Abstract
SnRK [SNF1 (sucrose non-fermenting-1)-related protein kinase] 2.6 [open stomata 1 (OST1)] is well characterized at molecular and physiological levels to control stomata closure in response to water-deficit stress. OST1 is a member of a family of 10 protein kinases from Arabidopsis thaliana (SnRK2) that integrates abscisic acid (ABA)-dependent and ABA-independent signals to coordinate the cell response to osmotic stress. A subgroup of protein phosphatases type 2C binds OST1 and keeps the kinase dephosphorylated and inactive. Activation of OST1 relies on the ABA-dependent inhibition of the protein phosphatases type 2C and the subsequent self-phosphorylation of the kinase. The OST1 ABA-independent activation depends on a short sequence motif that is conserved among all the members of the SnRK2 family. However, little is known about the molecular mechanism underlying this regulation. The crystallographic structure of OST1 shows that ABA-independent regulation motif stabilizes the conformation of the kinase catalytically essential α C helix, and it provides the basis of the ABA-independent regulation mechanism for the SnRK2 family of protein kinases.
Keywords: kinase regulation, protein structure, crystallography, plant abiotic stress, signaling
Introduction
The identification of the mechanism underlying the plant cell response to osmotic stress is central to produce improved crops able to survive under stress conditions. Protein kinases in the SnRK [SNF1 (sucrose non-fermenting-1)-related protein kinase] 2 family are activated by osmotic stress.1 They are involved in the regulation of signaling pathways that lead to the activation of protective processes such as the stimulation of stomata closure or the accumulation of osmolytes to maintain cell turgor and cell membrane integrity. This response triggers the production of second messengers, the regulation of ion channels and some changes in gene expression.2–4 The identification and characterization of all molecular species involved in such processes are crucial to understand and, ultimately, to control the cellular response to stress.
One important response to osmotic stress is the accumulation of the phytohormone abscisic acid (ABA), which plays critical roles in plant adaptation to stress, as well as other essential physiology-related processes such as growth and development.5–9 Some of the SnRK2 family members are also activated by the increase in the ABA intracellular concentration; hence, this protein family has been proposed as key component that merges the ABA-dependent and ABA-independent pathways for osmotic stress responses. Biochemical, cellular and structural studies have shown that the ABA signaling pathway relies on three protein classes: the pyrabactin resistance (PYR)/RCAR (regulatory component of ABA receptor) family of ABA receptors, a group of type 2C protein phosphatases (PP2Cs) and a subgroup of SnRK2s (SnRK2.2, SnRK2.3 and SnRK2.6 in Arabidopsis thaliana).10–12 In this model, protein–protein interactions, among the members of a signal–receptor complex, control the phosphorylation state of SnRK2 and are fundamental for the regulation of the system. SnRK2 is constitutively active in its phosphorylated state.13 However, under non-stress conditions, SnRK2 kinases are kept inactive by the PP2Cs through physical interaction and dephosphorylation.14,15 According to this model, during stress, the PYR receptor binds ABA. This complex interacts with PP2C proteins, abolishing phosphatase activity and allowing autophosphorylation and activation of the kinase. Subsequently, SnRK2 kinase phosphorylates the target proteins, triggering the cellular response. Hence, a balance between the activities of negative (PP2C) and positive (SnRK2) regulators fine-tunes the cell’s response in an ABA-dependent manner.
Structural studies have shown that ABA sensing involves a molecular rearrangement of the PYR receptor that creates a site for interaction with the PP2C protein and, thereby, blocks the PP2C active site.16,17 Biochemical studies show that the other known component of the signaling pathway, SnRK2, has a molecular architecture that includes the catalytic domain plus a C-terminal regulatory domain. The regulatory domain can be divided functionally into domains I and II (DI and DII). DI is highly conserved among the members of the SnRK2 family; it is essential for kinase activity and is responsible for ABA-independent activation in response to osmotic stress.18–20 On the other hand, DII classifies the family into two groups of kinases according to their ability to be activated in an ABA-dependent manner. DII includes the minimum interaction region for recognition of the PP2C phosphatases.
Open stomata 1 (OST1; also called SnRK2E or SnRK2.6) is well characterized as a key component to control the stomatal response to ABA in A. thaliana.21,20 OST1 works together with SnRK2.2 and SnRK2.3 to form a major hub in the ABA signaling network. OST1 physically interacts with PP2C family members HAB1, ABI1, ABI2 and PP2CA.14,15 It has been shown that these PP2Cs inactivate OST1 by dephosphorylation of Ser175 residue at the activation loop of the kinase. However, it is known that a truncated version of OST1, lacking the PP2C binding domain DII, can be also activated under low humidity stress.19 This suggests the existence of an additional regulatory mechanism other than the ABA-dependent inactivation of the PP2C. Active OST1 targets the slow anion channel-associated 115 and the potassium channel in A. thaliana KAT1.22 It also phosphorylates the transcription factors involved in transcriptional responses associated with ABA, such as ABF2/AREB1.13
Despite all the known information about SnRK2 kinases, little is known about the molecular basis of the ABA-independent regulation at present. To understand this issue and the molecular architecture of the SnRK2, we have determined the X-ray crystallographic structure of the OST1 kinase. The model includes the catalytic domain and the ABA-independent regulatory domain DI. The latter forms an α-helix that binds to a hydrophobic patch of the catalytic domain and stabilizes the catalytically essential α C helix conformation. The structure provides the molecular basis of the ABA-independent activation of SnRK2 family of kinases.
