Abstract

A novel series of histone deacetylase inhibitors combining N-hydroxycinnamamide bioactive fragment and indole bioactive fragment was designed and synthesized. Several compounds (17c, 17g, 17h, 17j, and 17k) exhibited comparable, even superior, total HDACs inhibitory activity and in vitro antiproliferative activities relative to the approved drug SAHA. A representative compound 17a with moderate HDACs inhibition was progressed to isoform selectivity profile, Western blot analysis, and in vivo antitumor assay. Although HDACs isoform selectivity of 17a was similar to that of SAHA, our Western blot results indicated that intracellular effects of 17a at 1 μM were class I selective. It was noteworthy that the effect on histone H4 acetylation of SAHA decreased with time, while the effect on histone H4 acetylation of 17a was maintained and even increased. Most importantly, compound 17a exhibited promising in vivo antitumor activity in a U937 xenograft model.
Keywords: histone deacetylases, inhibitor, N-hydroxycinnamamide, indole
The epigenetic regulation of histone and DNA plays an important role in chromatin structure and control of gene expression. Altered patterns of epigenetic modification are very common in many diseases, including cancer.1 As with genetic information, epigenetic modifications are heritable; in contrast, however, they are reversible and catalyzed by pairs of enzymes with converse activity. Among these epigenetic enzymes, histone deacetylases (HDACs) have now emerged as a promising new class of therapeutic targets.2
Four classes were identified in the HDACs family, characterized by different cellular localization and substrate specificity. The common feature of classes I (HDAC1–3 and -8), II (HDAC4–7, -9, and -10) and IV (HDAC11) active sites are characterized by the presence of a catalytic Zn2+ ion, and the class III HDACs (sirtuins 1–7) are NAD+-dependent. These HDACs isoforms perform their multiple functions by either epigenetic mechanism (deacetylation of histones) or nonepigenetic mechanism (deacetylation of nonhistone substrates). It has been revealed that Zn2+-dependent isozymes, especially class I and class II, are closely related to tumorigenesis and development.3 In the past 10 years, over 490 clinical trials of more than 20 histone deacetylase inhibitor (HDACI) candidates have been initiated, culminating in the approval of two antitumor drugs, vorinostat (SAHA) and romidepsin (FK228) (Figure 1).4,5
Figure 1.

Approved and clinical HDACIs with the N-hydroxycinnamamide fragment highlighted in red.
N-Hydroxycinnamamide (Figure 1) is a very common active fragment in HDACI design.6,7 This fragment could not only form bichelation with the active site Zn2+ by its hydroxamic acid group but also form a sandwichlike π–π interaction by inserting its vinyl benzene group into two parallel phenylalanine residues of HDAC. Currently, there are three HDACIs containing N-hydroxycinnamamide fragment in clinical trials (Figure 1). Indole is recognized as a privileged structure in drug design,8,9 and its derivatives have been found to exhibit anticancer activity by interacting with different targets.10 Kinds of natural and synthetic HDACIs containing an indole scaffold, especially in the cap group, possessed promising in vitro and in vivo potency.11−14 Recently, comprehensive HDACIs structure–activity relationship (SAR) studies revealed that N-hydroxycinnamamide-based compounds were more stable than their straight chain analogues, and compounds having indole groups exhibited the best in vivo efficacy and tolerability.15 On the basis of the aforementioned information, we designed a novel series of N-hydroxycinnamamide-based HDACIs with an indole-containing cap group.
Compounds 14a, 14b, and 15 were synthesized following the procedures described in Scheme 1. Methyl ester protection and tert-butyloxycarbonyl (Boc) protection of l-tryptophan (1) followed by LiAlH4 reduction provided the intermediate 9. Methyl ester protection of ferulic acid (10) afforded compound 11, which was hydrogenized to give compound 12. Both 11 and 12 could be connected with 9 under Mitsunobu reaction conditions to get 13a and 13b, which were converted to corresponding hydroxamic acid compounds 14a and 14b, respectively. Subsequent N-deprotection of 14a afforded compound 15.
Scheme 1. Synthesis of Compounds 14a, 14b, and 15.
