30) | 7 (30) | |
|
| |||
| Co-mutation | |||
| Yes | 9 (39) | 7 (30) | 0.67 |
| No | 3 (13) | 4 (18) | |
|
| |||
| Smoking | |||
| Never | 1 (4) | 7 (30) | 0.009 |
| Former/Current | 11 (48) | 4 (18) | |
Discussion
Consistent with the published literature, we have confirmed that PIK3CA mutations occur in ~2% of patients with lung adenocarcinoma. While single cases of adenocarcinoma harboring PIK3CA mutations and co-mutations have been previously reported(27–30), we found that the majority of tumors with PIK3CA mutations had another driver mutation as well (Figure 2). This is in contrast to the mutual exclusivity of driver oncogene mutations seen in adenocarcinoma of the lung harboring EGFR, KRAS and ALK, raising the possibility of tumor heterogeneity, though the high frequency of co-exiting mutations, lack of two evident primary tumors microscopically and the data regarding PIK3CA mutation in other diseases(31–33) make the explanation of tumor heterogeneity unlikely. While 2 cases of PIK3CA mutation acquisition have been reported after the development of acquired resistance to erlotinib in EGFR-mutant lung cancer(34), in this series the mutations were present before treatment with targeted therapies. Additional data are required to characterize the effect of concurrent PIK3CA mutations on responses to erlotinib and crizotinib in patients harboring EGFR mutations and ALK rearrangements.
Mutation detection in this study makes use of a mass-spectrometry based system (Sequenom). This assay was designed to detect known `hotspot' mutations in specific oncogenes based on the published literature and available databases. Some mutations are confirmed by direct Sanger sequencing but full sequencing is not performed on all specimens. While this directed approach may miss unknown point mutations that would be detected by direct sequencing, it is optimal for detecting recurrent oncogene mutations that are known to be activating and minimizes the identification of new mutations of unclear significance. Our panel detects >90% of the PI3KCA mutations reported in all histologies of lung cancer in the COSMIC database.
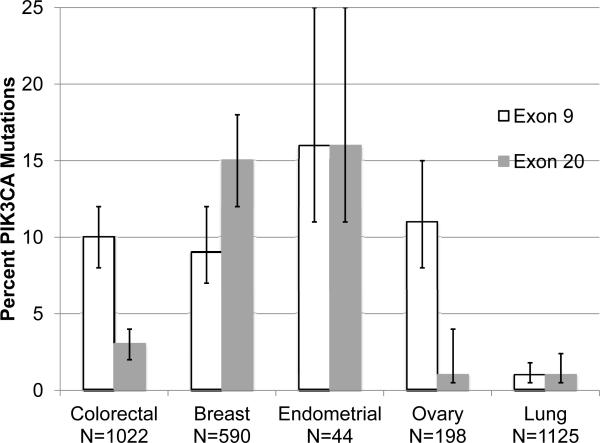
Beyond lung cancer, PIK3CA mutations have been identified in breast, ovarian, endometrial, and colorectal carcinomas. While the frequency of these mutations has been defined (Figure 3), the influence of PIK3CA mutations on pathogenesis, prognosis, and response to therapy is not uniform across disease types or between studies. An example of this is the effect of PIK3CA mutation status on the efficacy of cetuximab in metastatic colorectal cancer. While Prennen, et al found no effect of PIK3CA mutation on response to cetuximab with or without irinotecan(35), DeRoock reported an inferior response rate to cetuximab plus chemotherapy in patients harboring mutations in PIK3CA exon 20 but not the more commonly mutated exon 9(32). In breast cancer, response to trastuzumab in PIK3CA-mutant HER2+ breast cancer cell lines is inferior to PIK3CA wild-type HER2+ cell lines only with coincident PTEN loss (36) but not with intact PTEN(37) The co-existence of PIK3CA-mutations with mutations in KRAS, NRAS and BRAF has been demonstrated in colorectal adenocarcinoma(32) and with KRAS in pancreatic adenocarcinoma and ovarian carcinoma(31). Initial reports in endometrial carcinoma claimed that mutations in PIK3CA and KRAS were mutually exclusive(38), but more recent studies have found both KRAS mutations and PTEN loss in patients with PIK3CA mutant endometrial carcinoma(33). In the absence of comprehensive mutational profiling, this heterogeneity between diseases and mutational profiles, may explain the seemingly contradictory clinical outcomes described above.
Figure 3.