Results and Discussion
Overall structure of OST1
The crystal structures of two catalytically inactive OST1 mutants were solved by single anomalous diffraction (SAD) phasing: D160A at 2.5 Å resolution, which prevents Mg2+ binding, and the double mutant D160A/S175D at 2.8 Å resolution, which mimics a phosphorylated protein (Fig. 1a) (see Table 1 and Materials and Methods). Both structures are nearly identical [Cα backbone root-mean-square deviation (RMSD) of 0.3 Å]; hence, unless otherwise stated, all the results presented herein will refer to the OST1 D160A mutant. In both cases, the crystallographic analysis revealed two OST1 molecules in the asymmetric unit. However, analytical ultracentrifugation experiments, as well as gel-filtration chromatography, suggest that OST1 is monomeric at 25 μM concentration; hence, it seems that the crystal packing interactions between independent molecules are not physiologically relevant (Supplementary Fig. 1).
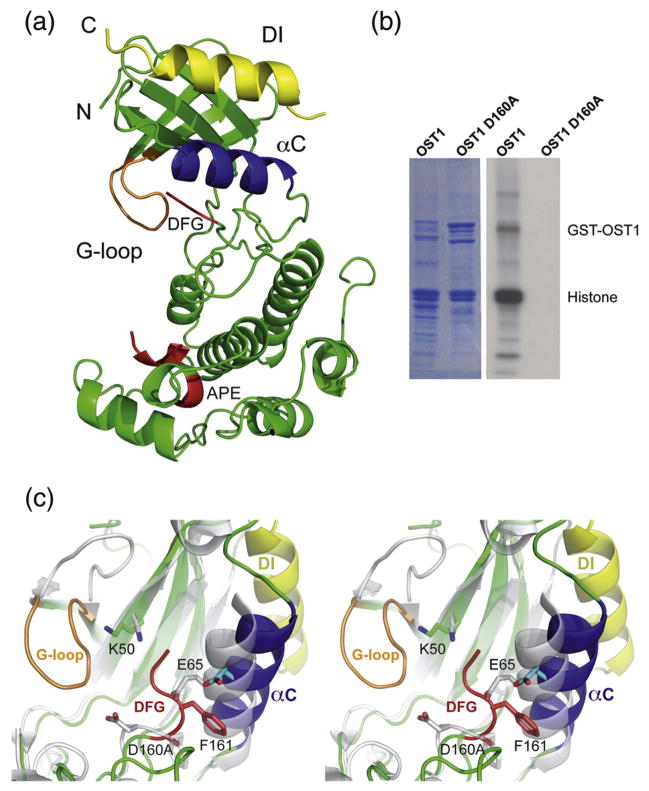
Fig. 1.
The crystal structure of OST1. (a) A ribbon representation of the OST1 D160A structure. Helix α C is displayed in blue, the G-loop is displayed in orange, the DI domain is displayed in yellow and the residues of the activation loop (from DFG to APE motives) are displayed in red. N-term and C-term are labeled. (b) OST1 D160A mutation abolishes recombinant kinase activity in vitro. The top band corresponds to autophosphorylation, and the bottom band corresponds to phosphorylation of histone III-S. (c) Structural comparison of the inactive OST1 and a related active SNF1. Stereo view of the superimposition of the ribbon diagrams corresponding to the active-site section of the OST1 D160A and the active yeast SNF1 (PDB ID: 3DAE). OST1 helix α C is displayed in blue, the G-loop is displayed in orange, the DI domain is displayed in yellow and the residues of the DFG motif are displayed in red. The active form of yeast SNF1 is displayed in white.
Table 1.
