Abstract
The daily rhythm of PERIOD protein (PER) expression is an integral component of the circadian clock, which is found among a broad range of animal species including fruit flies, marine mollusks and even humans. The use of antibodies directed against PER has provided a helpful tool in the discovery of PER homologues and the labeling of putative pacemaker cells, especially in animals for which an annotated genome is not readily available. In this study, DrosophilaPER antibodies were used to probe for PER in the American lobster, Homarus americanus. This species exhibits robust endogenous circadian rhythms but the circadian clock has yet to be located or characterized. PER was detected in the eyestalks of the lobster but not in the brain. Furthermore, a significant effect of the LD cycle on daily PER abundance was identified, and PER was significantly more abundant at mid dark than in early light or mid light hours. Our results suggest that PER is a part of the molecular machinery of the circadian clock located in the eyestalk of the lobster.
Keywords: Lobster, Homarus americanus, biological clock, circadian, PERIOD protein, PER, daily, rhythms, eyestalk
Introduction
The daily coordination of many behavioral and physiological processes to the external environment is a common necessity for nearly all living organisms. Clocks endogenous to the organism are often responsible for controlling this function. One such clock ubiquitous to animals is the circadian clock, which controls biological rhythms on a daily frequency. For instance, the fruit fly Drosophila melanogaster, exhibits a daily, bimodal rhythm of locomotor activity which is synchronized by light and is driven by an endogenous circadian clock (Veleri et al. 2003). In Drosophila and several other invertebrates, the circadian clock is composed of an integral set of proteins including PERIOD (PER), CLOCK (CLK), CYCLE (CYC), and TIMELESS (TIM) (Bell-Pederson et al., 2005; Hardin, 2009). These proteins interact with each other to form a double-negative feedback loop, and their expression is cyclical, with PER and TIM having the highest expression during the night, and CLK and CYC having the highest expression during the day.
PER appears to be conserved as a part of the circadian clock in a wide variety of animals ranging from insects (Hardin et al., 1990) and mollusks (Siwicki et al. 1989) to mammals (Tei et al. 1997). PER has been detected in several other invertebrates besides Drosophila, such as the Giant Silkmoth, Antherea pernyi (Sauman and Reppert 1996), the mollusks Bulla gouldiana and Aplysia californica (Siwicki et al. 1989), and the crayfish, Procambarus clarkii (Arechiga and Rogriguez-Sosa 1998), which suggests that it may be an evolutionarily conserved clock protein among the invertebrates. Its cyclic expression also appears to be conserved in some of these species (Sauman and Reppert 1996; Siwicki et al. 1989)
While PER has been found in many organisms representative of a diverse range of classes, it has not been well studied in many species within those classes. For example, the crayfish is the only known crustacean for which PER has been detected (Arechiga and Rogriguez-Sosa 1998). Another crustacean, the American lobster, Homarus americanus, expresses many physiological and behavioral circadian rhythms including locomotion (Jury et al. 2005), heart rate (Chabot and Webb 2008), neurogenesis (Goergen et al. 2002), and serotonin release (Wildt et al. 2004). All of these processes are thought to be driven by one or more endogenous circadian clocks, but surprisingly, PER has not yet been detected in this species. PER has been found the crayfish (Arechiga and Rogriguez-Sosa 1998), and there is a recent report of its apparent modulation by time of day and the circadian system (Escamilla-Chimal et al., 2010). Based on the prevalence of PER among many different animal species, we hypothesize that PER is also an integral component of the circadian clock in the lobster. Further, because PER is likely a part of the circadian clock, we predict that PER will vary by time of day such that levels will be higher at night versus the day. Because the circadian clock has been localized to either the eye or the brain in several species including another crustacean, the crayfish. P. clarkii (Arechiga and Rogriguez-Sosa 1998) one would predict that either or both of these tissues would exhibit rhythms of PER concentration. Some evidence suggests that in the lobster that the circadian clock resides in the eyestalk (Arechiga et al. 1993; Harzsch et al. 2009). In this study we used Western blotting to detectr PER reactivity in the eyestalk and the brain. We report here significant changes in PER in the eyestalk over time suggesting that PER is indeed part of the circadian clock located in the eyestalk in the American lobster.
