Abstract
Heat shock protein 70 (Hsp70) plays critical roles in proteostasis and is an emerging target for multiple diseases. However, competitive inhibition of the enzymatic activity of Hsp70 has proven challenging and, in some cases, may not be the most productive way to redirect Hsp70 function. Another approach is to inhibit Hsp70’s interactions with important co-chaperones, such as J proteins, nucleotide exchange factors (NEFs) and tetratricopeptide repeat (TPR) domain-containing proteins. These co-chaperones normally bind Hsp70 and guide its many diverse cellular activities. Complexes between Hsp70 and co-chaperones have been shown to have specific functions, such as pro-folding, pro-degradation and pro-trafficking. Thus, a promising strategy may be to block protein-protein interactions between Hsp70 and its co-chaperones or to target allosteric sites that disrupt these contacts. Such an approach might shift the balance of Hsp70 complexes and re-shape the proteome and it has the potential to restore healthy proteostasis. In this review, we discuss specific challenges and opportunities related to those goals. By pursuing Hsp70 complexes as drug targets, we might not only develop new leads for therapeutic development, but also discover new chemical probes for use in understanding Hsp70 biology.
Keywords: heat shock protein 70, molecular chaperones, J proteins, nucleotide exchange factors, tetratricopeptide repeat-containing proteins, protein complex
INTRODUCTION
Diversity of Hsp70 Functions
Heat shock protein 70 (Hsp70) is a molecular chaperone that plays a central role in protein quality control [1, 2]. Hsp70 binds to protein substrates to assist with their folding [3, 4], degradation [5–7], transport [8], regulation [9, 10] and aggregation prevention [11]. The capacity of Hsp70 to carry out these widely divergent functions arises, in part, from three features. First, evolution has given rise to multiple homologous Hsp70 genes [12, 13]. These Hsp70s populate the major subcellular compartments. For example, the cytosol of human cells has two major Hsp70 paralogs, a stress-inducible form (Hsp72/HSPA1A/B) and a constitutive form (Hsc70/HSPA8). Additionally, BiP (HSPA5) is the Hsp70 paralog in the endoplasmic reticulum, while mortalin (HSPA9) is found in the mitochondria. For the purposes of this review, “Hsp70” will often be used to broadly refer to these chaperones because they are thought to, in many cases, have similar biochemical properties. Another source of functional diversity is Hsp70’s cooperation with other chaperones, such as the heat shock proteins Hsp90 and Hsp60 [4]. Cooperation between Hsp70 and Hsp90, for example, is critical to the function of nuclear hormone receptors [8]. Finally, the full diversity of Hsp70’s activities is achieved by collaborating with a large network of co-chaperones [1, 14], including J proteins, nucleotide exchange factors (NEFs), and tetratricopeptide repeat (TPR) domain-containing proteins [15]. These factors bind to Hsp70 and guide its many chaperone activities. In addition, the Hsp70 system is further diversified by the fact that each co-chaperone class contains multiple members (Figure 1).
Figure 1.
Hsp70 forms the core of a multi-protein complex and associates with numerous co-chaperones. Three distinct classes of co-chaperones, NEFs, J proteins, and TPR domain-containing proteins, interact with Hsp70 and regulate its activities. The J proteins and NEFs interact with the NBD, while the TPR domain-containing proteins bind the C-terminal region. Representative structures from each class are shown with the corresponding PDB code. Images were prepared in PyMol.
Hsp70 as a Therapeutic Target
Hsp70 has been implicated in multiple diseases, such as neurodegenerative disorders [16], cancer [17], and infectious disease [18] and the evidence linking Hsp70 to disease has been recently reviewed [19–21]. Despite this strong connection, relatively little progress has been made in bringing Hsp70 inhibitors to the clinic. One of the contributing factors to this lack of translational progress is that Hsp70’s functional promiscuity makes it difficult to predict potential off-target effects. As discussed above, Hsp70 is involved in many key processes in the cell; thus, it is not clear how therapeutics could be used to rebalance some pathological Hsp70 functions without impacting global proteostasis. One attractive possibility may be to target the interactions between Hsp70 and its co-chaperones because these factors are thought to diversify Hsp70’s functions.
A major focus of this review is to explore the structure and function of Hsp70 multi-protein complexes and evaluate recent progress in identifying compounds that selectively target the assembly/disassembly of these complexes. The underlying model is that each complex composed of an Hsp70 (e.g. Hsc70, Bip, etc) bound to a specific set of co-chaperones (e.g. J protein, NEF, or TPR domain-containing protein) might be involved in a discrete aspect of chaperone biology (e.g. clathrin uncoating, protein folding, degradation, etc.). Thus, if small molecules selectively disrupted an interaction between Hsp70 and a specific co-chaperone, then only a subset of Hsp70 biology might be impacted. In other words, the complexity of this chaperone network provides a unique opportunity to influence specific subsets of protein quality control while leaving the rest unperturbed. The challenge of this strategy is that it has been notoriously difficult to target protein-protein interactions [22–24], such as those between Hsp70 and its co-chaperones. However, new advances in high throughput screening (HTS) methodology are rapidly changing the landscape of discovery in this area. In fact, Hsp70 might be a particularly attractive target for deploying these methods, owing to its high number of protein-protein contacts that are important in guiding Hsp70 biology.
Structure and Function of Hsp70 and Its Complexes
Hsp70 consists of two domains, a 45 kDa N-terminal nucleotide binding domain (NBD) and a 25 kDa C-terminal substrate binding domain (SBD), which are connected by a short flexible linker [25]. The NBD of Hsp70 is further divided into two subdomains, lobes I and II, which are each divided into an “A” and “B” region (Figure 2). These lobes form a cleft that binds ATP with a nucleotide binding cassette that is related to hexokinase and actin [26]. Hsp70’s SBD is composed of a 15 kDa β-sandwich subdomain with a hydrophobic groove for polypeptide binding and a 10 kDa α-helical region which forms a “lid” over the polypeptide binding site [27]. Hsp70 preferentially binds hydrophobic regions of proteins and can therefore bind newly synthesized linear peptides or exposed regions on partially unfolded proteins [3, 28]. Additionally, a lack of strong sequence specificity allows Hsp70 to bind a variety of client proteins including signal transduction proteins, clathrin, nuclear hormone receptors, and cytoskeletal proteins [29, 30].
Figure 2.
Structure and ATPase cycle of Hsp70. (A) Hsp70 is composed of a 45 kDa N-terminal nucleotide binding domain (NBD) connected to a 25 kDa substrate binding domain (SBD) by a short hydrophobic linker. The SBD is composed of a β-sandwich and an α-helical “lid” domain. The structure of the prokaryotic Hsp70, DnaK, is shown (PDB code 2KHO), but the general architecture appears to be conserved amongst prokaryotic and eukaryotic family members. (B) Schematic of ATP hydrolysis and the role of co-chaperones. Substrate binding in the SBD coupled with J-domain co-chaperone interactions in the NBD promotes ATP hydrolysis. Conformational changes associated with ATP conversion close the “lid” and enhance affinity for the substrate. The cycle is completed when a nucleotide exchange factor interacts with the NBD and assists with ADP release.
The ATPase cycle of Hsp70 has been largely studied for the prokaryotic DnaK ortholog. In this chaperone, ATP hydrolysis involves critical allostery between the NBD and SBD. In the ATP-bound state, Hsp70 has a low affinity for substrate and retains an “open” substrate-binding cleft, but conversion to the ADP-bound state causes the α-helical lid region to “close” (Figure 3) [31]. In DnaK, this crosstalk between the NBD and SBD appears to be bidirectional, because substrate binding also promotes nucleotide hydrolysis [31, 32]. Thus, ATP hydrolysis in Hsp70 is thought to be a major determinant of chaperone function. For example, mutations in the ATP binding cassette have dramatic effects on chaperone function in vitro and in vivo [33]. However, recent mutagenesis studies have further shown that the relationship between ATP hydrolysis and chaperone function is indirect [33]. For example, some mutations in DnaK that dramatically reduce ATP turnover have only modest effects on luciferase refolding. In the context of this review, these observations suggest that inhibiting the ATPase activity of Hsp70 might not always directly lead to proportional changes in functional outcomes, such as reduced client stability. Rather, modifying the interactions with co-chaperones might have a more predictable effect on chaperone functions [33].
Figure 3.
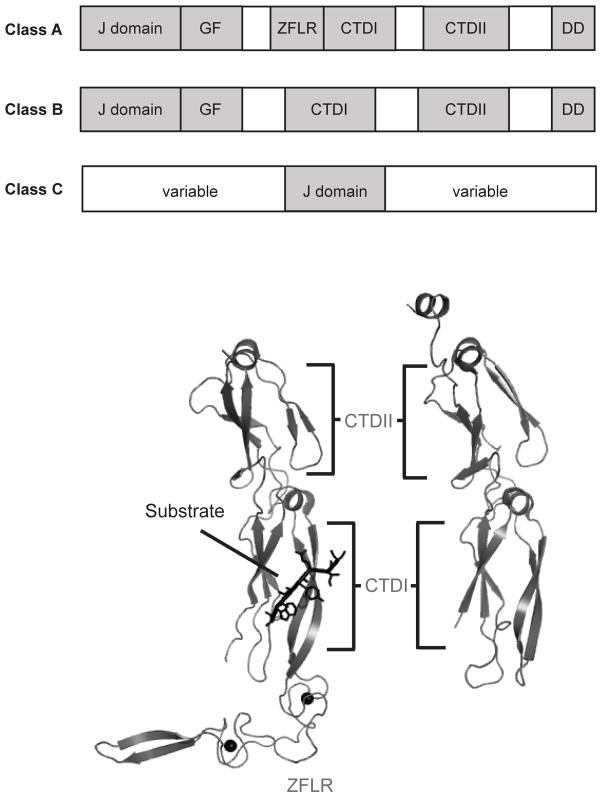
J protein co-chaperones fall into three structural classes. (A) The domain architecture of each class of J protein is depicted as a schematic beginning with the N-terminus to the left. The domain types are J domain, GF (glycine-phenylalanine rich region), ZFLR (zinc finger-like region), CTDI and II (C-terminal domain) and DD (dimerization domain). (B) The crystal structures of the C-terminal portions of Ydj1 (yeast class A J protein) and Sis1 (yeast class B J protein) are shown with corresponding PDB codes. Images were prepared in PyMol.