Data collection and refinement statistics
| OST1 D160A | OST1 D160A+S175D | |
|---|---|---|
| Data collection | ||
| Space group | P2221 | P2221 |
| Cell dimensions | ||
| a, b, c (Å) | 77.27, 99.19, 107.83 | 77.57, 99.02, 108.45 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 41.74–2.50 (2.64–2.50) | 77.57–2.70 (2.85–2.70) |
| Rsym, Rpim | 0.06 (0.52), 0.03 (0.27) | 0.08 (0.8), 0.02 (0.22) |
| I/σ(I) | 13.3 (2.8) | 22.6 (4.1) |
| Completeness (%) | 99.7 (99.7) | 100.0 (100.0) |
| Redundancy | 4.5 (4.6) | 14.2 (14.7) |
| Refinement | ||
| Resolution (Å) | 41.74–2.50 (2.59–2.50) | 77.57–2.70 (2.9–2.7) |
| Number of reflections | 29,227 | 20,110 |
| Rwork/Rfree | 19.7/25.1 (32.6/40.0) | 23.8/27.9 (32.1/33.3) |
| Number of atoms | ||
| Protein | 4523 | 4506 |
| Gold atoms | — | 1 |
| Water molecules | 59 | 48 |
| Average B-factors | 64.7 | 68.3 |
| RMSD | ||
| Bond lengths (Å) | 0.0105 | 0.0100 |
| Bond angles (°) | 1.0708 | 1.1500 |
| Ramachandran plot statistics | 96.4% in the core 3.2% in the allowed 0.4% outliers |
92.0% in the core 7.5% in the allowed 0.5% outliers |
The highest-resolution shell is shown in parenthesis.
Rsym = Σhkl|Ihkl − 〈Ihkl〉|/ΣhklIhkl.
The crystallographic model comprises residues from 12 to 319; thus, it includes the catalytic domain plus a single α-helix that constitutes the ABA-independent regulatory DI (Supplementary Fig. 2). We have not been able to see any of those amino acids that comprise the ABA-dependent regulatory DII. Mass spectrometry analysis of protein sample obtained from the crystals shows two species of 33,586 Da and 37,529 Da. The larger one is consistent with the molecular weight of a fragment slightly longer than the crystallographic model. This means that DII residues are cleaved since we overexpressed the intact kinase.
Overall, the structure of catalytic OST1 displays the canonical Ser/Thr protein kinase fold, similar to other SNF1 kinase domains [Protein Data Bank (PDB) IDs: 3DAE and 3FAM, with RMSD less than 2 Å; Dali server].23 In summary, the protein folds into two separate subdomains. The N-terminal domain consists of a five-stranded β-sheet plus an α-helix called helix α C. The C-terminal domain is larger and mainly helical. All the residues involved in catalysis, the ATP and metal binding sites and the substrate binding pocket are placed in between these domains.
The OST1 D160A and D160A/S175D display an open inactive conformation
SNF1 kinase activation implies a conformational rearrangement of the relative position of the N-lobe with respect to the C-lobe, which yields a transition from an “open” inactive state to a “closed” active state (Supplementary Fig. 3).24,25 This movement is maximal at helix α C and implies the productive interaction of the conserved lysine in the N-lobe, a glutamic residue at α C helix and the catalytic aspartic residue from the DFG motif (K50, E65 and D160 in OST1). In addition, the activation requires the stabilization of ATP by a conserved loop connecting the first two β-strands at the N-lobe (G-loop) and the phosphorylation of some residues at the activation loop, which adopts an extended conformation compatible with substrate binding.
In vitro kinase assay of a recombinant protein (Fig. 1b) shows that OST1 D160A has no detectable kinase activity. According to this, the comparison of the OST1 D160A structure with those structures of an active and a self-inhibited SnRK24 shows that OST1 displays an inactive conformation (Fig. 1c). First, the F161 side chain from the DFG motif26 lies below helix α C and stabilizes the N-lobe into an open inactive kinase state. Second, the OST1 G-loop displays an unproductive ATP binding conformation, and third, the unphosphorylated OST1 activation loop lies between catalytic residues, thus preventing catalysis.
Interestingly, the same open conformation is observed for the pseudophosphorylated OST1 D160A/S175D double mutant. This is in agreement with biochemical data showing that this mutant does not display catalytic activity and does not complement the srk2e stomatal phenotype.18 Ng et al. showed that p21-activated kinases require direct interactions between a basic residue at the N-terminus of the α C and the phosphate moiety of the phosphoserine residue to promote a network of interactions that bring together N-lobe and C-lobe and stabilize the proper positioning of the helix α C to bind cofactors and substrate.27 A topologically equivalent residue is also present in all the SnRK2s. This suggests a similar activation mechanism for p21-activated kinases and SnRK2s (Supplementary Fig. 4).
The regulation of OST1
The characterization of the molecular basis underlying the SnRK2 regulation is important since these kinases are activated in response to osmotic stress. Available data have shown that the cell response can be divided between ABA-dependent and ABA-independent pathways. Accordingly, the molecular organization of the C-terminal regulatory domain of SnRK2s is divided functionally into ABA-independent DI and ABA-dependent DII.