Materials and Methods
Animal and Environmental Conditions
Experiment 1: PER Eyestalk and Brain Pilot
Two adult American lobsters (Homarus americanus; appx 470g) were purchased from a local supplier, and placed in 2 aquarium tanks with water kept at 17 °C, pH = 8, and salinity = 31±1 psu. To ensure synchronization of endogenous clocks to our artificial LD cycle, the lobsters were exposed to a 12:12 light/dark (LD) cycle for ten days, and for procedural ease, one lobster was exposed to a normal light/dark cycle in one room and one to a reverse light/dark cycle in a separate room. Locomotor activity was recorded using a “racetrack” technique (Jury et al. 2005), which involved attaching a magnet to the dorsal side of each lobster with epoxy and cyanoacrylate and then placing the lobsters in individual activity chambers (76cm X 31 cm). Three stacked bricks were aligned centrally in the chamber to create an outer “racetrack,” and two magnetic reed switches were placed on opposite lateral sides of the track. When a lobster passed by a reed switch, the magnet attached to the lobster caused switch closure, and this event was sent to a computer for storage and later analysis using ClockLab Collection and Analysis computer program (Actimetrics, Evanston, IL).
Detection of PER Eyestalk and Brain
Prior to tissue sample collection, actograms were generated using the ClockLab software to verify daily activity patterns. After ten days, the two lobsters were dissected under ice anesthesia to remove the brain and eyestalks. Extraction of the brain and eyestalks occurred at two-time points: one at mid-light (ZT 6: N=1) and one at mid-dark (ZT 18: N=1). Both dissections were performed under fluorescent light (25 lux) and took less than 15 minutes. Eyestalks were removed before brain dissection by grasping the external eye firmly with tissue forceps and cutting the stalk with a pair of fine scissors. After dissection, brain and eyestalk tissue samples were immediately hand-homogenized separately for 1 – 2 minutes and cells were lysed into Triton-X 100 protein extraction buffer containing 1% Triton-X 100, 0.01 M Tris and 0.14 M NaCl. The mixture was allowed to incubate for 30 minutes at room temperature, and then centrifuged at 9,500 g for 10 minutes. The supernatant was collected, aliquotted and stored at −80°C. Following protein extraction, the DC Protein Assay (Bio-Rad, Hercules, CA) was used to determine the concentration of total protein in each of the samples.
Western blot. Fifty ug of protein from each sample and an equal volume of 2X Laemmlie sample buffer were loaded into individual wells of 10 % Tris-HCl polyacrylamide gels. Proteins were separated based on size by gel electrophoresis, which ran for 30 minutes at 200 volts, and were then transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA) for 15 hours at 20V. Membranes were blocked in 5% blotting grade milk protein (Bio-Rad, Hercules, CA) with 1X Tris-buffered saline containing 0.1% Tween-20 (TBST buffer) for 30 minutes. They were subsequently washed 5 times for 5 minutes each in TBST. For immunodetection of PER, the membrane was incubated for 3 hours in a dilution of 1:2,000 polyclonal rabbit anti-Drosophila-PER primary antibody (US Biological, Swampscott, MA), washed 3X for 5 minutes in TBST, and followed with a 35-minute incubation in 1:5000 dilution of an anti-rabbit horseradish peroxidase-conjugated (HRP) secondary antibody (Affinity Bioreagants, Golden, CO). After several final washes in TBST, the blots were incubated in a chemiluminscent HRP substrate (Millipore, Billerica, MA) for 3 minutes and then exposed to radiography film (Kodak Eastman Co, Rochester, NY) for 1 minute. The film was developed using developer and fixer photoprocessing chemicals as outlined in manufacturer’s protocol (Kodak Eastman Co, Rochester, NY).