PIK3CA Mutation Incidence with 95% Confidence intervals in Various Malignancies Colorectal(32), Breast(39), Endometrial(38), Ovary(45), and Lung Adenocarcinoma
Independent of treatment efficacy, mutations in distinct domains of PIK3CA may impart unique biologies. Studies in breast cancer have found clinical differences between patients with mutations in the helical and kinase domains with fewer lymph node metastases in individuals with mutations in the kinase domain(39) and inferior overall survival in those with mutations in the helical domain(40). These observations are further supported by findings in soft tissue sarcoma, where downstream activation of AKT is higher in tumors with helical domain mutations than those with kinase domain mutations(41). Our cohort of PIK3CA-mutant lung cancer patients has too many coexisting mutations to allow for comparison of outcomes between domain-specific mutation populations, although we can conclude that the never smokers were more likely to have mutations in the kinase domain. Interestingly, similar to the findings in TP53-mutant and KRAS-mutant adenocarcinoma of the lung, where never smokers were more likely to harbor transition mutations (substitution purine for purine, or pyrimidine for pyrimidine) and not transversion mutations (substitution pyrimidine for purine, or purine for pyrimidine) (42, 43), the PIK3CA kinase domain mutations, more commonly identified in never smokers, are transition and not transversion mutations.
Interestingly, in this small sample of patients with PIK3CA-mutant advanced disease, the presence of a co-existing oncogene mutation was correlated with an inferior outcome. Only one patient with a PIK3CA mutation received an experimental agent targeting the PI3K pathway, therefore we cannot base the above average survival in this arm on effective targeted therapies. While this sample size is too small to draw any conclusions, the survival findings are thought provoking.
Despite the uncertain effect of PIK3CA mutations on prognosis and response to standard therapies, it is an important target for drug development. We and others recommend the testing of agents specifically targeting PIK3CA only in patients with tumors that have evidence of dependency on PI3K pathway (PIK3CA mutation or PTEN loss). Cell lines with PIK3CA mutations are sensitive to downstream inhibitors such as everolimus, an inhibitor of mTOR, although this sensitivity can be abrogated by coincident mutation in KRAS(44). This is an expected yet important observation in light of the high frequency of coincident mutations found in this study. A recent report demonstrated a 7-fold increase in response rate (35% vs. 5%) of PI3K pathway targeted agents in patients with evidence of PIK3CA mutation in tumor specimens(31). These data call into question the utility and appropriateness of testing PI3K pathway targeted agents in patients whose tumors lack evidence of PI3K dependency and accentuates the importance of comprehensive genotyping of tumor specimens in all patients under consideration for molecularly targeted therapies.
Our data indicate that the majority of patients with lung adenocarcinoma harboring mutations in PIK3CA have coexisting mutations in other oncogenes. A shortcoming of this study is that the high throughput system used did not allow for testing of PTEN or TP53 loss; this is a step that we feel is essential moving forward and plan to incorporate into future studies to fully understand the effect of PI3K pathway alterations in lung adenocarcinoma. The timing of acquisition of PIK3CA mutation (and/or PTEN loss) in relation to that of other oncogenes and the contribution to tumor biology and response to therapy is unclear. As agents targeting various pathways including PI3K are in development, comprehensive (and perhaps sequential) mutation profiling should be carried out on tumor specimens from all patients to assess the impact of coincident mutations on the response to the targeted agents.
Acknowledgements
We thank L. Borsu for assistance with Sequenom assays. We thank D. Ang for assistance with Sequenom data review and E. Brzostowski and M. Pilloff for Sequenom data management. The MSKCC Sequenom facility was supported by the Anbinder Fund. The mutation data was obtained from the Sanger Institute Catalogue Of Somatic Mutations In Cancer web site, http://www.sanger.ac.uk/cosmic Bamford et al (2004) The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer, 91,355–358.
Financial Support:
Kris - P01 CA129243, RC2 CA148394
Chaft - Conquer Cancer Foundation of ASCO Young Investigator Award
Abbreviations list
- PIK3CA
phosphoinositide-3-kinase catalytic alpha polypeptide
- PI3K
phosphatidylinositol 3-kinase
- EGFR
epidermal growth factor receptor
- KRAS
Kirsten rate sarcoma viral oncogene homolog
- BRAF
v-Raf murine sarcoma viral oncogene homolog B1
- AKT1
v-akt murine thymoma vial oncogene homolog 1
- NRAS
v-ras neuroblastoma viral oncogene homolog
- MEK1
dual specificity mitogen-activated protein kinase kinase 1
- ALK
anaplastic lymphoma kinase
- CI
confidence interval
- NSCLC
non-small cell lung cancer
- mTOR
mammalian target of rapamycin
- AJCC
American joint commission on cancer
- PCR
polymerase chain reaction
Footnotes
Disclosures: None
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Camidge DR, Bang Y, Kwak EL, Shaw AT, Iafrate AJ, Maki RG, et al. Progression-free survival from a phase I study of crizotinib (PF-02341066) in pathients with ALK-positive non-small cell lung cancer. J Clin Oncol. 2011;29s abstr 2501. [Google Scholar]
- 7.Crino L, Kim D, Riely GJ, Janne PA, Blackhall FH, Camidge DR, et al. Initial phase II results with crizotinib in advanced ALK-positive non-small cell lung cancer (NSCLC): PROFILE 1005. ASCO Meeting Abstracts. 2011;29:7514. [Google Scholar]