Co-Chaperones Regulate Hsp70 Structure and Activity
The major families of co-chaperones bind to distinct interaction surfaces on Hsp70 (Figures 1 and 2). The J protein co-chaperones bind protein substrates and interact with Hsp70 at lobes IA and IIA of the NBD. This interaction results in an accelerated rate of ATP hydrolysis [34]. The NEF co-chaperones bind lobes IB and IIB of Hsp70’s NBD and facilitate the release of ADP, which has also been shown to accelerate Hsp70’s ATPase rate [35]. TPR domain-containing co-chaperones bind Hsp70’s C-terminus and have been shown to modulate the fates of Hsp70 substrates [36]. Thus, these major families of co-chaperones bind Hsp70 to regulate its enzymatic activity, its choice of substrates and its triage decisions. These systems will be discussed in more detail below.
Approaches to Targeting Hsp70
What is the best way to chemically target Hsp70? One possible approach is to inhibit ATPase activity with competitive nucleotide analogs[20], as has been done with Hsp90 inhibitors [37]. The nucleotide binding cleft of Hsp70 is well defined and relatively deep, suggesting that it might be suitable for development of inhibitors. However, Hsp70 has a relatively tight affinity (mid-nanomolar) for nucleotide, 300-fold better than Hsp90 [38–41]. Because the cellular concentration of ATP is typically 1–5 mM, protein targets with a high affinity for ADP and ATP are much more difficult to inhibit than those with a lower affinity. Further, the ATP binding cassette in Hsp70 is highly homologous in actin and other abundant proteins. Thus, selectivity for the chaperone might be challenging. Despite these challenges, innovative work performed by Vernalis has produced competitive, orthosteric inhibitors of Hsp70, using structure-based design [42]. Consistent with their design, these compounds inhibit cancer cell viability [42] and this group has even been successful at selectively targeting BiP [43]. However, Massey has reported that the path towards orthostatic competitive inhibitors of Hsp70 is quantitatively more challenging than the parallel path to other related targets, such as Hsp90 [41]. Given these hurdles, it seems prudent to pursue additional routes to the design and discovery of potent and selective small molecule modulators targeting Hsp70.
Targeting the substrate binding cleft of Hsp70 is the next logical avenue, given the depth of the site and its known affinity for relatively low molecular mass peptides. This approach has been taken by Chaperone Technologies in their development of antibiotics. For example, a series of 18–20 amino acid peptides, including drosocin, pyrrhocoricin, and apidaecin, are known to interact with DnaK [18]. Of these peptides, pyrrhocoricin exhibited broad-spectrum antibacterial activity. Competition experiments indicated that this peptide has two binding sites on DnaK, one of which is thought to be adjacent to the substrate binding pocket. Interestingly, pyrrhocoricin has activity against bacteria but not mammalian cells [44], suggesting that the SBD could be leveraged to gain selectivity between different homologs of Hsp70. While this work highlights the usefulness of SBD-targeted compounds as antibiotics, it is unclear whether this strategy could be implemented in the development of therapeutics for different Hsp70 related diseases. Of particular interest is whether enough selectivity could be generated in the peptide binding groove to avoid widespread disruption of the proteome.
One promising, unbiased approach has been recently reported by Garrido and colleagues, in which they used a yeast-two hybrid experiment to identify peptide aptamers that bind either the NBD or SBD of Hsp70. These aptamers sensitize cancer cells to anti-cancer drugs in vivo [45, 46], strongly suggesting the potential of this approach. Thus far, it isn’t clear whether these aptamers compete with nucleotide or peptide substrates or whether they have another mechanism of action. Given that these molecules were identified in a cell-based screen, it seems likely that they do not directly compete with the abundant nucleotide or substrates.
Given the significant challenges associated with the targeting of either the nucleotide or substrate binding regions of Hsp70, additional strategies are worth pursuing. A number of additional Hsp70 inhibitors have been identified, but their mechanisms are not known yet [47–49]. To supplement this collection of compounds, targeting the PPIs between Hsp70 and its many co-chaperones may be an effective approach. In the following sections, we discuss the each co-chaperone class in more detail and outline some of the successes and challenges associated with targeting these PPIs.
OPPORTUNITIES FOR DRUG DISCOVERY IN THE HSP70 COMPLEX
J Proteins
J proteins are a class of Hsp70 co-chaperones whose diversity in structure and function are crucial to the flexibility of the Hsp70 machinery. Evolution has dramatically expanded the cellular repertoire of J proteins relative to Hsp70s, such that humans have over 40 J protein encoding genes, but only 13 Hsp70 genes [50, 51]. Moreover, the coexistence of many J proteins within the cytosol and nucleus suggests that they have evolved for distinct functions [52, 53]. All J proteins share a conserved J domain but they diverge in other regions, perhaps providing the functional diversity needed to recruit Hsp70 into many different cellular activities. Accordingly, various J proteins have been linked extensively with a wide array of pathological conditions including cancer, neurodegeneration, muscular dystrophy, and viral infection [54–58]. Thus, J proteins may be interesting pharmacological targets because they have the potential to impact a subset of Hsp70-dependent functions.
The J domain is a highly conserved structure that consists of four α-helices (Figure 1). The J domain interacts directly with the NBD of Hsp70 to stimulate ATP hydrolysis and allosteric conversion into a high affinity substrate binding conformation [59–61]. For the bacterial DnaJ-DnaK interaction, the interface consists of the positively charged helix II of the J domain interacting electrostatically with the negatively charged NBD in lobes IA and IIA [34, 62–64]. Additionally, J domains include an invariant His-Pro-Asp (HPD) motif in the loop between helices II and III that is required for function. Though the overall four-helix architecture of the J domain is largely conserved among J proteins, subtle structural differences suggest that some functional diversity may arise from J domain interactions with Hsp70 [65]. For example, mutants in the NBD of the yeast BiP disrupt interactions with only a subset of available J proteins [66, 67]. Although speculative, these findings suggest that it might be possible to independently target specific J domains at the contact surface with Hsp70.
J proteins have been traditionally grouped into three classes based on structural homology to the Escherichia coli DnaJ (Figure 3A). Class A consists of an N-terminal J domain, a glycine-phenylalanine (G/F) rich region, a zinc finger-like region (ZFLR), a barrel topology C-terminal domain (CTD) and a dimerization domain [68, 69]. Class B has the N-terminal J domain and G/F region, lacks a ZFLR and is more structurally variable at the C-terminus, but often contains two CTDs (CTDI and CTDII) [51]. Class C, the largest class, consists of proteins containing a J domain and no other structural homology to DnaJ. More recently, Kampinga and Craig have provided a revised classification system based more closely on function [69]. This classification represents an important new paradigm in thinking about J proteins and it highlights the major contribution of J proteins to directing the activity of Hsp70.
Specific functions have been described for only some of the individual J proteins and much more work is needed to clarify this area. However, some convincing and illustrative examples include auxilin (DNAJC6), which has a C-terminal J domain and a clathrin-binding domain. This J protein is exclusively involved in the Hsp70-dependent uncoating of clathrin-coated vesicles [70–72], an activity not readily redundant with other J proteins. Similarly, DNAJC7 interacts with both Hsp70 and Hsp90 and seems to play a “recycling” role in the chaperoning of specific substrates, such as the progesterone receptor [73]. In the ER, ERdj3 (DNAJB11) works with BiP to assist with ER-associated degradation (ERAD) [74, 75]. These and other examples [76] lead to a speculative model in which individual J proteins might be responsible for each of Hsp70’s specific functions. In support of this idea, a systematic study of human J proteins found that a subset are able to refold luciferase, while others inhibit aggregation of heat-denatured luciferase [53], further suggesting that these co-chaperones may be specialized.
One prevailing model is that J proteins may bind to substrates and present them to Hsp70. While this concept is likely oversimplified when applied to the large family of J proteins, the interaction of these co-chaperones with substrates seems to play a crucial role in some cases. For example, Lu and coworkers deleted the J domain of Ydj1 (yeast DNAJA1) and found that the remaining portion suppresses rhodanese aggregation on its own [77]. Later work identified a shallow hydrophobic depression on the CTDI of Sis1 (yeast DNAJB1) and found that four point mutants in this domain inhibited luciferase binding and refolding [78]. These studies suggest that J proteins can bind directly to substrates. Further insight into how J proteins bind to their substrates has largely been gained from peptide microarray experiments. These studies have revealed that the prokaryotic DnaJ binds ~8mer peptides enriched in hydrophobic residues [79]. Interestingly, DnaJ does not discriminate between L-peptides and D-peptides, indicating that peptide binding involves side chain interactions [79, 80]. However, a crystal structure of the Ydj1 C-terminus bound to the peptide GWLYEIS suggests that the peptide forms a β-strand alongside a β-sheet in CTDI and several contacts are made with the peptide backbone [81] (Figure 3B). This discrepancy may be due to species differences and the fact that the general rules for J protein-substrate interactions are not yet clear. However, it is reasonable to hypothesize that formation of Hsp70-J protein-substrate complexes may be important in directing Hsp70 to “choose” specific substrates.