Experiment 2: Daily PER levels in the Eyestalk
A total of 18 American lobsters (Homarus americanus) were purchased from a local supplier (Mean weight– 460 g; males and females). Equal numbers of lobsters were group housed in two separate recirculating tanks and exposed to a 12:12 LD cycle for at least ten days. Nine lobsters were exposed to a normal light/dark cycle and nine to a reverse light/dark cycle, again for procedural ease. All environmental conditions were as described above in the Pilot experiment, but since activity was not monitored, they were allowed to roam freely.
Detection of Daily PER levels in the Eyestalk
Since results from Experiment 1 indicated the presence of PER in the eyestalks but not in the brain (data not shown), only eyestalks were harvested and used for PER detection in this subsequent experiment. After ten days of 12:12 LD exposure, dissection of lobster eyestalks occurred every four hours (n=3) in a 24-hour period, and individual eyestalk extractions took no more than 5 minutes to complete. During the dark phase of the cycle, dissection was conducted under dim red light (590+ nm) in an otherwise dark room. Proteins were extracted and quantified by Western blot using the same procedures described above. However, the following modifications to the protocol were used to optimize the results: 15 ug of protein from each sample was loaded in order to visualize changes in PER protein abundance better over time; a 7.5% Tris-HCl polyacrylamide gel was used for better band migration and separation; and, for immunodetection of PER, the membrane was incubated for 2 hours in a dilution of 1:1000 of the primary antibody, followed by a 45-minute incubation in 1:2500 dilution of a secondary antibody, using the same antibodies as previously. PER was again visualized using the chemilunimenescent detection method as described previously.
Data Analysis
KODAK Molecular Imaging Software (version 4.0, Eastman Kodak Co., Rochester, NY) was used to analyze the Western blots. First, bands of PER were located and the molecular weight was estimated by using either a pre-stained molecular weight ladder (Experiment 1; Bio-Rad, Hercules, CA) or a chemiluminescent-visualized molecular weight ladder (Experiment 2; Bio-Rad, Hercules, CA). PER quantification was also determined using the same program. First, each band detected was given a net intensity value, which was normalized against a Gaussian-fit background value. Next, the bands’ relative intensities were calculated based on the most intense staining band, which was assigned a relative value of 1. For the second experiment, a one-way ANOVA (Excel, Microsoft Corp., Redmond, WA) was used to assess the effects of PER abundance among time-points during a 24 hour day (p<0.05) and a post-hoc Bonferroni/Dunn (p<0.05) analysis was applied to determine if differences existed between means.
Results
Experiment 1: PER Eyestalk and Brain Pilot
Typical locomotor activity records of lobsters in an LD cycle are presented in Figure 1. Note that there was significant daily rhythm as indicated by both visual (Jury et al. 2005; Chabot and Webb 2008) and periodogram analyses (data not shown). Activity was almost wholly confined to the dark portion of the LD cycle. Western blot analysis of PER in brain tissue from these lobsters indicated a lack of dPER binding (Fig. 2). In the eyestalk, the Drosophila-PER (dPER) reacting protein had a mean estimated size of 73 kDa (n=2).
Figure 1.

A. Locomotor activity actograms of lobsters in a 12:12 LD cycle. Open boxes = hours of light. Filled boxes = hours of dark. For better visualization of rhythmic locomotor activity, actograms were double plotted. Movement is represented by black marks. * = time of Eyestalk and Brain tissue collection.
Figure 2.
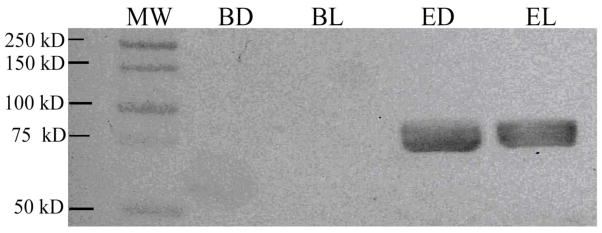
Western blot of PER-reacting protein in brain and eyestalk of lobsters. BD = Brain dissected at mid-Dark. BL = Brain dissected at mid-Light. ED = Eyestalk dissected at mid-Dark. EL = Eyestalk dissected at mid-Light. A molecular weight ladder (MW) is shown on the left-hand side of the blot. ED and EL stained positive for PER (n=1). However, no staining of PER was detected in BL or BD tissue (n=1).