Reagents and conditions: (a) SOCI2, CH3OH, 98%. (b) (Boc)2O, Et3N, DCM, 80%. (c) LiAIH4, anhydrous THF, 0 °C, 86%. (d) PTSA, CH3OH, 80 °C, 96%. (e) H2, 10% Pd–C, CH3OH. (f) PPh3, DEAD, anhydrous THF, 0 °C, 75% for 8a, 79% for 8b. (g) NH2OK, CH3OH, 51% for 9a, 49% for 9b. (h) HCI, anhydrous EtOAc, 90%.
The preliminary HDACs inhibitory assay was tested against HeLa cell nuclear extract, mainly containing HDAC1 and HDAC2. Results listed in Table 1 revealed that compound 14a (IC50 = 0.65 μM) was more potent than its analogues 14b (IC50 = 2.09 μM) and 15 (IC50 = 2.91 μM), which indicated that both the vinyl benzene group and the substituent located on the amine atom (the Boc group in 14a) were advantageous to HDACs inhibition. Therefore, using 14a as lead, we kept its N-hydroxycinnamamide group unchanged and derivatized its Boc group to other functional groups. Such derivatizations were performed according to the methods in Scheme 2. N-Deprotection of 13a with trifluoroacetic acid (TFA) and subsequent amide condensation or sulfonylation gave intermediates 16a–u, which were treated with NH2OK to get target compounds 17a–u.
Table 1. HeLa Cell Nuclear Extract Inhibitory Activity.
| compd | IC50 (μM)a | compd | IC50 (μM)a |
|---|---|---|---|
| 14a | 0.65 ± 0.12 | 17j | 0.18 ± 0.03 |
| 14b | 2.09 ± 0.31 | 17k | 0.14 ± 0.03 |
| 15 | 2.91 ± 0.34 | 17l | 0.32 ± 0.05 |
| 17a | 1.08 ± 0.20 | 17m | 0.43 ± 0.05 |
| 17b | 0.49 ± 0.09 | 17n | 1.17 ± 0.21 |
| 17c | 0.17 ± 0.04 | 17o | 0.80 ± 0.17 |
| 17d | 0.62 ± 0.13 | 17p | 1.12 ± 0.23 |
| 17e | 0.57 ± 0.09 | 17q | 0.73 ± 0.16 |
| 17f | 0.38 ± 0.05 | 17r | b |
| 17g | 0.16 ± 0.04 | 17s | 1.06 ± 0.20 |
| 17h | 0.18 ± 0.04 | 17t | 1.47 ± 0.36 |
| 17i | 0.43 ± 0.05 | 17u | 0.71 ± 0.19 |
| SAHA | 0.19 ± 0.03 |
Assays were performed in replicate (n ≥ 2). Data are shown as means ± SDs.
Compound 17r was undissolved under our test condition.
Scheme 2. Synthesis of Compounds 17a–u.
Reagents and conditions: (a) (i) TFA, Et3N, DCM; (ii) R′COOH, TBTU, Et3N, THF, 52–75% for two steps. (b) (i) TFA, Et3N, DCM; (ii) R′SO2CI, Et3N, DCM, 60–76% for two steps. (c) NH2OK, CH3OH, 31–54%.
As shown in Table 1, we found that compounds 17b–n with N-Boc-protected natural α-amino acid residues as R group except 17n exhibited more potent activity than their parent compound 14a. Most importantly, the total HDACs inhibitory activities of compounds 17c, 17g, 17h, 17j, and 17k were comparable, even more potent than that of SAHA. However, replacing the Boc group of 14a with valproyl group (17a), unnatural amino acid residues (17o–q) and sulfuryl group (17r–u) were detrimental to activity.
To characterize HDACs isoform selectivity of these analogues, representative compound 17a with moderate total HDACs inhibitory activity was tested against HDAC1, HDAC2, HDAC3, and HDAC6 using acetylated substrate. Besides, the class IIa inhibitory activity was evaluated against MDA-MB-231 cell lysate using class IIa-specific triflouroacetylated substrate.16 Results in Table 2 showed that 17a displayed modest preference for HDAC1 and HDAC3 over HDAC2 and HDAC6 but exhibited no obvious inhibition against class IIa HDACs up to 10 μM. The overall selectivity profile of 17a was similar to that of SAHA, which was in line with literature information.17
Table 2. HDACs Isoform Selectivity of 17a and SAHAa.
| class I |
class IIb | class IIa | |||
|---|---|---|---|---|---|
| compd | HDAC1 | HDAC2 | HDAC3 | HDAC6 | cell lysate |
| 17a | 0.39 ± 0.12 | 1.42 ± 0.06 | 0.28 ± 0.13 | 0.94 ± 0.14 | NAb |
| SAHA | 0.076 ± 0.011 | 0.256 ± 0.003 | 0.028 ± 0.011 | 0.118 ± 0.012 | NAb |
Assays were performed in replicate (n ≥ 2). IC50 (μM) values are shown as means ± SDs.