The interaction between J proteins and substrates appears to important for several disease-relevant proteins [82]. For example, DNAJB1 and DNAJB6 inhibit the aggregation and toxicity of mHtt, which is involved in Huntington’s disease [83–85]. However, another J protein, DNAJA1, co-localizes with mHtt aggregates [86] and its over-expression increases mHtt aggregation [87]. These observations suggest that individual J proteins, such as DNAJB1 and DNAJA1, might have unique roles in protein quality control. This concept is further illustrated by studies on the Hsp70 substrate, tau [88], in which DNAJB1 inhibits aggregation of tau in vitro [89], while DNAJA1 over-expression causes the proteasomal degradation of tau [90]. Together, these observations suggest that the interactions between J proteins and their substrates might be interesting drug targets, but that more information is needed about how this network is assembled.
While the interactions between J proteins and their substrates have not been pharmacologically targeted, several compounds impact the ability of J proteins to act on Hsp70s. This PPI between Hsp70 and its J proteins is attractive because of the importance of co-chaperone in regulating ATP turnover. The first of the compounds to interfere with this PPI was 15-deoxyspergualin (DSG), a modified natural product that stimulates cytosolic Hsp70 ATP hydrolysis [91–93]. Chemical screens for structurally similar molecules identified R/1, a compound that specifically inhibits the J protein-stimulated ATPase activity of the yeast cytosolic Hsp70, Ssa1 [94] (Figure 4). These findings suggested that drug-like molecules could be identified that alter the allostery between J proteins and Hsp70s. In further support of this idea, an unrelated class of molecules, the sulfogalactosyl ceramide (SGC) mimics, was developed. SGC is a cell surface receptor that binds the NBD of multiple members of the Hsp70 family [95, 96]. Park and coworkers developed a soluble mimic of SGC called adamantylSGC (AdaSGC). AdaSGC inhibits the J protein-stimulated ATPase activity of Hsp70, but not its intrinsic (i.e. unstimulated) activity, suggesting that it may directly inhibit the J domain-Hsp70 interaction [97].
Figure 4.
Structures of chemical modulators of the Hsp70-J protein system.
More recent high throughput screening (HTS) efforts have identified a broader range of compounds that specifically influence J protein-stimulated Hsp70 ATPase activity. For example, screening of a collection of dihydropyrimidines identified three examples, including MAL3-101, which had no effect on intrinsic Hsp70 ATP turnover, but inhibited J protein-stimulated turnover [98]. Subsequent screening and structural studies confirmed this outcome and showed that the dihydropyrimidines bind to a region at the J protein-Hsp70 interface [99–101]. Moreover, these studies also found that some dihydropyrimidines promote J protein activity, while others are inhibitory. For example, 115-7c is able to stimulate the ATPase activity of Hsp70 synergistically with DnaJ [99]. 115-7c binds better to the DnaJ-DnaK complex than DnaK alone and nuclear magnetic resonance (NMR) studies found that 115-7c binds directly adjacent to the J domain-binding site on DnaK. However, the related compound 116-9e, which (similar to MAL3-101) has a diphenyl substitution on the dihydropyrimidine ring, inhibits DnaJ stimulation of ATPase activity, without impacting NEF function [99]. Interestingly, MAL3-101 seems to discriminate between J proteins because it inhibits Ssa1 stimulation by SV40 large T Antigen (TAg), a viral J protein, but had less potent activity against the combination of Ssa1 and Ydj1. This finding suggests that it may be possible to achieve J protein-specific inhibition even by targeting the J protein-Hsp70 interface. MAL3-101 was subsequently found to have potent anti-cancer effects in a multiple myeloma cell line and mouse model [102], while other dihydropyrimidines have been found to control stability of other Hsp70 substrates, including tau, polyglutamines and Akt [48, 73, 103, 104]. This growing body of work suggests that targeting the Hsp70-J protein interface may be a productive approach for guiding Hsp70 functions. Importantly, these compounds are not generally cytotoxic and they do not activate a stress response [48, 103, 104], consistent with the idea that disrupting PPIs in the Hsp70 complex may be relatively well tolerated.
Other chemical series also appear to have activity against the Hsp70-J protein interaction and, interestingly, some of these compounds use mechanisms different than the one used by the dihydropyrimidines. For example, an HTS effort against the DnaK-DnaJ pair identified the flavinoid myricetin, which inhibits DnaJ-stimulated ATPase and substrate binding activities, without affecting intrinsic or NEF stimulated activity [48, 105]. NMR revealed that myricetin binds the NBD in a region between the IB and IA subdomains, which is a more than 20 Å away from the J domain-binding site [105]. However, myricetin blocks binding of DnaJ to DnaK, suggesting that it acts across a long-distance allosteric pathway. Additional HTS efforts have shown that methylene blue (MB) blocks J stimulation of ATP turnover in vitro. However, like myricetin, MB’s effects in cells and animals are complex and it is likely to have targets other than Hsp70s [48, 73, 106]. Despite this complexity, MB and myricetin have clearly shown Hsp70-dependent effects on pathological substrates in cellular and animal models [48, 73, 107] and they reduce Akt levels in cancer cells [104]. Interestingly, these effects are blocked by co-administration of 115-7c, the dihydropyrimidine activator of J protein function [48], further suggesting that the Hsp70-J protein contact is critical. Finally, a larger HTS effort using more than 55,000 compounds identified zafirlukast as an inhibitor of the DnaK-DnaJ combination [108] and a screen of more than 300,000 compounds identified an inhibitor of TAg [109]. The binding site of these molecules is not yet known, but it shows that a screening strategy employing reconstituted chaperone complexes can be used to identify specific inhibitors of a PPI in the Hsp70 system. Like the dihydropyrimidines, some of these are likely orthostatic, while others may be allosteric, like myricetin.
The effects of small molecules on disease-relevant Hsp70 substrates are an initial indication that this is a promising avenue of investigation. However, J protein biology is complex and more work is needed to rationally refine these studies to focus on specific J protein-Hsp70 pairs. More specifically, if a discrete Hsp70-J protein pair can be clearly attributed to a distinct pathobiology, then HTS approaches might be employed to selectively disrupt (or even promote) the key protein-protein interactions.
Nucleotide Exchange Factors (NEFs)
Nucleotide exchange factors (NEF) provide another potential “handle” for targeting the Hsp70 chaperone complex. NEFs bind Hsp70 and help to facilitate the exchange of ADP for ATP. The biochemisty of the NEF family of co-chaperones has classically been investigated using the prokaryotic NEF, GrpE, as a model [110]. However, the eukaryotic cytosol does not contain a GrpE homolog. Rather, there are three main sub-classes of human NEFs: Hsp110, HspBP1, and the BAG proteins, all of which are structurally distinct with little to no sequence homology. Consistent with their diverse structures, they also differ in their mode of binding to Hsp70s and their roles in guiding Hsp70 biology. For example, BAG2 is associated with proteasomal degradation of the Hsp70 substrate, tau, while BAG1-Hsp70 is linked to increased tau stability [111, 112]. These observations suggest that the formation of specific NEF-Hsp70 complexes may help decide the fate of Hsp70-bound substrates. Additionally, these findings illustrate that differential disruption of specific Hsp70-NEF contacts might be beneficial in disease. For example, members of the NEF family are differentially expressed in multiple diseases, including cancer, Alzheimer’s, cardiomyopathies, and ischemia [113–116], highlighting the rationale for developing chemical modulators of NEF-Hsp70.
Hsp110 was originally observed and classified as a heat shock protein based on the appearance of a 110 kDa band in the lysates of Chinese Hamster Ovary (CHO) cells upon heat shock [117]. In humans the major cytosolic Hsp110 protein is called Hsp105 (HSPH1) and it has two major isoforms α and β [118]. Hsp105β results from alternative splicing at exon 12 and lacks 43 amino acids from its C-terminus. Recently, a mutant of Hsp110 that skips exon 9 and results in a truncated form of Hsp110, Hsp110ΔE9, has also been described [119]. This truncated Hsp110ΔE9 is able to act as a dominant negative mutant, abrogating Hsp110 chaperone activity and sensitizing cancer cells to chemotherapy treatments [119]. Since Hsp110 has been shown to protect cancer cells against apoptotic death [120], strategies to block its function or its interactions with Hsp70 could be promising cancer therapies.
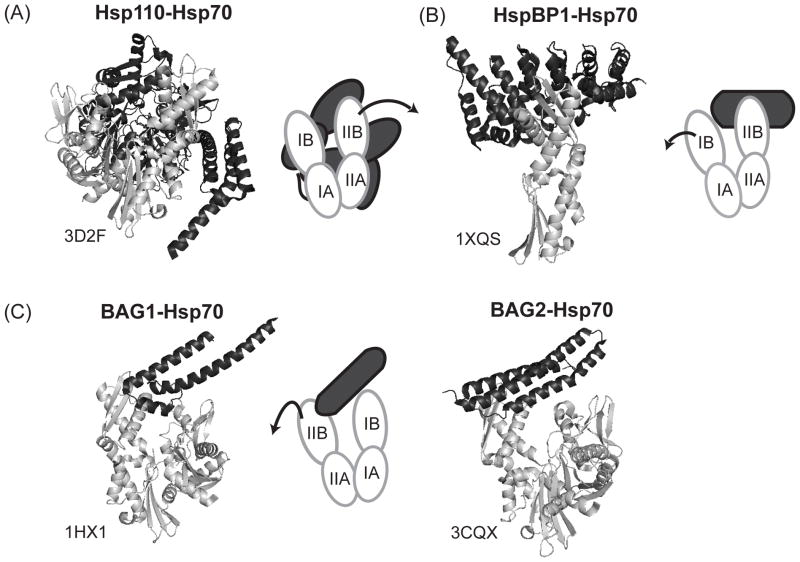
Hsp110 is an evolutionary relative of the Hsp70 family and therefore it has very similar domain architecture, with the main differences including a longer acidic loop region between the β-sandwich and α-helical lid of the SBD and a larger unstructured C-terminal extension [121, 122]. Despite the structural similarity, Hsp110 only functions as a holdase and has no ability to refold substrates without the help of the Hsp70 machinery [122–126]. Furthermore, while Hsp110 homologs bind nucleotide, this function seems to be dispensable for their NEF activity [127]. The crystal structure of the complex between Hsp70 and yeast Hsp110, Sse1, shows that the interaction covers a large surface area involving their respective NBDs [128, 129]. This interaction between Hsp70 and Hsp110 causes several rotations in Hsp70’s NBD, especially in lobe IIB [130], allowing ADP release (Figure 5).