Experiment 2: Daily PER levels in the Eyestalk
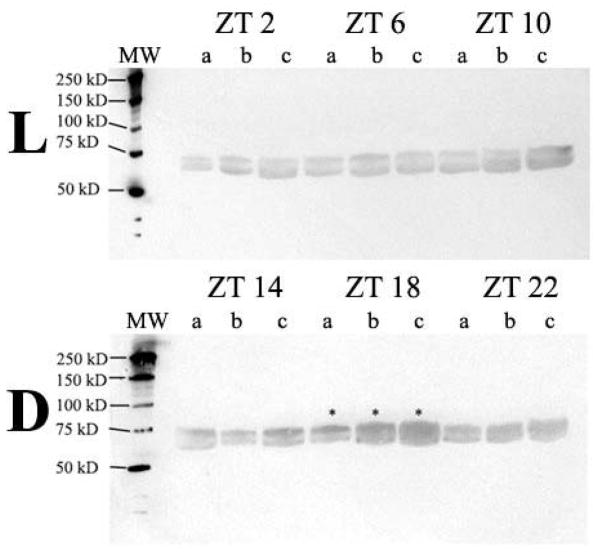
Western blots illustrating bands of a dPER reacting protein detected in lobster eyestalks (n=3) removed every 4-hours in a 24-hour period are presented in Figure 3. In this experiment, the PER-reacting protein had an estimated mean size of 71 kDa. Reactivity to PER was detected at every time-point, and the bands at ZT 18 were noticeably more intense in staining than at all other time-points.
Figure 3.
Western blots of PER-reacting protein in eyestalks removed from lobsters (n=18) over a 24-hour period in LD (Lobster Daily Blot). Collection of eyestalk tissue occurred every 4 hours (n=3, each labeled “a”, “b” and “c”) starting at Zeitgeber Time (ZT – in hours) 2 → 22 (ZT “0”= lights-on, ZT “12”= lights-off). A molecular weight ladder (MW) is shown on the left-hand side of both blots. L – Light; D – Dark.
Quantified PER abundances levels by time of day are presented in Figure 4. A significant effect of the LD cycle on PER abundance was detected (F(5,12) =6.21, P<0.005), with a sharp peak appearing at ZT 18. Bonferroni post-hoc analysis indicated that the ZT 18 peak represents a significantly greater abundant than PER detected in samples at ZT 2 (P<0.002) or ZT 6 (P<0.001).
Figure 4.
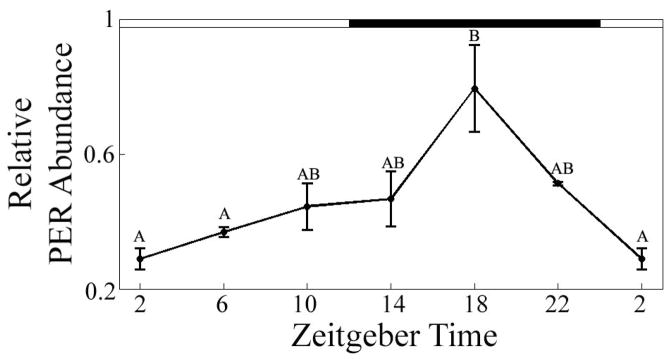
Relative amounts of a DrosophilaPER-reacting protein detected in the eyestalks of lobsters over a 24-hour time period in LD (F(5, 12) = 6.21, p<0.005). Open boxes = hours of light. Filled boxes = hours of dark. Data points represent the relative staining intensity of dPER-reacting bands (mean ± SEM of bands, n=3). Means with different letters above are significantly different (p<0.05).