The ABA-dependent regulation of OST1 is well understood at both cellular and molecular levels. In a non-stressed situation, OST1 is kept inactive by the direct interaction of PP2C with the DII N-terminal residues (321–331) and by the dephosphorylation of Ser175 at the activation loop.14 In an osmotic stress situation, an increase in the ABA intracellular concentration yields to the inactivation of the PP2C phosphatase through its interaction with the PYR/ RCAR ABA receptor.14,13,28 The steric hindrance between the PP2C–PYR complex and OST1 necessarily implies the release of the complex from the kinase active site. In this situation, OST1 becomes active by autophosphorylation. Although we were not able to obtain structural information about DII, our structural work provides some additional information to this model. OST1 structure defines the framework for an efficient interaction with PP2Cs since the C-terminal end of the DI domain and, consequently, the N-terminal residues of DII point toward the OST1 activation loop at the active site (Supplementary Fig. 5).
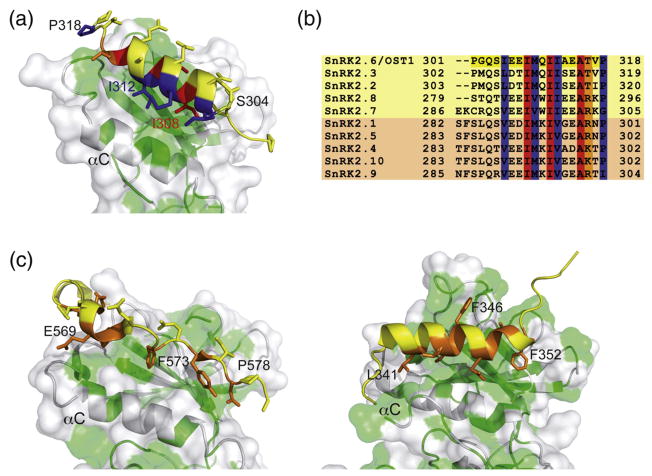
On the other hand, the molecular basis of the ABA-independent regulation of OST1 is not clear. Biochemical and cellular studies have shown that OST1 needs to be activated to trigger cell response to osmotic stress.1,18,19 This implies the stabilization of a “closed” active conformation and the subsequent phosphorylation of the activation loop despite its interaction with PP2C. The regulation of many kinases relies on the stabilization of either the active or the inactive conformation depending on cell requirements. This is achieved by intermolecular interactions involving other molecules or the oligomerization of the kinase or by intramolecular interactions involving the regulatory domains of the kinase. The analysis of the OST1 structure supports that the ABA-independent regulatory mechanism depends on the interaction of the regulatory DI and the catalytic domain. The structure shows that DI forms a single α-helix that is packed against the N-lobe (Figs. 1a and 2a). It fills a hydrophobic pocket formed between the α C helix and the central β-sheet. It is largely stabilized by hydrophobic interactions between DI and the kinase domain since more than 70% of the buried surface area is formed by nonpolar atoms (494 Å2 out of 694 Å2).29 A sequence analysis of the SnRK2 family reveals that the residues involved in the stabilization of the DI domain at the N-lobe (I305, I308, M309, I311, I312 and A315) are highly conserved (Fig. 2b). This is consistent with a common role of DI on the ABA-independent regulation for all the SnRK2 family members. The activation by the intramolecular interaction of an N-terminal or a C-terminal α-helix to a hydrophobic patch in the N-lobe is a common feature of several kinases.30 These include PKA (protein kinase A)31 and other AGC (cAMP dependent, cGMP dependent, protein kinase C) protein kinases32 and ERK2 (extracellular signal-regulated kinase 2).33 In these protein families, the regulation mechanism consists of the binding of a segment of the protein, located immediately after the C-terminal end of the kinase domain, to a structurally equivalent OST1 hydrophobic pocket (Fig. 2c). The structural basis of this regulation involves the stabilization of the helix α C to facilitate its optimal alignment and to promote catalysis. In support to this notion, it has been shown that the binding of the regulatory segment to the catalytic domain produces an overall thermal stabilization of the kinase of around 8–10 °C.34,35 To test if OST1 shares similar regulatory mechanism, we monitored the thermal stability of the pseudophosphorylated inactive but ATP-bound OST1 S175D and the dead OST1 D160A and OST1 D160A/S175D by circular dichroism spectroscopy (Materials and Methods; Fig. 3). Although the measurements in far UV on mutant proteins revealed no significant differences in the secondary structure composition, our results show an increase of around 6 °C in thermal stability for the dead mutants with respect to the ATP-bound OST1 S175D. Hence, we suggest that the ABA-independent regulation mechanism could be associated with a conformational change and/or the release of DI at the hydrophobic pocket that renders to the stabilization of the protein. The mechanism is also supported by the fact that a recombinant truncated OST1 protein lacking domain DI has no detectable in vivo or in vitro kinase activity, although it is correctly folded.18,19 The joined analysis of the available biochemical and structural data suggests that filling the hydrophobic pocket by DI is necessary to activate the kinase but is not sufficient since, as it is the case of OST1 D160A, phosphorylation of the kinase is essential to achieve the active conformation.