Figure 5.
Structures of Hsp70-NEF complexes. (A) Crystal structure of yeast Hsp110, Sse1, and human Hsp70 NBD. Complex formation between Hsp70 and Hsp110 leads to a rotation in lobe IIB allowing nucleotide release. (B) Crystal structure of HspBP1 and lobe II of Hsp70’s NBD. HspBP1 wraps around lobe IIB displacing lobe I and opening the nucleotide cleft. (C) Crystal structures of Hsp70 NBD in complex with the BAG domain of BAG1 and BAG2. Association between Hsp70 and the BAG proteins cause an outward rotation of lobe II, promoting nucleotide exchange. In all figures Hsp70 is colored in light grey and NEFs are colored in dark grey with PDB codes indicated. Images were prepared in PyMol.
The large buried surface area between Hsp70 and Hsp110 may make targeting this interaction difficult. The problem in PPI systems like this is that binding energy is often distributed across a large and complex topology, precluding easy inhibition by small (<500 Da) molecules. However, inhibiting PPIs with large surface areas is not unprecedented and compounds with potency values in the low nM range have been reported [131]. A common feature of previous successful strategies is that the small molecules tend to target so-called “hotspots” of the PPI, meaning the inhibitor binds in a region on one partner containing a small number of residues that are responsible for the majority of the binding strength [132, 133]. Thus, it will be important to identify residues that are critical to the Hsp70-NEF interaction. Another common feature of successful PPI inhibitors is that they bind in allosteric sites to impact the topology of protein-protein contact surfaces from a distance [133]. This approach lets the small molecule bind in a relatively concise pocket and impact larger surfaces to block PPIs. It seems likely that similar mechanisms will need to be employed to target the Hsp110-Hsp70 interaction.
Similar issues are important in considering the potential for inhibition of the other major classes of NEFs. For example, HspBP1 is a 40 kDa protein that is composed of two structural domains, a largely unstructured N-terminal domain and a C-terminal domain that is mostly α-helical and is responsible for HspBP1 binding to Hsp70 [134]. This C-terminal region has been shown to be sufficient for eliciting Hsp70 nucleotide release [134, 135] and co-crystal structures suggest that HspBP1’s C-terminal domain interacts with lobe II of Hsp70’s NBD (Figure 5) [135]. Importantly, this interaction is not the same as the PPI between Hsp70 and Hsp110, suggesting that this contact might be selectively inhibited. This goal might be attractive because of HspBP1’s links to cancer and chemotherapy resistance [136].
Additional lessons about how to potentially target the Hsp70-NEF interaction are illustrated by the BAG family of co-chaperones, which includes BAG1-6. BAG proteins are defined by a characteristic C-terminal BAG domain that binds lobe IB and IIB of Hsp70’s NBD and facilitates nucleotide release [137, 138]. This BAG domain typically consists of 110 to 124 amino acids and forms a three-helix bundle with the second and third helices providing the binding interface for Hsp70 [35, 139]. The association between the BAG domain and Hsp70 causes a 14° rotation in lobe II, which results in an opening of the nucleotide binding cleft and promotes ADP release (Figure 5) [35]. Interestingly, while all BAG proteins interact with Hsp70 through their conserved BAG domains, their N-terminal region is highly variable. This diversity is likely to be key for pathway specificity and BAG proteins may use these domains to determine the timing and location of nucleotide-dependent delivery of Hsp70-bound cargo.
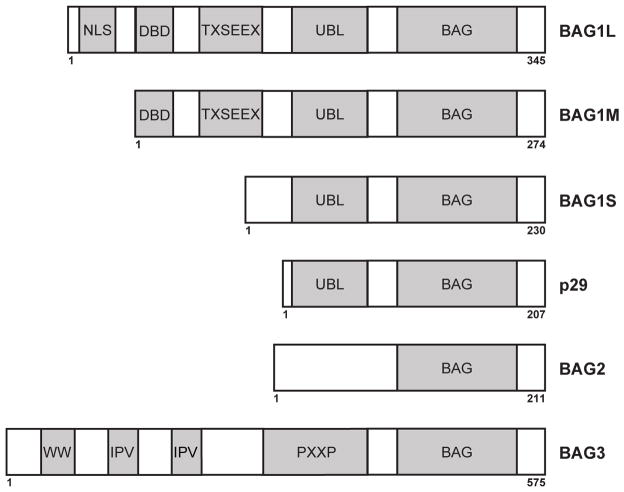
BAG1 is the founding member of the BAG protein family. It was initially discovered by two independent research groups using immunochemical screening methods to identify interacting partners of the anti-apoptotic protein Bcl-2 and the glucocorticoid receptor, respectively [140, 141]. The former researchers entitled their protein Bcl-2-associated AnthanoGene-1 (BAG1) [140]. Four human BAG1 isoforms are expressed through alternative initiation sites and are designated BAG1L (p50, Hap50), BAG1M (p46, Rap46, Hap46), BAG1S (p36, Hap33), and p29 (Hap29) [142]. BAG1S is the most abundant isoform expressed in cells, followed by BAG1L and BAG1M, while p29 is not consistently detected [143]. BAG1 isoforms share a common C-terminus, containing the BAG domain and an Ubiquitin-like (UBL) domain, while their N-termini differ based on the translation initiation site (Figure 6). Besides their BAG and UBL domains, longer isoforms of BAG1 (M & L) also contain TXSEEX repeats, a DNA-binding domain (DBD), and BAG1L contains a nuclear localization signal (NLS). These various domains help to dictate interacting partners as well as cellular function and localization of each BAG1 isoform (for review see [144]).
Figure 6.
Domain architecture of the BAG family of co-chaperones.
BAG1 regulates the fate of Hsp70-bound substrates. For example, the UBL domain of BAG1 allows for BAG1-Hsp70 complexes to associate with the proteasome and promotes the degradation of specific substrates such as the glucocorticoid receptor, BCR-ABL and Htt [145–147]. However, BAG1 has also been shown to inhibit proteasomal degradation of other Hsp70 substrates, such as tau [112]. These observations suggest that chemically targeting BAG1-Hsp70 complexes could be used to reshape the proteome. Work towards that goal been reported by Sharp et. al, in which they performed a screen for inhibitors of the BAG1-Hsp70 interaction using GST pulldowns. After hit validation, NSC71948 (Thioflavin S), was selected for further study [148]. This compound inhibits ERK phosphorylation and growth of ZR-75-1 human breast cancer cells. Another promising scaffold was identified by Leu et. al, in which they identified PES in a screen for anti-cancer compounds that impact p53-mediated apoptosis. Later, they found that PES disrupts Hsp70 co-chaperone complexes including its association with BAG1 [149]. The preliminary successes of these screening approaches suggest that targeting a BAG-Hsp70 complex is both feasible and beneficial, however, further studies are still needed. For example, the binding sites and mechanisms of these molecules are not yet clear.
The BAG family members BAG2 and BAG3 were identified in a yeast two hybrid screen with the NBD of Hsp70 as bait and were named based on their structural and functional similarity to BAG1 [137]. The crystal structure of its BAG domain revealed that BAG2 does not adopt the canonical three-helix bundle and instead forms a dimeric structure with each monomer consisting of only two long antiparallel helices [150]. Intriguingly BAG2 has also been shown to independently bind an Hsp70 client substrate, CFTR (cystic fibrosis transmembrane conductance regulator), and inhibit its proteasomal degradation [151]. Like BAG1, however, BAG2 regulation of Hsp70 function is substrate specific. BAG2 has also been shown to increase the proteasomal degradation of tau in an ubiquitin-independent manner [111].
BAG3 is one of the largest BAG proteins and it contains a WW domain, a proline rich region containing multiple PXXP motifs, and two IPV motifs, which have recently been shown to facilitate interactions between BAG3 and certain small heat shock proteins (Figure 6). In comparison to other BAG proteins, BAG3 is unique in that it is the only member induced under stress conditions, mainly through activation of heat shock factor 1 (HSF1) [152]. HSF1 is required for tumor initiation and maintenance in a variety of cancer models, which suggests a role for BAG3 in tumor formation [153]. In support of this notion, it has been shown that the BAG3-Hsp70 complex stabilizes a number of key oncogenes, suppressing apoptosis [154–158]. Accordingly, silencing of BAG3 in multiple tumor lines sensitizes the cells to chemotherapy, suggesting that the BAG3-Hsp70 complex is an especially attractive drug target [157].
One of the major questions in this field is whether the structural differences between the major NEF classes can be exploited to produce selective inhibitors of the various families (Figure 5). Similarly, can different members of the BAG family be individually targeted? Further, it is not yet clear how many NEF functions are dependent on Hsp70 and how many are independent.
Tetratricopeptide Repeat (TPR) Domain-Containing Proteins
Hsp70 also cooperates with a number of TPR domain-containing proteins. The TPR motif is defined by a degenerate 34 amino acid sequence that forms an amphipathic antiparallel α-helix [38, 159–162] and a TPR domain is typically assembled from 3 to 16 tandem TPR motifs. Although first identified in subunits of the anaphase promoting complex [163, 164], the TPR domain has since been found to be a common feature of protein-protein interactions, including those with Hsp70 co-chaperones.