- 8.Miller VA, Kris MG, Shah N, Patel J, Azzoli C, Gomez J, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–9. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 9.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, et al. Clinical Characteristics of Patients With Lung Adenocarcinomas Harboring BRAF Mutations. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arcila ME, Chaft JE, Nafa K, Kris MG, Zakowski MF, Ladanyi M. Molecular and clinicopathologic characteristics of HER2-mutant lung adenocarcinoma. J Clin Oncol. 2011;29s abstr 10596. [Google Scholar]
- 11.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 12.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 13.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 14.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–9. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samuels Y, Diaz LA, Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–73. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–4. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 17.Okudela K, Suzuki M, Kageyama S, Bunai T, Nagura K, Igarashi H, et al. PIK3CA mutation and amplification in human lung cancer. Pathol Int. 2007;57:664–71. doi: 10.1111/j.1440-1827.2007.02155.x. [DOI] [PubMed] [Google Scholar]
- 18.Bianconi F, Pistola L, Chiari R, Minotti V, Colella R, Giuffrida D, et al. Phosphoinositide-3-Kinase Catalytic Alpha and KRAS Mutations are Important Predictors of Resistance to Therapy with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients with Advanced Non-small Cell Lung Cancer. J Thorac Oncol. 2011 doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 19.Lee SY, Kim MJ, Jin G, Yoo SS, Park JY, Choi JE, et al. Somatic mutations in epidermal growth factor receptor signaling pathway genes in non-small cell lung cancers. J Thorac Oncol. 2010;5:1734–40. doi: 10.1097/JTO.0b013e3181f0beca. [DOI] [PubMed] [Google Scholar]
- 20.Kawano O, Sasaki H, Okuda K, Yukiue H, Yokoyama T, Yano M, et al. PIK3CA gene amplification in Japanese non-small cell lung cancer. Lung Cancer. 2007;58:159–60. doi: 10.1016/j.lungcan.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Rekhtman N, Paik PK, Tafe L, Riely GJ, Miller VA, Kris MG, et al. Screening for EGFR, KRAS, and PIK3CA mutations in well characterized, immunohistochemically confirmed squamous cell carcinoma of the lung. J Clin Oncol. 2011;29s abstr e21143. [Google Scholar]
- 22.Kris MG, Lau CY, Ang D, Brzostowski E, Riely GJ, Rusch VW, et al. Initial results of LC-MAP: An institutional program to routinely profile tumor specimens for the presence of mutations in targetable pathways in all patients with lung adenocarcinoma. J Clin Oncol. 2010;28:7009. Meeting Abstracts. [Google Scholar]
- 23.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–14. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 26.Rizvi NA, Rusch V, Pao W, Chaft JE, Ladanyi M, Miller VA, et al. Molecular Characteristics Predict Clinical Outcomes: Prospective Trial Correlating Response to the EGFR Tyrosine Kinase Inhibitor Gefitinib with the Presence of Sensitizing Mutations in the Tyrosine Binding Domain of the EGFR Gene. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sequist LV, Heist RS, Shaw AT, Fidias P, Temel JS, Lennes IT, et al. SNaPshot genotyping of non-small cell lung cancers in clinical practice. J Clin Oncol. 2011;29s doi: 10.1093/annonc/mdr489. abstr 7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endoh H, Yatabe Y, Kosaka T, Kuwano H, Mitsudomi T. PTEN and PIK3CA expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol. 2006;1:629–34. [PubMed] [Google Scholar]
- 29.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–21. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludovini V, Bianconi F, Pistola L, Chiari R, Minotti V, Colella R, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2011;6:707–15. doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 31.Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, et al. PIK3CA Mutations in Patients with Advanced Cancers Treated with PI3K/AKT/mTOR Axis Inhibitor. Mol Cancer Ther. 2011 doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 33.Rudd ML, Price JC, Fogoros S, Godwin AK, Sgroi DC, Merino M, et al. A unique spectrum of somatic PIK3CA (p110a) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75–26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prenen H, De Schutter J, Jacobs B, De Roock W, Biesmans B, Claes B, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009;15:3184–8. doi: 10.1158/1078-0432.CCR-08-2961. [DOI] [PubMed] [Google Scholar]
- 36.Kataoka Y, Mukohara T, Shimada H, Saijo N, Hirai M, Minami H. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann Oncol. 2010;21:255–62. doi: 10.1093/annonc/mdp304. [DOI] [PubMed] [Google Scholar]
- 37.Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, Lanchbury JS, et al. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177:1647–56. doi: 10.2353/ajpath.2010.090885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang S, Seo SS, Chang HJ, Yoo CW, Park SY, Dong SM. Mutual exclusiveness between PIK3CA and KRAS mutations in endometrial carcinoma. Int J Gynecol Cancer. 2008;18:1339–43. doi: 10.1111/j.1525-1438.2007.01172.x. [DOI] [PubMed] [Google Scholar]
- 39.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–59. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 40.Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, Del Grammastro M, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–9. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 41.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Taniere P, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076–83. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 43.Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–4. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120:2858–66. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–8. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]