Fig. 2.
Interactions between DI and the OST1 catalytic kinase domain and related structures. (a) A section of the OST1 structure showing the residues involved in the relevant interactions between DI and the kinase domain. A cartoon representation of the kinase N-lobe is displayed together with a semitransparent surface. Hydrophobic residues of the kinase domain are highlighted in green. Residues buried in the interaction with the kinase domain are colored according to their conservation degree among the members of the family [see (b)]. (b) A comparison of the DI domain amino acids in A. thaliana SnRK2s. The kinases boxed in yellow or light orange belong to the ABA-dependent or ABA-independent regulation subgroup, respectively. (c) An equivalent section to the OST1 structure showed in (a) of the atypical protein kinase C (PDB ID: 1ZRZ) (left) and the ERK2 protein kinase (PDB ID: 1ERK) (right). Residues buried in the interaction with the kinase domain are colored orange.
Fig. 3.
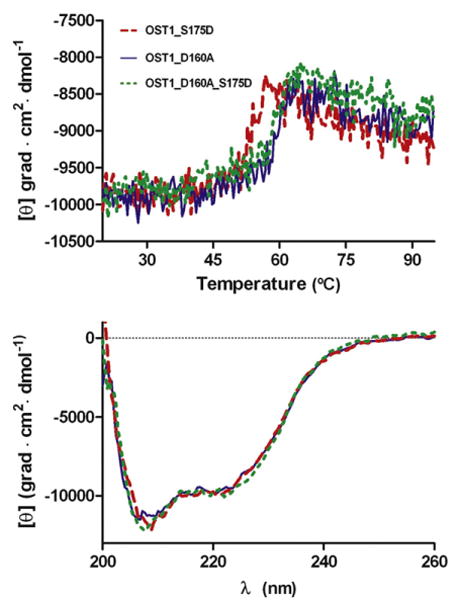
Comparison of the thermal stability profiles monitored by circular dichroism (top) and far-UV spectra (bottom) for OST1 S175D, OST1 D160A and OST1 D160A/S175D.
For some of the ACG kinases, this regulation mechanism is triggered by the phosphorylation of a conserved Ser or Thr on the regulatory segment.32 In this sense, we tested if the phosphorylation at DI domain provides a regulation mechanism for OST1. The comparison of the DI domain amino acid sequence of A. thaliana SnRK2s shows that the OST1 Ser304 is conserved among all the family members (Fig. 2b). To evaluate if the phosphorylation of this residue had any effect on the OST1 activity, we performed kinase activity assays for the S304D and S304A mutants. In both cases, the recombinant mutated protein obtained was active (Supplementary Fig. 6a). Therefore, we conclude that the phosphorylation of OST1 Ser304 is not involved in the kinase regulation mechanism.
Even though SnRK2 and SnRK3 families of kinases do not function in the same pathway, it is worth noting that there are some structural similarities between them. As OST1, the SnRK3 SOS (salt overly sensitive) 2 is regulated by a single helical segment (FISL) located C-terminal to the kinase domain, which is followed by a PP2C-interacting domain.36–42 It is also known that the activation of SOS2 and PP2C binding cannot occur simultaneously and implies the release of the FISL from the kinase domain.43 This is triggered by the direct integrations of the FISL motif with the calcium binding protein SOS3.36,43 Although it has been shown that SOS3-like proteins do not interact with SnRK2s,13 it is tempting to speculate that FISL and DI share the same hydrophobic pocket on their kinase domains and, consequently, that they also share a similar regulation mechanism (Fig. 4). The identification of the molecular components responsible of the ABA-independent SnRK2 regulation remains to be investigated and will be central to produce a complete picture of the osmotic stress signaling pathway.
Fig. 4.
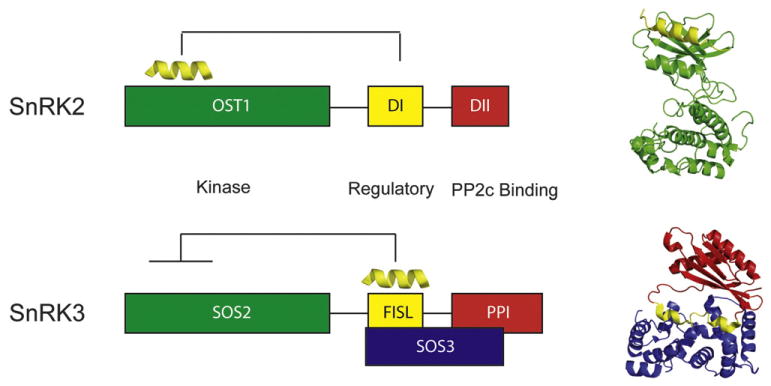
Comparison of the domain structure of SnRK2s and SnRK3s. Ribbon representations of the structure of the representative kinase domain plus the regulatory DI (yellow) of SnRK2 (OST1) and the structure of the calcium sensor SOS3 plus the representative regulatory domain FISL (yellow) and the PP2C binding domain of SnRK3 (SOS2).