Members of the family of TPR co-chaperones, as a whole, share little homology outside their TPR domains and they typically have regions involved in functions unrelated to Hsp70/Hsp90 binding [38, 159–162]. For example, the TPR co-chaperone CHIP (carboxyl terminus of Hsc70 interacting protein) is an ubiquitin E3 ligase with an effector Ubox domain [165]. This co-chaperone directs ubiquitination of Hsp70-bound substrates, marking them for proteasome-mediated degradation [166, 167]. In contrast, the TPR co-chaperone Hop (Hsp70/Hsp90 organizing protein) has three TPR domains: TPR1, TPR2A, and TPR2B. Of these domains, TPR1 and TPR2A mediate the association with Hsp70 and Hsp90, respectively [168, 169]. Thus, Hop facilitates the coordination of Hsp70 and Hsp90, ultimately allowing for the transfer of substrate between these two chaperone systems [170, 171]. This coordination allows Hop to play a central role in the folding of non-native protein substrates, such as nuclear hormone receptors [172, 173]. Thus, when Hop and CHIP compete for binding to Hsp70 through their TPR domains, they establish a choice between two opposing fates: folding vs. degradation. These findings clarify our understanding of the combinatorial assembly of Hsp70 complexes, in which mutually exclusive binding of Hsp70 to specific co-chaperones dictates the fate of substrates [174–176]. Taken together, these features suggest that chaperone complexes may have the potential to be chemically modulated in order to “tune” the proteome.
TPR co-chaperones interact with the intrinsically disordered C-terminus of Hsp70. Mutagenesis studies [169, 177, 178] and co-crystal structures of the TPR domains of Hop and CHIP with Hsp70 C-terminal peptides [168, 179] illustrate the importance of the C-terminal EEVD-COOH amino acids in mediating these PPIs [177, 180]. Based on these findings, the EEVD motif of Hsp70 has been generalized as the minimal binding site for TPR co-chaperones. This motif is also present in the extreme C-terminus of the evolutionarily unrelated molecular chaperone Hsp90, but not in the prokaryotic DnaK, mitochondrial or ER-resident Hsp70 homologs. These observations highlight the role of the EEVD motif as a recruitment element that anchors TPR co-chaperones to the cytoplasmic Hsp70 and Hsp90 chaperone systems. However, there is not much known about how TPR co-chaperones “compete” for binding to Hsp70. Thus, compounds that block the EEVD-TPR interaction might be exciting probes for understanding chaperone biology and these compounds may serve as leads for drug discovery.
The importance of EEVD-TPR domain contacts in facilitating PPIs between Hsp70/90 and TPR co-chaperones is well appreciated within the chaperone field. However, much less attention has been paid to interaction surfaces outside this canonical binding site. Immunoprecipitation experiments as well as in vitro binding studies performed on the Hsp70-Hop complex, demonstrate that binding involves secondary contacts outside the EEVD motif [169, 181]. Additionally, sequences outside the TPR domain of Hop, CHIP, and other TPR co-chaperones cause differential binding to Hsp90 mutants [182, 183]. Together, these findings suggest that interactions between TPR proteins and Hsp70 are more complex than the minimal TPR-EEVD interactions. Thus, many key fundamental questions remain unanswered: What are the molecular interactions between Hsp70 and TPR co-chaperones? Do these interactions differ among TPR co-chaperones? What molecular events influence the choice to bind one TPR protein over another? Because TPR co-chaperone structures are divergent in nature, additional contacts outside the EEVD-TPR binding site may provide an avenue for the development of chemical probes that can modulate specific TPR-chaperone interactions. Such compounds would be useful in further dissecting the complex mechanisms of Hsp70 and individual TPR co-chaperones in protein quality control.
The development of small molecule modulators of Hsp70-TPR complexes is still in its infancy. However, in the Hsp90 system, Yi and co-workers have targeted the TPR domain of Hop and identified pyrimidotriazinediones as inhibitors of that PPI [184]. Additionally, derivatives of the natural product sansalvamide A have been shown to modulate Hsp90 interactions with TPR co-chaperones [185, 186]. Taken together, this work suggests that the Hsp70-TPR interactions may also be amenable to inhibition. However, further studies are still needed because the binding sites and mechanisms of these molecules are not yet clear. Compared to the other PPIs (e.g. J proteins and NEFs), the interactions between TPR domains and Hsp70s are relatively more concise, which might accelerate discovery in that area. The challenges will be in understanding how to engender selectivity and guide the “choice” of TPR partner.
CONCLUSIONS
There are compelling reasons to target the PPIs between Hsp70 and its co-chaperones. These contacts help shape Hsp70 activities and, as such, they might be targeted to re-direct the protein quality control system. Molecules that disrupt the assembly and disassembly of the Hsp70 complex might supplement other types of Hsp70 inhibitors, such as competitive inhibitors of ATP and substrate binding, providing a more complete suite of chemical probes and potential therapeutics. However, the number of PPIs in the Hsp70 complex means that there are a large number of contacts yet to be explored.
PPIs are notoriously difficult to inhibit and the specific interactions involved in binding to Hsp70 are particularly challenging, given their large buried surface areas. What strategies might be used to disrupt these contacts? Based on growing evidence from other PPI inhibitors discovery programs [22–24], it seems likely that compounds that are able to bind to allosteric sites might be in the best position to target the types of PPIs in the Hsp70 system. Another key tool will likely be the development of HTS platforms that are specifically suited to finding inhibitors of PPIs. Recent developments in this area, including AlphaLisa, flow cytometry protein interaction assay (FCPIA) and gray box screening [105, 108], might lower the barrier to uncovering suitable compounds. Also, the creation of chemical libraries enriched for more complex small molecules (e.g. natural product-like, etc) may further accelerate discovery in this area [187]. A clever combination of these methods might overcome the challenges associated with targeting the Hsp70 complex.
One major question that looms large over this field is how the global proteome will respond to inhibitors of Hsp70 (both orthostatic and allosteric). This concept has not been rigorously tested and it remains uncertain how cells will respond to different types of Hsp70 inhibitors. What will happen to protein stability and turnover when Hsp70 function is blocked or even “tuned”? The answers to this question may depend on how the molecule works (e.g. competitive inhibitor of ATP binding, allosteric inhibitor of J proteins, etc.) and whether it is selective for specific Hsp70 paralogs. It seems likely that the only way to address these significant concerns is to develop potent inhibitors and then use them to develop empirical models.
Literature Cited
- 1.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–51. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Ann Rev Biochem. 2001;70:603–47. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 5.Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–10. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 6.Arndt V, Rogon C, Hohfeld J. To be, or not to be--molecular chaperones in protein degradation. Cell Mol Life Sci. 2007;64:2525–41. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kettern N, Dreiseidler M, Tawo R, Hohfeld J. Chaperone-assisted degradation: multiple paths to destruction. Biol Chem. 2010;391:481–9. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 8.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228:111–33. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 9.Gamer J, Bujard H, Bukau B. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell. 1992;69:833–42. doi: 10.1016/0092-8674(92)90294-m. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez F, Arsene-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol Cell. 2008;32:347–58. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–88. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 12.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–10. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 13.Brocchieri L, Conway de Macario E, Macario AJ. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol. 2008;8:19. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–90. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meimaridou E, Gooljar SB, Chapple JP. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 16.Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583:2647–53. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 17.Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–18. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- 18.Otvos L, OI, Rogers ME, Consolvo PJ, Condie BA, Lovas S, Bulet P, Blaszczyk-Thurin M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000;39:14150–9. doi: 10.1021/bi0012843. [DOI] [PubMed] [Google Scholar]
- 19.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem. 2010;53:4585–602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodsky JL, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6:1215–25. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 21.Patury S, Miyata Y, Gestwicki JE. Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem. 2009;9:1337–51. doi: 10.2174/156802609789895674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gestwicki JE, Marinec PS. Chemical control over protein-protein interactions: beyond inhibitors. Comb Chem High Throughput Screen. 2007;10:667–75. doi: 10.2174/138620707782507296. [DOI] [PubMed] [Google Scholar]
- 23.Berg T. Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed Engl. 2003;42:2462–81. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 24.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 25.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci. 2009;106:8471–6. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci. 1992;89:7290–4. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Kurochkin AV, Pang Y, Hu W, Flynn GC, Zuiderweg ER. NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction. Biochemistry. 1998;37:7929–40. doi: 10.1021/bi9800855. [DOI] [PubMed] [Google Scholar]