Discussion
Our results indicate for the first time that a PER-like protein is present in the eyestalks of the American lobster. Although PER has been detected in a wide variety of organisms and appears to be widely conserved among animals as a molecular component of a circadian clock this is only the third report of its presence in a crustacean (crayfish: Arechiga and Rodriguez-Sosa 1998; Daphnia pulex – Tilden et al., 2011). The mechanism of the clock has been especially well described in the fruit fly, Drosophila melanogaster. In this species the clock is driven by negative feedback loops involving PER and TIM (Zheng and Sehgal 2008). Transcription of per and tim is initiated by the binding of CLK-CYC heterodimers to the promoter region of each protein (Taylor and Hardin, 2008). After PER and TIM are translated, these two proteins dimerize with each other and translocate to the nucleus where they down regulate their own transcription via their binding to CLK and CYC. The impact of light on this clock appears to be mediated by cryptochrome (cry) which co-localizes to clock cells (Emery et al. 1998). CRY is activated by light, where-upon it interacts with TIM and so disrupts the PER-TIM heterodimer. This leads to the degradation of both PER and TIM (Busza et al. 2004). While the function or presence of these proteins is generally not known in lobsters, CRY has been detected in the protocerebrum in crayfish (Fanjul-Moles et al. 2004) and apparently cycles by time of day and circadian time (Escamilla-Chimal and Fanjul-Moles 2008). Notably, these four core proteins have been found in the genome of another crustacean, Daphnia along with several “accessory” clock proteins (Tilden et al., 2011). Although there is an EST database for Homarus americanus published on NCBI (www.ncbi.nlm.nih.gov), PER like proteins have not been annotated.
Importantly, we detected a significant effect of the LD cycle on the rhythm of PER expression with nocturnal peaks of PER. Since PER appears to be part of the circadian clock mechanism in all animals in which it is found, significant changes in the concentration of this protein were expected. Similar increases in PER levels were seen in the well-studied model of the Drosophila circadian clock (Bell-Pedersen et al. 2005) An interesting difference is that, in lobsters, the expression peak of PER occurred at midnight, slightly earlier than that characterized in Drosophila, which is most abundant during the late night hours (Zerr et al. 1990) The phasing between locomotion and PER levels is also very different. Lobsters, are generally nocturnally active (Jury et al. 2005) and so exhibit a positive correlation with PER levels while Drosophila are generally diurnal or crepuscular (Grima et al. 2004; Stoleru et al. 2004) and exhibit a negative correlation with PER levels (Rieger et al. 2006). However, there may be many downstream pathways that mediate signaling caused by changes in PER and other clock proteins, and locomotor activity does not appear to be directly coupled to PER abundance in all species. For instance, nocturnal peak abundance of PER is seen in at least one other nocturnal animal, the giant silkmoth (Sauman and Reppert 1996), and additionally, in an animal that does not exhibit robust circadian locomotor rhythms, the German cockroach (Lin et al. 2002). Interestingly, although much is known about the inner-workings of the circadian clock in Drosophila, much less is known about how signals are transmitted from the clock cells to regulate motor circuits (see Taghert and Shafer 2006).
Furthermore, there are several other explanations to suggest why peak PER abundance in the lobster may not be directly correlated to low locomotor activity. First of all, there is a high degree of variability in the locomotor activity patterns of lobsters (Jury et al. 2005); not all lobsters are nocturnal, and some will display diurnal bouts of locomotor activity or even show no preference for either day nor night (Golet et al. 2006). Several other activities thought to be under circadian control do not occur during the night, such as serotonin release, which is highest at pre-dusk (Wildt et al. 2004), and neurogenesis, which is highest at dusk (Goergen et al. 2002). Finally, the bands we observed in the Western blot appear thick and at times, similar to a doublet (Fig.3). Although we did not analyze for phosphorylation, we speculate that the appearance of the wide, doublet-like bands may be representative of PER regulation via phosphorylation, with the differing band sizes representative of different states of phosphorylation. Phosporylation is an important post-translational modification of PER, and in Drosophila, it is known to regulate its accumulation and localization (Price et al. 1998; Suri et al. 2000; Nawathean et al. 2007). Similarly, we suspect that phosphorylation may further serve to regulate PER localization and function in the lobster. Importantly, several proteins known to phosphorylate PER and other clock proteins have been found in the genome of Daphnia (Tilden et al., 2011). In sum, it seems that the expression rhythm we observed in PER shows that it is affected by the daily light/dark cycle, but the way in which PER functions in the coordination and timing output of different downstream behaviors cannot yet be determined.