Materials and Methods
SnRK2.6 gene cloning and site-directed mutagenesis
The OST1 gene from A. thaliana, encoding OST1 full length, was cloned into a pGEX4T1 (GE Healthcare) expression plasmid, and OST1 D160A mutant was produced as previously described.44 Other mutants (OST1 S175D, OST1 D160A/S175D double mutant, OST1 S304D and OST1 S304A) were produced using standard site-directed mutagenesis techniques.45 Primers used in the mutagenesis were as follows: S175D Forward 5′ GCA ACC AAA AGA CAC TGT TGG AAC TCC 3′, OST1 S175D Reverse 5′ CCA ACA GTG TCT TTT GGT TGC GAA TG 3′, S304D Forward 5′ CG GGC CAA GAC ATA GAA GAA ATT ATG CAG ATC ATT GC 3′, S304D Reverse 5′ CAT TTC TTC TAT GTC TTG GCC CGG TTG ATC C3′, S304A Forward 5′ CG GGC CAA GCC ATA GAA GAA ATT ATG CAG ATC ATT GC 3′ and S304A Reverse 5′ CAT TTC TTC TAT GGC TTG GCC CGG TTG ATC C3′.
Protein preparation, crystallization and data collection
OST1 D160A and OST1 D160A/S175D were obtained from cultures of Escherichia coli Rosetta (DE3) pLys and purified to homogeneity as described previously.44 To summarize, we purified the overexpressed glutathione S-transferase (GST)-tagged proteins using glutathione Sepharose beads (GE Healthcare) according to manufacturer’s instructions and cleaved them from GST using 7.5 units of thrombin protease (Novagen) per milligram of GST-tagged protein. A final polishing step was performed using a Superdex 200 16/60 (Amersham Biosciences Limited, UK). OST1 D160A and OST1 D160A/S175D were concentrated to a final concentration of 12 mg ml−1 in 50 mM Tris–HCl (pH 7.5) and 50 mM NaCl. The final sample purity was examined by SDS-PAGE and mass spectrometry.
Prior to crystallization, the protein samples were incubated with DTT, MgCl2 and adenylyl imidodiphosphate to reach the final additive concentration of 1 mM, 2 mM and 2 mM, respectively. Crystallization experiments were carried out using the hanging-drop vapor-diffusion method at 291 K by mixing drops of 2 μl protein solution with 1 μl precipitant solution. OST1 D160A crystals were grown using 0.1 M Hepes (pH 6.5), 12% polyethylene glycol 10,000 and 12% ethylene glycol as precipitant. OST1 D160A/ S175D crystals were grown from a similar crystallization solution buffered at pH 7.0 and containing 14% polyethylene glycol 10,000. Crystals were mounted in fiber loops, transferred to the cryoprotectant solution and flash-frozen at 100 K in a nitrogen gas stream. The cryoprotectant solution resulted from the increase in the ethylene glycol concentration from 12% to 18% on the crystal mother liquor. Some OST1 D160A/S175D crystals were subjected to a 5- to 10-s flash soak in cryoprotectant solution containing 100 mM gold (I) potassium cyanide [KAu(CN)2]. Afterwards, the crystals were washed back in the cryoprotectant solution without the heavy atom for 5 s.
Native OST1 D160A and SAD OST1 D160A/S175D X-ray diffraction data were collected in a CCD detector using the European Synchrotron Radiation Facility (Grenoble, France) radiation source at 0.93-Å and 1.04-Å wavelengths at the ID14.1 and ID23.1 beamlines, respectively. Diffraction data were processed with XDS46 and scaled with SCALA from the CCP4 package (Collaborative Computational Project, Number 4, 1994). A summary of the data collection statistics is given in Table 1.
Structure determination
The X-ray structure of OST1 D160A was solved by molecular replacement with the program MolRep47 using the coordinates from the protein kinase domain of the yeast AMP-activated protein kinase SNF1(PDB ID: 3HYH).48 The model shows a sequence identity of 42% with respect to OST1 (BLAST),49 and it yields a conclusive solution that includes two molecules in the asymmetric unit. However, the calculated electron density map was noisy and strongly biased. We carried out SAD experiments with crystals soaked in heavy-atom derivatives [KAu(CN)2] to avoid the model bias. The OST1 D160A/S175D structure was solved by SAD using the anomalous information corresponding to the gold atoms. The structure was solved using the MRSAD methods as implemented in Auto-Rickshaw, the European Molecular Biology Laboratory Hamburg automated crystal structure determination platform.50 Refinement of the gold substructure and phasing were performed using SHELX.51 Subsequent solvent flattening was performed with DM,52 yielding an electron density map good enough to automatically trace an initial model (Buccaneer)53 that was first refined using PHENIX.54 Several cycles of restrained refinement with REFMAC555 and PHENIX54 and iterative model building with Coot56 were carried out. The water structure was also modeled. The refined structure of OST1 D160A/S175D was used to phase and refine the OST1 D160A structure using similar protocols. Calculations were performed using CCP4 programs.57 The comparison of the OST1 structure with respect to the model used for the molecular replacement procedure explains why this method did not produce the expected results. The structural alignment between them renders to only 206 Cα backbone residues with an RMSD of 1.9 Å. This is due to the different relative orientation of N-terminal lobes with respect to the C-terminal lobes of the kinases.