- 28.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–14. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young JC, Barral JM, Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends in biochemical sciences. 2003;28:541–7. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Gaestel M. Molecular chaperones in signal transduction. Handbook of Experimental Pharmacology. 2006:93–109. doi: 10.1007/3-540-29717-0_4. [DOI] [PubMed] [Google Scholar]
- 31.Mayer MP, Schroder H, Rudiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol. 2000;7:586–93. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- 32.Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006;281:38705–11. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 33.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK) J Biol Chem. 2010;285:21282–91. doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B, Bertelsen EB, Zuiderweg ER. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc Natl Acad Sci. 2011;108:18966–71. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–7. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 36.Liu FH, Wu SJ, Hu SM, Hsiao CD, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem. 1999;274:34425–32. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- 37.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 38.Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress & Chaperones. 2011;16:353–67. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borges JC, Ramos CHI. Spectroscopic and thermodynamic measurements of nucleotide-induced changes in the human 70-kDa heat shock cognate protein. Arch Biochem Biophys. 2006;452:46–54. doi: 10.1016/j.abb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Williamson DS, Borgognoni J, Clay A, Daniels Z, Dokurno P, Drysdale MJ, Foloppe N, Francis GL, Graham CJ, Howes R, Macias AT, Murray JB, Parsons R, Shaw T, Surgenor AE, Terry L, Wang YK, Wood M, Massey AJ. Novel Adenosine-Derived Inhibitors of 70 kDa Heat Shock Protein, Discovered Through Structure-Based Design. J Med Chem. 2009;52:1510–3. doi: 10.1021/jm801627a. [DOI] [PubMed] [Google Scholar]
- 41.Massey AJ. ATPases as drug targets: insights from heat shock proteins 70 and 90. J Med Chem. 2010;53:7280–6. doi: 10.1021/jm100342z. [DOI] [PubMed] [Google Scholar]
- 42.Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, Macias AT, Daniels Z, Geoffroy S, Dopson M, Lavan P, Matassova N, Francis GL, Graham CJ, Parsons R, Wang Y, Padfield A, Comer M, Drysdale MJ, Wood M. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother Pharmacol. 2011;66:535–45. doi: 10.1007/s00280-009-1194-3. [DOI] [PubMed] [Google Scholar]
- 43.Macias AT, Williamson DS, Allen N, Borgognoni J, Clay A, Daniels Z, Dokurno P, Drysdale MJ, Francis GL, Graham CJ, Howes R, Matassova N, Murray JB, Parsons R, Shaw T, Surgenor AE, Terry L, Wang Y, Wood M, Massey AJ. Adenosine-derived inhibitors of 78 kDa glucose regulated protein (Grp78) ATPase: insights into isoform selectivity. J Med Chem. 2010;54:4034–41. doi: 10.1021/jm101625x. [DOI] [PubMed] [Google Scholar]
- 44.Kragol G, Lovas S, Varadi G, Condie BA, Hoffmann R, Otvos L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry. 2001;40:3016–26. doi: 10.1021/bi002656a. [DOI] [PubMed] [Google Scholar]
- 45.Rerole AL, Gobbo J, De Thonel A, Schmitt E, Pais de Barros JP, Hammann A, Lanneau D, Fourmaux E, Deminov O, Micheau O, Lagrost L, Colas P, Kroemer G, Garrido C. Peptides and aptamers targeting HSP70: a novel approach for anticancer chemotherapy. Cancer Res. 2011;71:484–95. doi: 10.1158/0008-5472.CAN-10-1443. [DOI] [PubMed] [Google Scholar]
- 46.Rerole AL, Jego G, Garrido C. Hsp70: anti-apoptotic and tumorigenic protein. Methods Mol Biol. 2011;787:205–30. doi: 10.1007/978-1-61779-295-3_16. [DOI] [PubMed] [Google Scholar]
- 47.Williams DR, Ko SK, Park S, Lee MR, Shin I. An apoptosis-inducing small molecule that binds to heat shock protein 70. Angew Chem Int Ed Engl. 2008;47:7466–9. doi: 10.1002/anie.200802801. [DOI] [PubMed] [Google Scholar]
- 48.Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, O’Leary J, Morgan D, Lee DC, Shults CL, Rousaki A, Weeber EJ, Zuiderweg ER, Gestwicki JE, Dickey CA. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J Neurosci. 2009;29:12079–88. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cellitti J, Zhang Z, Wang S, Wu B, Yuan H, Hasegawa P, Guiney DG, Pellecchia M. Small molecule DnaK modulators targeting the beta-domain. Chem Biol Drug Des. 2009;74:349–57. doi: 10.1111/j.1747-0285.2009.00869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–70. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–11. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–55. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 53.Hageman J, van Waarde MA, Zylicz A, Walerych D, Kampinga HH. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem J. 2011;435:127–42. doi: 10.1042/BJ20101247. [DOI] [PubMed] [Google Scholar]
- 54.Sterrenberg JN, Blatch GL, Edkins AL. Human DNAJ in cancer and stem cells. Cancer Lett. 2011;312:129–42. doi: 10.1016/j.canlet.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 55.Kalia SK, Kalia LV, McLean PJ. Molecular chaperones as rational drug targets for Parkinson’s disease therapeutics. CNS Neurol Disord Drug Targets. 2010;9:741–53. doi: 10.2174/187152710793237386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitra A, Shevde LA, Samant RS. Multi-faceted role of HSP40 in cancer. Clin Exp Metastasis. 2009;26:559–67. doi: 10.1007/s10585-009-9255-x. [DOI] [PubMed] [Google Scholar]
- 57.Harms MB, Sommerville RB, Allred P, Bell S, Ma D, Cooper P, Lopate G, Pestronk A, Weihl CC, Baloh RH. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann Neurol. 2012;71:407–16. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knox C, Luke GA, Blatch GL, Pesce ER. Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle. Virus Res. 2011;160:15–24. doi: 10.1016/j.virusres.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 59.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–9. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 60.Wall D, Zylicz M, Georgopoulos C. The conserved G/F motif of the DnaJ chaperone is necessary for the activation of the substrate binding properties of the DnaK chaperone. J Biol Chem. 1995;270:2139–44. doi: 10.1074/jbc.270.5.2139. [DOI] [PubMed] [Google Scholar]
- 61.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A. 1994;91:10345–9. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci U S A. 1998;95:6108–13. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci U S A. 1998;95:15229–34. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci U S A. 1998;95:15223–8. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14:1697–709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Awad W, Estrada I, Shen Y, Hendershot LM. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc Natl Acad Sci U S A. 2008;105:1164–9. doi: 10.1073/pnas.0702132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vembar SS, Jonikas MC, Hendershot LM, Weissman JS, Brodsky JL. J domain co-chaperone specificity defines the role of BiP during protein translocation. J Biol Chem. 2010;285:22484–94. doi: 10.1074/jbc.M110.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holstein SE, Ungewickell H, Ungewickell E. Mechanism of clathrin basket dissociation: separate functions of protein domains of the DnaJ homologue auxilin. J Cell Biol. 1996;135:925–37. doi: 10.1083/jcb.135.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xing Y, Bocking T, Wolf M, Grigorieff N, Kirchhausen T, Harrison SC. Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J. 2010;29:655–65. doi: 10.1038/emboj.2009.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bocking T, Aguet F, Harrison SC, Kirchhausen T. Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat Struct Mol Biol. 2011;18:295–301. doi: 10.1038/nsmb.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang AM, Morishima Y, Clapp KM, Peng HM, Pratt WB, Gestwicki JE, Osawa Y, Lieberman AP. Inhibition of hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J Biol Chem. 2010;285:15714–23. doi: 10.1074/jbc.M109.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakanishi K, Kamiguchi K, Torigoe T, Nabeta C, Hirohashi Y, Asanuma H, Tobioka H, Koge N, Harada O, Tamura Y, Nagano H, Yano S, Chiba S, Matsumoto H, Sato N. Localization and function in endoplasmic reticulum stress tolerance of ERdj3, a new member of Hsp40 family protein. Cell Stress Chaperones. 2004;9:253–64. doi: 10.1379/CSC-52.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan CY, Lee S, Ren HY, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell. 2004;15:761–73. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–8. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 78.Lee S, Fan CY, Younger JM, Ren H, Cyr DM. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J Biol Chem. 2002;277:21675–82. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- 79.Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–50. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feifel B, Schonfeld HJ, Christen P. D-peptide ligands for the co-chaperone DnaJ. J Biol Chem. 1998;273:11999–2002. doi: 10.1074/jbc.273.20.11999. [DOI] [PubMed] [Google Scholar]
- 81.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–83. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 82.Arawaka S, Machiya Y, Kato T. Heat shock proteins as suppressors of accumulation of toxic prefibrillar intermediates and misfolded proteins in neurodegenerative diseases. Curr Pharm Biotechnol. 2010;11:158–66. doi: 10.2174/138920110790909713. [DOI] [PubMed] [Google Scholar]
- 83.Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9:2009–18. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 84.Zhou H, Li SH, Li XJ. Chaperone suppression of cellular toxicity of huntingtin is independent of polyglutamine aggregation. J Biol Chem. 2001;276:48417–24. doi: 10.1074/jbc.M104140200. [DOI] [PubMed] [Google Scholar]
- 85.Chuang JZ, Zhou H, Zhu M, Li SH, Li XJ, Sung CH. Characterization of a brain-enriched chaperone, MRJ, that inhibits Huntingtin aggregation and toxicity independently. J Biol Chem. 2002;277:19831–8. doi: 10.1074/jbc.M109613200. [DOI] [PubMed] [Google Scholar]