Our results provide additional evidence that the circadian clock may be located in the eyestalk of the lobster and perhaps crustaceans in general. Previous studies have suggested that in crustaceans, the location of the circadian clock is in the optic peduncle at the base of the eyestalk (Arechiga and Rodriguez-Sosa 1998) and/or the supraesophageal ganglion of the central nerve cord (Thurman 2004) and there is evidence to support both locations. For instance, PER has also been detected in the eyestalk of the crayfish (Arechiga and Rodriguez-Sosa 1998) Additionally, when the supraesophageal ganglion and an intact eyestalk in the crayfish were isolated, the tissues continued to generate circadian rhythms in the electroretinograms (ERG) of the eye, suggesting that the circadian clock was located somewhere within them (Barrera-Mera and Block 1990). Furthermore, pigment dispersing hormone (PDH), which is regulated in the crayfish by a circadian pacemaker and induces changes in the photoreceptor’s sensitivity to light (and hence generates ERG rhythms), is produced in the X organ-sinus gland system, which is located in the optic peduncle at the base of the eyestalk (Verde et al. 2007). Importantly, PDH expression varies in concert with locomotor activity rhythms and has been implicated as a possible coupling agent between the circadian clock and locomotion in two species of crayfish (Sullivan et al., 2009). In Drosophila, this same hormone co-localizes with a subset of circadian clock cells (reviewed by (Taghert and Shafer 2006) and is thought to be an output factor of the circadian clock (Helfrich-Forster 2005). Thus, the above evidence all suggests that the circadian clock is indeed located in the base of the eyestalk. However, another circadian clock protein, CRY, is detected in both the base of the eyestalk and in the brain of the crayfish, and furthermore, it oscillates with a circadian abundance rhythm only in the brain (Fanjul-Moles et al. 2004). Intriguingly, PER was not detected in the brain in our studies. However, it should be noted that this may have been due to experimental procedures rather than an absolute lack of PER. It is possible, that because the brains from both lobsters were dissected under ambient lighting, there may have been time during the procedure for CRY signaling to initiate PER degradation. Additionally, it is possible that PER is expressed in much lower abundance in the brain relative to the eyestalk, and an increase in the amount of brain proteins or concentration of dPER antibodies used would have yielded a PER signal in this tissue. Indeed, PER has been found to be widely distributed in many tissues in some animals (Ivanchenko et al. 2001; Tanoue et al. 2004; Kohsaka and Bass 2007). Future immunohistochemical studies will help to identify PER containing cells and, with labeling of additional known clock proteins such as CRY, will help to identify the clock cells in this species.
Interestingly, the PER-like protein we detected is around 71–73 kDa in size, which is very similar to that reported in crayfish (Escamilla-Chimal et al., 2010) but smaller than published sizes of PER in many other invertebrates, including Drosophila (Bachleitner et al. 2007), the medfly (120kDa; (Mazzotta et al. 2005), the blowfly (110 kDa; (Warman et al. 2000), a marine gastropod, Bulla gouldiana (101 kDa; (Constance CM 2002) the silkmoth (94 kDa; (Reppert et al. 1994), and the German cockroach (132 kDa (Lin et al. 2002) but larger than that of the marine gastropod, Aplysia (48 kDa) (Siwicki et al. 1989), suggesting that a high amount of variability exists in the size of PER homologues from various organisms.