The stereochemistry of the models was verified with PROCHECK.58 Ribbon figures were produced using PyMOL.59 The accessible surface area calculations were calculated with the program NACCESS from the LIGPLOT package.29 The refinement statistics are summarized in Table 1.
Protein preparation for physicochemical characterization in solution (analytical ultracentrifugation, circular dichroism and mass spectrometry) and for in vitro kinase phosphorylation assays
OST1 D160A, OST1 S175D, OST1 D160A/S175D, OST1 S304A, OST1 D160A/S304A, OST1 S304D and OST1 D160A/S304D were obtained from cultures of E. coli Rosetta (DE3) pLys and purified to homogeneity as described above. The OST1 mutants used to the phosphorylation in vitro assay were not cleaved from GST; they were used as GST protein fusion.
Analytical ultracentrifugation: Sedimentation equilibrium
The experiments were performed in a Beckman Optima XL-A ultracentrifuge using a Ti50 rotor and six-channel centerpieces of Epon charcoal (optical pathlength=12 mm). Samples of OST1 were equilibrated against 50 mM Tris–HCl (pH 7.5) and 50 mM NaCl buffer and were centrifuged at 10,000 rpm, 12,000 rpm and 17,000 rpm. Conservation of mass in the cell was checked in all the experiments. After the equilibrium scans, a high centrifugation run (45,000 rpm) was performed to estimate the corresponding baseline offsets. Weight-average buoyant molecular weights were determined by fitting a single species model to the experimental data using the Hetero-Analysis program.60 The absolute molecular weight of OST1 was determined using the partial specific volume (0.7258 ml g−1), calculated for the amino acid composition, and the solvent density (1.0035 mg ml−1) was determined using the SEDNTERP program.
Circular dichroism spectra
Circular dichroism spectra were recorded in a JASCO J-810 spectropolarimeter. Far-UV spectra were recorded in a 0.1-cm-pathlength quartz cell at a protein concentration of 0.3 mg ml−1 (7.3×10−6 M). The observed ellipticities were converted to mean residue ellipticities [θ] using an average molecular mass per residue of 113.3 Da. Thermal denaturation was monitored at a wavelength of 220 nm by increasing the temperature from 20 °C to 95 °C at 30 °C h−1 and allowing the temperature to equilibrate for 1 min before recording the spectra.
Far-UV spectra and the thermal denaturation profiles were recorded for OST1 S175D, OST1 D160A and OST1 D160A/S175D.
Mass spectrometry
OST1 mutants were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry on an Ultraflex III mass spectrometer (Bruker Daltonics, Germany).
In vitro kinase phosphorylation assays
Phosphorylation in vitro assay was performed for different OST1 mutants as GST protein fusion (GST_OST1) by incubation for 30 min at 30 °C of 100 ng GST_OST1, purified as described above, and 200 ng histone (Sigma H4524) in 30 μl of 20 mM Tris (pH 7.5), 5 mM MgCl2, 1 mM DTT and 10 mM β-glycerophosphate with 5 μCi of [γ-33P]ATP (3000 Ci mmol−1). Reaction was stopped by adding 10 μl Laemmli 4× and heating at 95 °C for 3 min. Proteins were separated by SDS-PAGE using a 10% (w/v) acrylamide gel. Radioactivity was detected on the dry gels using Biomax Light-1 (Kodak) films.
Supplementary Material
Acknowledgments
We thank Dr. Laura Lagartera for the analytical ultracentrifugation data analysis, Dr. F. J. Quintero for the access to his laboratory and advice in the kinase activity measurements and Dr. John Klingler for critical reading of the manuscript. A.A. thanks the European Synchrotron Radiation Facility for the access to the synchrotron radiation source. This work was funded by the grant BFU2008-00368/BMC, BFU2001-25384 and “Factoría de Cristalización” Consolider-Ingenio 2010 of the Spanish “Plan Nacional” (Ministerio de Ciencia e Innovación) to A.A.