- 86.Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13:1389–405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 87.Wyttenbach A, Carmichael J, Swartz J, Furlong RA, Narain Y, Rankin J, Rubinsztein DC. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington’s disease. Proc Natl Acad Sci U S A. 2000;97:2898–903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miyata Y, Koren J, Kiray J, Dickey CA, Gestwicki JE. Molecular chaperones and regulation of tau quality control: strategies for drug discovery in tauopathies. Future Med Chem. 2011;3:1523–37. doi: 10.4155/fmc.11.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sahara N, Maeda S, Yoshiike Y, Mizoroki T, Yamashita S, Murayama M, Park JM, Saito Y, Murayama S, Takashima A. Molecular chaperone-mediated tau protein metabolism counteracts the formation of granular tau oligomers in human brain. J Neurosci Res. 2007;85:3098–108. doi: 10.1002/jnr.21417. [DOI] [PubMed] [Google Scholar]
- 90.Abisambra JF, Jinwal UK, Suntharalingam A, Arulselvam K, Brady S, Cockman M, Jin Y, Zhang B, Dickey CA. DnaJA1 Antagonizes Constitutive Hsp70-Mediated Stabilization of Tau. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nadler SG, Tepper MA, Schacter B, Mazzucco CE. Interaction of the immunosuppressant deoxyspergualin with a member of the Hsp70 family of heat shock proteins. Science. 1992;258:484–6. doi: 10.1126/science.1411548. [DOI] [PubMed] [Google Scholar]
- 92.Brodsky JL. Selectivity of the molecular chaperone-specific immunosuppressive agent 15-deoxyspergualin: modulation of Hsc70 ATPase activity without compromising DnaJ chaperone interactions. Biochem Pharmacol. 1999;57:877–80. doi: 10.1016/s0006-2952(98)00376-1. [DOI] [PubMed] [Google Scholar]
- 93.Nadeau K, Nadler SG, Saulnier M, Tepper MA, Walsh CT. Quantitation of the interaction of the immunosuppressant deoxyspergualin and analogs with Hsc70 and Hsp90. Biochemistry. 1994;33:2561–7. doi: 10.1021/bi00175a027. [DOI] [PubMed] [Google Scholar]
- 94.Fewell SW, Day BW, Brodsky JL. Identification of an inhibitor of hsc70-mediated protein translocation and ATP hydrolysis. J Biol Chem. 2001;276:910–4. doi: 10.1074/jbc.M008535200. [DOI] [PubMed] [Google Scholar]
- 95.Mamelak D, Mylvaganam M, Whetstone H, Hartmann E, Lennarz W, Wyrick PB, Raulston J, Han H, Hoffman P, Lingwood CA. Hsp70s contain a specific sulfogalactolipid binding site. Differential aglycone influence on sulfogalactosyl ceramide binding by recombinant prokaryotic and eukaryotic hsp70 family members. Biochemistry. 2001;40:3572–82. doi: 10.1021/bi001643u. [DOI] [PubMed] [Google Scholar]
- 96.Mamelak D, Lingwood C. The ATPase domain of hsp70 possesses a unique binding specificity for 3′-sulfogalactolipids. J Biol Chem. 2001;276:449–56. doi: 10.1074/jbc.M006732200. [DOI] [PubMed] [Google Scholar]
- 97.Park HJ, Mylvaganum M, McPherson A, Fewell SW, Brodsky JL, Lingwood CA. A soluble sulfogalactosyl ceramide mimic promotes Delta F508 CFTR escape from endoplasmic reticulum associated degradation. Chem Biol. 2009;16:461–70. doi: 10.1016/j.chembiol.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fewell SW, Smith CM, Lyon MA, Dumitrescu TP, Wipf P, Day BW, Brodsky JL. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J Biol Chem. 2004;279:51131–40. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 99.Wisen S, Bertelsen EB, Thompson AD, Patury S, Ung P, Chang L, Evans CG, Walter GM, Wipf P, Carlson HA, Brodsky JL, Zuiderweg ER, Gestwicki JE. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem Biol. 2010;5:611–22. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–76. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 101.Wisen S, Androsavich J, Evans CG, Chang L, Gestwicki JE. Chemical modulators of heat shock protein 70 (Hsp70) by sequential, microwave-accelerated reactions on solid phase. Bioorg Med Chem Lett. 2008;18:60–5. doi: 10.1016/j.bmcl.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 102.Braunstein MJ, Scott SS, Scott CM, Behrman S, Walter P, Wipf P, Coplan JD, Chrico W, Joseph D, Brodsky JL, Batuman O. Antimyeloma Effects of the Heat Shock Protein 70 Molecular Chaperone Inhibitor MAL3–101. J Oncol. 2011;2011:232037. doi: 10.1155/2011/232037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walter GM, Smith MC, Wisen S, Basrur V, Elenitoba-Johnson KS, Duennwald ML, Kumar A, Gestwicki JE. Ordered assembly of heat shock proteins, Hsp26, Hsp70, Hsp90, and Hsp104, on expanded polyglutamine fragments revealed by chemical probes. J Biol Chem. 2011;286:40486–93. doi: 10.1074/jbc.M111.284448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koren J, 3rd, Jinwal UK, Jin Y, O’Leary J, Jones JR, Johnson AG, Blair LJ, Abisambra JF, Chang L, Miyata Y, Cheng AM, Guo J, Cheng JQ, Gestwicki JE, Dickey CA. Facilitating Akt clearance via manipulation of Hsp70 activity and levels. J Biol Chem. 2010;285:2498–505. doi: 10.1074/jbc.M109.057208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang L, Miyata Y, Ung PM, Bertelsen EB, McQuade TJ, Carlson HA, Zuiderweg ER, Gestwicki JE. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol. 2011;18:210–21. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Congdon EE, Wu JW, Myeku N, Figueroa YH, Herman M, Marinec PS, Gestwicki JE, Dickey CA, Yu WH, Duff K. Methylthioninium chloride (methylene blue) induces autophagy and attenuates tauopathy in vitro and in vivo. Autophagy. 2012:8. doi: 10.4161/auto.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O’Leary JC, 3rd, Li Q, Marinec P, Blair LJ, Congdon EE, Johnson AG, Jinwal UK, Koren J, 3rd, Jones JR, Kraft C, Peters M, Abisambra JF, Duff KE, Weeber EJ, Gestwicki JE, Dickey CA. Phenothiazine-mediated rescue of cognition in tau transgenic mice requires neuroprotection and reduced soluble tau burden. Mol Neurodegener. 2010;5:45. doi: 10.1186/1750-1326-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miyata Y, Chang L, Bainor A, McQuade TJ, Walczak CP, Zhang Y, Larsen MJ, Kirchhoff P, Gestwicki JE. High-throughput screen for Escherichia coli heat shock protein 70 (Hsp70/DnaK): ATPase assay in low volume by exploiting energy transfer. J Biomol Screen. 2010;15:1211–9. doi: 10.1177/1087057110380571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seguin SP, Evans CW, Nebane-Akah M, McKellip S, Ananthan S, Tower NA, Sosa M, Rasmussen L, White EL, Maki BE, Matharu DS, Golden JE, Aube J, Brodsky JL, Noah JW. High-throughput screening identifies a bisphenol inhibitor of SV40 large T antigen ATPase activity. J Biomol Screen. 2012;17:194–203. doi: 10.1177/1087057111421630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harrison C. GrpE, a nucleotide exchange factor for DnaK. Cell Stress & Chaperones. 2003;8:218–24. doi: 10.1379/1466-1268(2003)008<0218:ganeff>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carrettiero DC, Hernandez I, Neveu P, Papagiannakopoulos T, Kosik KS. The cochaperone BAG2 sweeps paired helical filament- insoluble tau from the microtubule. J Neurosci. 2009;29:2151–61. doi: 10.1523/JNEUROSCI.4660-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Elliott E, Tsvetkov P, Ginzburg I. BAG-1 associates with Hsc70. Tau complex and regulates the proteasomal degradation of Tau protein. J Biol Chem. 2007;282:37276–84. doi: 10.1074/jbc.M706379200. [DOI] [PubMed] [Google Scholar]
- 113.Souza AP, Albuquerque C, Torronteguy C, Frasson A, Maito F, Pereira L, Duval da Silva V, Zerwes F, Raynes D, Guerriero V, Bonorino C. HspBP1 levels are elevated in breast tumor tissue and inversely related to tumor aggressiveness. Cell Stress & Chaperones. 2009;14:301–10. doi: 10.1007/s12192-008-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Elliott E, Laufer O, Ginzburg I. BAG-1M is up-regulated in hippocampus of Alzheimer’s disease patients and associates with tau and APP proteins. J Neurochem. 2009;109:1168–78. doi: 10.1111/j.1471-4159.2009.06047.x. [DOI] [PubMed] [Google Scholar]
- 115.Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–55. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 116.Nakamura J, Fujimoto M, Yasuda K, Takeda K, Akira S, Hatayama T, Takagi Y, Nozaki K, Hosokawa N, Nagata K. Targeted disruption of Hsp110/105 gene protects against ischemic stress. Stroke. 2008;39:2853–9. doi: 10.1161/STROKEAHA.107.506188. [DOI] [PubMed] [Google Scholar]
- 117.Subjeck JR, Sciandra JJ, Chao CF, Johnson RJ. Heat shock proteins and biological response to hyperthermia. British J Cancer Supplement. 1982;5:127–31. [PMC free article] [PubMed] [Google Scholar]
- 118.Ishihara K, Yasuda K, Hatayama T. Molecular cloning, expression and localization of human 105 kDa heat shock protein, hsp105. Biochim Biophys Acta. 1999;1444:138–42. doi: 10.1016/s0167-4781(98)00254-1. [DOI] [PubMed] [Google Scholar]
- 119.Dorard C, de Thonel A, Collura A, Marisa L, Svrcek M, Lagrange A, Jego G, Wanherdrick K, Joly AL, Buhard O, Gobbo J, Penard-Lacronique V, Zouali H, Tubacher E, Kirzin S, Selves J, Milano G, Etienne-Grimaldi MC, Bengrine-Lefevre L, Louvet C, Tournigand C, Lefevre JH, Parc Y, Tiret E, Flejou JF, Gaub MP, Garrido C, Duval A. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med. 2011;17:1283–9. doi: 10.1038/nm.2457. [DOI] [PubMed] [Google Scholar]
- 120.Hosaka S, Nakatsura T, Tsukamoto H, Hatayama T, Baba H, Nishimura Y. Synthetic small interfering RNA targeting heat shock protein 105 induces apoptosis of various cancer cells both in vitro and in vivo. Cancer Sci. 2006;97:623–32. doi: 10.1111/j.1349-7006.2006.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–20. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR. The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J Biol Chem. 1999;274:15712–8. doi: 10.1074/jbc.274.22.15712. [DOI] [PubMed] [Google Scholar]
- 123.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–28. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol Biol Celll. 2002;13:2760–70. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chemy. 1997;272:31636–40. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- 126.Yamagishi N, Yokota M, Yasuda K, Saito Y, Nagata K, Hatayama T. Characterization of stress sensitivity and chaperone activity of Hsp105 in mammalian cells. Biochem Biophys Res Commun. 2011;409:90–5. doi: 10.1016/j.bbrc.2011.04.114. [DOI] [PubMed] [Google Scholar]
- 127.Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J Biol Chem. 2004;279:21992–2001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- 128.Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, Jin S, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–43. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–79. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 130.Hendrickson WA, Liu Q. Exchange we can believe in. Structure. 2008;16:1153–5. doi: 10.1016/j.str.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 131.Wilson CG, Arkin MR. Small-molecule inhibitors of IL-2/IL-2R: lessons learned and applied. Current Topics Microbiol Immunol. 2011;348:25–59. doi: 10.1007/82_2010_93. [DOI] [PubMed] [Google Scholar]