Acknowledgments
The authors would like to thank Luciana Ferraris for help with the Western blotting and Win Watson for many useful conversations on lobster behavior and physiology. We also want to thank the NSF (IOS to CCC and WHW III) and the New Hampshire IDeA Network of Biological Research Excellence (NH-INBRE) with grants from the National Center for Research Resources (5P20RR030360-03) and the National Institute of General Medical Sciences (8P20GM103506-03), National Institutes of Health for support in writing the manuscript.
References
- Arechiga H, Fernandez-Quiroz F, Miguel FF, Rodriguez-Sosa L. The circadian system of crustaceans. Chronobiol Int. 1993;10:1–19. doi: 10.3109/07420529309064477. [DOI] [PubMed] [Google Scholar]
- Arechiga H, Rogriguez-Sosa L. Circadian clock function in isolated eyestalk tissue of crayfish. Proc R Soc Lond. 1998;265:1819–1823. doi: 10.1098/rspb.1998.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachleitner W, Kempinger L, Wulbeck C, Rieger D, Helfrich-Forster C. Moonlight shifts the endogenous clock of Drosophila melanogaster. PNAS. 2007;104:3538–3543. doi: 10.1073/pnas.0606870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Mera B, Block GD. Protocerebral circadian pacemakers in the crayfish: evidence for mutually coupled pacemakers. Brain Res. 1990;522:241–245. doi: 10.1016/0006-8993(90)91467-u. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian Rhythms from Multiple Osculators: Lessons from Diverse Organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the Two Drosophila CRYPTOCHROME Structural Domains in Circadian Photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Chabot CC, Webb LK. Circadian rhythms of heart rate in freely moving and restrained American lobsters, Homarus americanus. Mar Fresh Behav Physiol. 2008;41:1–13. [Google Scholar]
- Constance CMGC, Tei H, Block GD. Bulla gouldiana exhibits unique regualtion at the mRNA and Protein levels. J Biol Rhythms. 2002;17:413–427. doi: 10.1177/074873002237136. [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Escamilla-Chimal EG, Fanjul-Moles ML. Daily and circadian expression of cryptochrome during the ontogeny of crayfish. Comp Biochem Physiol. 2008;151:461–470. doi: 10.1016/j.cbpa.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Escamilla-Chimal EG, Velazquez-Amado RM, Fiordelisio T, Fanjul-Mole ML. Putative pacemakers of crayfish show clock proteins interlocked wth circadian oscillations. J Exp Biol. 2010;213:3723–3733. doi: 10.1242/jeb.047548. [DOI] [PubMed] [Google Scholar]
- Fanjul-Moles ML, Escamilla-Chimal EG, Gloria-Soria A, Hernandez-Herrera G. The crayfish Procambarus clarkii CRY shows daily and circadian variation. J Exp Biol. 2004;207:1453–1460. doi: 10.1242/jeb.00900. [DOI] [PubMed] [Google Scholar]
- Hardin PE. Molecular mechanisms of circadian timekeeping in Drosophila. Sleep Biol Rhythms. 2009;7:235–242. [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Harzsch S, Dircksen H, Beltz BS. Development of pigment-dispersing hormone-immunoreactive neurons in the American lobster: homology to the insect circadian pacemaker system? Cell Tissue Res. 2009;335:417–429. doi: 10.1007/s00441-008-0728-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Neurobiology of the fruit fly’s circadian clock. Genes Brain Behav. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Goergen EM, Bagay L, Rehm K, Beltz B. Circadian Control of Neurogenesis. Journal of Neurobiol. 2002;53:90–95. doi: 10.1002/neu.10095. [DOI] [PubMed] [Google Scholar]
- Golet WJ, Scopel DA, Cooper AB, Watson WH. Daily patterns of locomotion expressed by American lobsters (Homarus americanus) in their natural habitat. J Crust Biol. 2006;26:610–620. [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Ivanchenko M, Stanewsky R, Giebultowicz JM. Circadian photoreception in Drosophila: Functions of cryptochrome in peripheral and central clocks. J Biol Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- Jury SH, Chabot CC, Watson WH., III Daily and circadian rhythms of locomotor activity in the American Lobster, Homarus americanus. J Exp Mar Biol Ecol. 2005;318:61–70. [Google Scholar]