NA, not active at 10 μM.
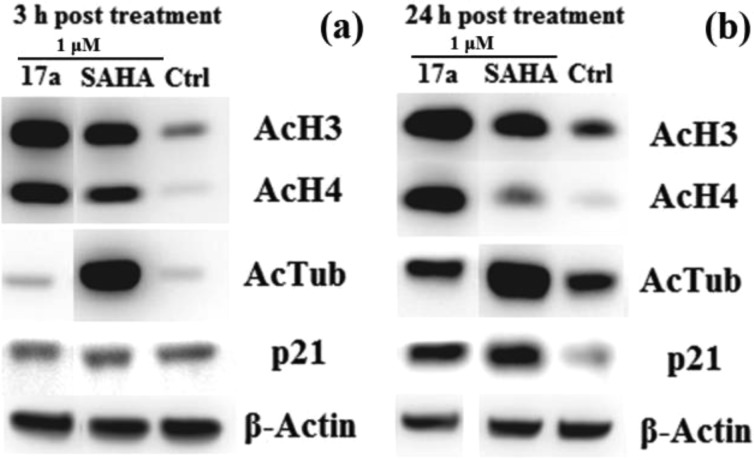
Compound 17a was confirmed to be cell permeable and able to inhibit intracellular and even nuclear HDACs by monitoring the acetylation levels of tubulin (target of HDAC6) and histones H3 and H4 (targets of HDAC1 and HDAC2) in the MDA-MB-231 cell line. Moreover, the effect on the expression level of the cyclin-dependent kinase (CDK) inhibitor p21 was also investigated (Figure 2). Although the inhibition against class I HDACs of 17a was inferior to that of SAHA (Table 2), its effects on the levels of histone acetylation and p21 expression at the concentration of 1 μM were comparable and even superior to SAHA, especially after 24 h of treatment. However, 17a had almost no effect on acetylated tubulin level as compared with SAHA at both time points. There was a possible explanation that the intracellular amide hydrolysis of 17a could release compound 15 and valproic acid (VPA), which is a clinical HDACI with class I selectivity.18 Therefore, 17a and its intracellular metabolites (15 and VPA) could exhibit synergistic inhibition against class I HDACs but not HDAC6. It was noteworthy that the effect on histone H4 acetylation of SAHA decreased with time, while the effect on histone H4 acetylation of 17a was maintained and even increased. This indicated that 17a might be promising in tumor prevention and treatment because it has been shown that global hypoacetylation of histone H4 is a common hallmark of cancer and changes in H4 acetylation happen early in tumorigenesis.19 The exact intracellular mechanism of 17a needs further research.
Figure 2.
Western blot analysis of acetylated tubulin, acetylated histone H3, acetylated histone H4, and p21 in MDA-MB-231 cell lines after 3 (a) and 24 h (b) of treatment with compounds at 1 μM. β-Actin was used as a loading control.
Compound 17a and five other analogues with comparable total HDACs inhibitory activity to SAHA were selected to test their effects on tumor cell viability. The results in Table 3 showed that the antiproliferative activities of these derivatives were similar to those of SAHA. It was intriguing that although total HDACs inhibition of 17a was inferior, its cellular potency was comparable and even superior, which was in line with our Western blot analysis results.
Table 3. In Vitro Antiproliferative Activity of Representative Compounds.
| IC50 (μM)a |
||||||
|---|---|---|---|---|---|---|
| compd | U937 | PC-3 | A549 | ES-2 | MDA-MB-231 | HCT116 |
| 17a | 1.8 | 3.7 | 4.4 | 5.4 | 3.1 | 5.5 |
| 17c | 3.1 | 10.5 | 11.8 | 29.2 | 7.2 | 6.0 |
| 17g | 2.2 | 10.4 | 4.2 | 25.1 | 4.5 | 3.8 |
| 17h | 2.2 | 5.8 | 1.6 | 4.4 | 6.8 | 5.9 |
| 17j | 2.7 | 5.4 | 7.0 | 8.9 | 7.2 | 2.4 |
| 17k | 3.9 | 8.2 | 13.5 | 9.5 | 11.7 | 9.4 |
| SAHA | 2.3 | 9.9 | 3.8 | 12.7 | 5.6 | 6.0 |
Values are the means of at least two experiments. The SD values are <20% of the mean.