- 132.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–83. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 133.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 134.McLellan CA, Raynes DA, Guerriero V. HspBP1, an Hsp70 cochaperone, has two structural domains and is capable of altering the conformation of the Hsp70 ATPase domain. J Biol Chem. 2003;278:19017–22. doi: 10.1074/jbc.M301109200. [DOI] [PubMed] [Google Scholar]
- 135.Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–79. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 136.Tanimura S, Hirano AI, Hashizume J, Yasunaga M, Kawabata T, Ozaki K, Kohno M. Anticancer drugs up-regulate HspBP1 and thereby antagonize the prosurvival function of Hsp70 in tumor cells. J Biol Chem. 2007;282:35430–9. doi: 10.1074/jbc.M707547200. [DOI] [PubMed] [Google Scholar]
- 137.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–6. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 138.Takayama S, Bimston DN, Matsuzawa S, Freeman BC, Aime-Sempe C, Xie Z, Morimoto RI, Reed JC. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–96. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Briknarova K, Takayama S, Brive L, Havert ML, Knee DA, Velasco J, Homma S, Cabezas E, Stuart J, Hoyt DW, Satterthwait AC, Llinas M, Reed JC, Ely KR. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol. 2001;8:349–52. doi: 10.1038/86236. [DOI] [PubMed] [Google Scholar]
- 140.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan JA, Reed JC. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80:279–84. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 141.Zeiner M, Gehring U. A protein that interacts with members of the nuclear hormone receptor family: identification and cDNA cloning. Proc Natl Acad Sci. 1995;92:11465–9. doi: 10.1073/pnas.92.25.11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang X, Chernenko G, Hao Y, Ding Z, Pater MM, Pater A, Tang SC. Human BAG-1/RAP46 protein is generated as four isoforms by alternative translation initiation and overexpressed in cancer cells. Oncogene. 1998;17:981–9. doi: 10.1038/sj.onc.1202032. [DOI] [PubMed] [Google Scholar]
- 143.Townsend PA, Cutress RI, Sharp A, Brimmell M, Packham G. BAG-1: a multifunctional regulator of cell growth and survival. Biochim Biophys Acta. 2003;1603:83–98. doi: 10.1016/s0304-419x(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 144.Gehring U. Multiple, but concerted cellular activities of the human protein Hap46/BAG-1M and isoforms. Int J Mol Sci. 2009;10:906–28. doi: 10.3390/ijms10030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chemy. 2000;275:4613–7. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- 146.Tsukahara F, Maru Y. Bag1 directly routes immature BCR-ABL for proteasomal degradation. Blood. 2010;116:3582–92. doi: 10.1182/blood-2009-10-249623. [DOI] [PubMed] [Google Scholar]
- 147.Sroka K, Voigt A, Deeg S, Reed JC, Schulz JB, Bahr M, Kermer P. BAG1 modulates huntingtin toxicity, aggregation, degradation, and subcellular distribution. J Neurochem. 2009;111:801–7. doi: 10.1111/j.1471-4159.2009.06363.x. [DOI] [PubMed] [Google Scholar]
- 148.Sharp A, Crabb SJ, Johnson PW, Hague A, Cutress R, Townsend PA, Ganesan A, Packham G. Thioflavin S (NSC71948) interferes with Bcl-2-associated athanogene (BAG-1)-mediated protein-protein interactions. J Pharm Exp Med. 2009;331:680–9. doi: 10.1124/jpet.109.153601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu Z, Page RC, Gomes MM, Kohli E, Nix JC, Herr AB, Patterson C, Misra S. Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat Struct Mol Biol. 2008;15:1309–17. doi: 10.1038/nsmb.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell. 2005;16:5891–900. doi: 10.1091/mbc.E05-07-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Franceschelli S, Rosati A, Lerose R, De Nicola S, Turco MC, Pascale M. Bag3 gene expression is regulated by heat shock factor 1. J Cell Physiol. 2008;215:575–7. doi: 10.1002/jcp.21397. [DOI] [PubMed] [Google Scholar]
- 153.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chiappetta G, Ammirante M, Basile A, Rosati A, Festa M, Monaco M, Vuttariello E, Pasquinelli R, Arra C, Zerilli M, Todaro M, Stassi G, Pezzullo L, Gentilella A, Tosco A, Pascale M, Marzullo L, Belisario MA, Turco MC, Leone A. The antiapoptotic protein BAG3 is expressed in thyroid carcinomas and modulates apoptosis mediated by tumor necrosis factor-related apoptosis-inducing ligand. J Clin Endocrinol Metabolism. 2007;92:1159–63. doi: 10.1210/jc.2006-1712. [DOI] [PubMed] [Google Scholar]
- 155.Jacobs AT, Marnett LJ. HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J Biol Chem. 2009;284:9176–83. doi: 10.1074/jbc.M808656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang HQ, Liu BQ, Gao YY, Meng X, Guan Y, Zhang HY, Du ZX. Inhibition of the JNK signalling pathway enhances proteasome inhibitor-induced apoptosis of kidney cancer cells by suppression of BAG3 expression. British J Pharmacol. 2009;158:1405–12. doi: 10.1111/j.1476-5381.2009.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Festa M, Del Valle L, Khalili K, Franco R, Scognamiglio G, Graziano V, De Laurenzi V, Turco MC, Rosati A. BAG3 protein is overexpressed in human glioblastoma and is a potential target for therapy. Am J Path. 2011;178:2504–12. doi: 10.1016/j.ajpath.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu P, Xu B, Li J, Lu H. BAG3 gene silencing sensitizes leukemic cells to Bortezomib-induced apoptosis. FEBS letters. 2009;583:401–6. doi: 10.1016/j.febslet.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 159.Goebl M, Yanagida M. The TPR Snap Helix - a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–7. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 160.Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–9. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 161.Blatch GL, Lässle M, Zetter BR, Kundra V. Isolation of a mouse cDNA encoding mSTI1, a stress-inducible protein containing the TPR motif. Gene. 1997;194:277–82. doi: 10.1016/s0378-1119(97)00206-0. [DOI] [PubMed] [Google Scholar]
- 162.D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–62. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 163.Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–17. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 164.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: Essential repeats in S. pombe nuclear protein nuc2 + Cell. 1990;60:319–28. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 165.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–38. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 166.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–6. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 167.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–5. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 169.Brinker A, Scheufler C, von der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. Ligand discrimination by TPR domains -Relevance and selectivity of EEVD-recognition in Hsp70 center dot Hop center dot Hsp90 complexes. J Biol Chem. 2002;277:19265–75. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 170.Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 171.Morishima Y, Murphy PJ, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor. hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J Biol Chem. 2000;275:18054–60. doi: 10.1074/jbc.M000434200. [DOI] [PubMed] [Google Scholar]
- 172.Hutchison KA, Dittmar KD, Czar MJ, Pratt WB. Proof that hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with hsp90. J Biol Chem. 1994;269:5043–9. [PubMed] [Google Scholar]
- 173.Smith DF. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–29. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- 174.Young JC, Barral JM, Hartl FU. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28:541–7. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 175.Meimaridou E, Gooljar SB, Chapple JP. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 176.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Reports. 2001;2:885–90. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wu SJ, Liu FH, Hu SM, Wang C. Different combinations of the heat-shock cognate protein 70 (hsc70) C-terminal functional groups are utilized to interact with distinct tetratricopeptide repeat-containing proteins. Biochem J. 2001;359:419–26. doi: 10.1042/0264-6021:3590419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ballinger CA, Connell P, Wu YX, Hu ZY, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–45. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang L, Liu Y-T, Hao R, Chen L, Chang Z, Wang H-R, Wang Z-X, Wu J-W. Molecular Mechanism of the Negative Regulation of Smad1/5 Protein by Carboxyl Terminus of Hsc70-interacting Protein (CHIP) J Biol Chem. 2011:286. doi: 10.1074/jbc.M110.201814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu FH, Wu SJ, Hu SM, Hsiao CD, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem. 1999;274:34425–32. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- 181.Carrigan PE, Nelson GM, Roberts PJ, Stoffer JN, Riggs DL, Smith DF. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J Biol Chem. 2004;279:16185–93. doi: 10.1074/jbc.M314130200. [DOI] [PubMed] [Google Scholar]
- 182.Cheung-Flynn J, Roberts PJ, Riggs DL, Smith DF. C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to hsp90. J Biol Chem. 2003;278:17388–94. doi: 10.1074/jbc.M300955200. [DOI] [PubMed] [Google Scholar]
- 183.Chen SY, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress & Chaperones. 1998;3:118–29. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yi F, Zhu PJ, Southall N, Inglese J, Austin CP, Zheng W, Regan L. An AlphaScreen (TM)-Based High-Throughput Screen to Identify Inhibitors of Hsp90-Cochaperone Interaction. J Biomol Screen. 2009;14:273–81. doi: 10.1177/1087057108330114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Vasko RC, Rodriguez RA, Cunningham CN, Ardi VC, Agard DA, McAlpine SR. Mechanistic Studies of Sansalvamide A-Amide: An Allosteric Modulator of Hsp90. ACS Med Chem Lett. 2010;1:4–8. doi: 10.1021/ml900003t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ardi VC, Alexander LD, Johnson VA, McAlpine SR. Macrocycles That Inhibit the Binding between Heat Shock Protein 90 and TPR-Containing Proteins. ACS Chem Biol. 2011;6:1357–66. doi: 10.1021/cb200203m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–9. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]