Abbreviations used
- ABA
abscisic acid
- PP2C
protein phosphatase type 2C
- OST1
open stomata 1
- PYR
pyrabactin resistance
- PDB
Protein Data Bank
- SAD
single anomalous diffraction
- GST
glutathione S-transferase
Footnotes
Accession numbers
The coordinates and structure factor amplitudes of OST1 D160A and OST1 D160A/S175D have been deposited in the PDB (PDB IDs: 3ZUT and 3ZUU, respectively).
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/ j.jmb.2011.09.041
References
- 1.Fujii H, Verslues PE, Zhu JK. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol. 2007;10:296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 4.Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 6.Hetherington AM. Guard cell signaling. Cell. 2001;107:711–714. doi: 10.1016/s0092-8674(01)00606-7. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering of drought hardiness in plants. Nature. 2001;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- 8.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 10.Halford NG, Hey SJ. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J. 2009;419:247–259. doi: 10.1042/BJ20082408. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 12.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signalling in plant guard cells. Proc Natl Acad Sci USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, et al. Structural mechanism of abscisic acid binding and signalling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 18.Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signalling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 21.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Jiao ZH, Zheng LS, Zhang YY, Xie ST, Wang ZX, Wu JW. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 25.Tsigelny I, Greenberg JP, Cox S, Nichols WL, Taylor SS, Ten Eyck LF. 600 ps molecular dynamics reveals stable substructures and flexible hinge points in cAMP dependent protein kinase. Biopolymers. 1999;50:513–524. doi: 10.1002/(SICI)1097-0282(19991015)50:5<513::AID-BIP5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 26.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 27.Ng YW, Raghunathan D, Chan PM, Baskaran Y, Smith DJ, Lee CH, et al. Why an A-loop phospho-mimetic fails to activate PAK1: understanding an inaccessible kinase state by molecular dynamics simulations. Structure. 2010;18:879–890. doi: 10.1016/j.str.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Hirayama T, Umezawa T. The PP2C–SnRK2 complex: the central regulator of an abscisic acid signalling pathway. Plant Signal Behav. 2010;5:160–163. doi: 10.4161/psb.5.2.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 30.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knighton DR, Zheng JH, Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 32.Gold MG, Barford D, Komander D. Lining the pockets of kinases and phosphatases. Curr Opin Struct Biol. 2006;16:693–701. doi: 10.1016/j.sbi.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 34.Biondi RM, Komander D, Thomas CC, Lizcano JM, Deak M, Alessi DR, van Aalten DM. High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J. 2002;21:4219–4228. doi: 10.1093/emboj/cdf437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komander D, Kular G, Deak M, Alessi DR, van Aalten DM. Role of T-loop phosphorylation in PDK1 activation, stability, and substrate binding. J Biol Chem. 2005;280:18797–18802. doi: 10.1074/jbc.M500977200. [DOI] [PubMed] [Google Scholar]
- 36.Halfter U, Ishitani M, Zhu JK. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for salt tolerance. Plant Cell. 2001;13:1383–1399. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu JK. Genetic analysis of plant salt tolerance using Arabidopsis thaliana. Plant Physiol. 2000;124:941–948. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang C, et al. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell. 2004;16:435–449. doi: 10.1105/tpc.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan WZ, Lee SC, Che YF, Jiang YQ, Luan S. Mechanistic analysis of AKT1 regulation by the CBL–CIPK–PP2CA interactions. Mol Plant. 2011;4:527–536. doi: 10.1093/mp/ssr031. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez-Barrena MJ, Martinez-Ripoll M, Zhu JK, Albert A. The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. J Mol Biol. 2005;345:1253–1264. doi: 10.1016/j.jmb.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Yunta C, Mantinez-Ripoll M, Albert A. SnRK2.6/OST1 from Arabidopsis thaliana: cloning, expression, purification, crystallization and preliminary X-ray analysis of K50N and D160A mutants. Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2011;67:364–368. doi: 10.1107/S1744309110053807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 46.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 47.Vagin AA, Isupov MN. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr, Sect D: Biol Crystallogr. 2001;57:1451–1456. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- 48.Rudolph MJ, Amadeo GA, Bai Y, Tong L. Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1. Biochem Biophys Res Commun. 2005;337:1224–1228. doi: 10.1016/j.bbrc.2005.09.181. [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Panjikar S, Parthasarathy V, Lamzin V, Weiss MS, Tucker PA. On the combination of molecular replacement and single anomalous diffraction phasing for automated structure determination. Acta Crystallogr, Sect D: Biol Crystallogr. 2009;65:1089–1097. doi: 10.1107/S0907444909029643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheldrick GM. A short history of SHELX. Acta Crystallogr, Sect A: Found Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 52.Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr, Sect D: Biol Crystallogr. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 53.Cowtan K. The Buccaneer software for automated model building. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 59.DeLano WI. The PyMOL Users Manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 60.Cole JL. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–323. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.