- Kohsaka A, Bass J. A sense of time: how molecular clocks organize metabolism. Trends Endo Metab. 2007;18:4–11. doi: 10.1016/j.tem.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GG, Liou RF, Lee HJ. The period gene of the German cockroach and its novel linking power between vertebrate and invertebrate. Chronobiol Int. 2002;19:1023–1040. doi: 10.1081/cbi-120015961. [DOI] [PubMed] [Google Scholar]
- Mazzotta GM, Sandrelli F, Zordan MA, Mason M, Benna C, Cisotto P, Rosato E, Kyriacou CP, Costa R. The clock gene period in the medfly Ceratitis capitata. Genet Res. 2005;86:13–30. doi: 10.1017/S0016672305007664. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh-Hilfiker A, Young MW. double-time Is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Tsai T, Roca AL, Sauman I. Cloning of a structural and functional homolog of the circadian clock gene period from the giant silkmoth Antheraea pernyi. Neuron. 1994;13:1167–1176. doi: 10.1016/0896-6273(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Rieger D, Shafer OT, Tomioka K, Helfrich-Forster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci. 2006;26:2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauman I, Reppert S. Circadian clock neurons in the silkmoth Antheraea pernyi, novel mechanisms of PERIOD protein regulation. Neuron. 1996;17:889–900. doi: 10.1016/s0896-6273(00)80220-2. [DOI] [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Siwicki KK, Strack S, Rosbash M, Hall JC, Jacklet JW. An antibody to the Drosophila period protein recognizes circadian pacemaker neurons in Aplysia and Bulla. Neuron. 1989;3:51–58. doi: 10.1016/0896-6273(89)90114-1. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Suri V, Hall JC, Rosbash M. Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J Neurosci. 2000;20:7547–7555. doi: 10.1523/JNEUROSCI.20-20-07547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Genco MC, Marlow ED, Benton JL, Beltz BS, Sandeman DC. Brain photoreceptor pahways control circadian rhythmicity in crayfish. Chronobiol Intl. 2009;26:1136–1168. doi: 10.3109/07420520903217960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Shafer OT. Mechanisms of clock output in the Drosophila circadian pacemaker system. J Biol Rhythms. 2006;21:445–457. doi: 10.1177/0748730406293910. [DOI] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Taylor P, Hardin PE. Rhythmic E-Box binding by CLK-CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol. 2008;28:4642–4652. doi: 10.1128/MCB.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H, OH, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Thurman CL. Unravelling the ecological significance of endogenous rhythms in intertidal crabs. Biol Rhythm Res. 2004;35:43–67. [Google Scholar]
- Tilden AR, McCoole MD, Haromon SM, Baer KN, Christie AE. Genomic identification of a putative circadian system in the cladoceran crustacean Daphni pulex. Comp Biochem Physiol D. 2011;6:282–309. doi: 10.1016/j.cbd.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Brandes C, Helfrich-Forster C, Hall JC, Stanewsky R. A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr Biol. 2003;13:1758–1767. doi: 10.1016/j.cub.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Verde MA, Barriga-Montoya C, Fuentes-Pardo B. Pigment dispersing hormone generates a circadian response to light in the crayfish, Procambarus clarkii. Comp Biochem Physiol A Mol Integr Physiol. 2007;147:983–992. doi: 10.1016/j.cbpa.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Warman GR, Newcomb RD, Lewis RD, Evans CW. Analysis of the circadian clock gene period in the sheep blow fly Lucilia cuprina. Genet Res. 2000;75:257–267. doi: 10.1017/s0016672399004425. [DOI] [PubMed] [Google Scholar]
- Wildt M, Goergen EM, Benton JL, Sandeman DC, Beltz BS. Regulation of serotonin levels by multiple light-entrainable endogenous rhythms. J Exp Biol. 2004;207:3765–3774. doi: 10.1242/jeb.01205. [DOI] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of PERIOD protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]