Among these tested tumor cell lines, human leukemic monocyte lymphoma (U937) was the most sensitive to our HDACI. To preliminarily investigate if our newly designed compounds were active in vivo, representative compound 17a was progressed to a U937 xenograft model using Tamibarotene as a positive control. Mice were treated once daily by oral gavage for 3 weeks. The tumor growth curve depicted in Figure 3 and the final tumor tissue size visualized in Figure 4 explicitly showed that compound 17a exhibited potent oral antitumor activity. Tumor growth inhibition (TGI) and relative increment ratio (T/C) were used as the indicators to evaluate the antitumor effects in tumor weight and tumor volume, respectively. Our calculated results revealed that the in vivo antitumor activity of 17a (TGI = 53%, T/C = 42%) was statistically significant (P < 0.05), while the potency of tamibarotene (TGI = 33%, T/C = 85%) was not statistically significant. In the mice group treated by 17a, no significant body weight loss and no evident toxic signs in liver and spleen were detected.
Figure 3.

Growth curve of implanted U937 xenograft in nude mice (seven mice per group). Data are expressed as the mean ± SD.
Figure 4.

Picture of dissected U937 tumor tissues.
In conclusion, we designed and synthesized a novel series of N-hydroxycinnamamide-based HDACIs with an indole-containing cap group, among which compounds 17c, 17g, 17h, 17j, and 17k exhibited similar HDACs inhibition and in vitro antitumor potency to SAHA. Our further research focused on 17a revealed several interesting results, which deserve a detailed mechanism study. Importantly, although its HDACs inhibitory activity was moderate among these analogues, 17a exhibited potent in vitro and in vivo antitumor activity. Currently, a detailed activity evaluation and mechanism study of 17a and other more potent analogues are underway in our laboratory.
Glossary
Abbreviations
- HDACs
histone deacetylases
- HDACIs
histone deacetylase inhibitors
- SAR
structure–activity relationship
- Boc
tert-butyloxycarbonyl
- DCM
dichloromethane
- THF
tetrahydrofuran
- PTSA
p-toluenesulfonic acid
- DEAD
diethyl azodicarboxylate
- TFA
trifluoroacetic acid
- TBTU
O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate
- CDK
cyclin-dependent kinase
- VPA
valproic acid
Supporting Information Available
Experimental procedures for compound synthesis, HDACs inhibition fluorescence assay, in vitro antiproliferative assay, Western blot analysis, in vivo antitumor assay, and characterization data for new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
⊥ These two authors contributed equally.
This work was supported by the National Scientific and Technological Major Project of Ministry of Science and Technology of China (Grant No. 2011ZX09401-015), National Natural Science Foundation of China (Grant No. 21172134), Doctoral Foundation of Ministry of Education of China (Grant No. 20110131110037), the National Center for Research Resources (Grant No. 5 P20 RR024485-02), and the National Institute of General Medical Sciences (Grant No. 8 P20 GM103542-02) from the National Institutes of Health.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007, 8, 286–298. [DOI] [PubMed] [Google Scholar]
- Best J. D.; Carey N. Epigenetic opportunities and challenges in cancer. Drug Discovery Today 2010, 15, 65–70. [DOI] [PubMed] [Google Scholar]
- Witt O.; Deubzer H. E.; Milde T.; Oehme I. HDAC family: What are the cancer relevant targets?. Cancer Lett. 2009, 277, 8–21. [DOI] [PubMed] [Google Scholar]
- Gryder B. E.; Sodji Q. H.; Oyelere A. K. Targeted cancer therapy: Giving histone deacetylase inhibitors all they need to succeed. Future Med. Chem. 2012, 4, 505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini G.; Cabri W.; Fattorusso C.; Rodriquez M. Histone deacetylase inhibitors in the treatment of cancer: overview and perspectives. Future Med. Chem. 2012, 4, 1439–60. [DOI] [PubMed] [Google Scholar]
- Miller T. A.; Witter D. J.; Belvedere S. Histone deacetylase inhibitors. J. Med. Chem. 2003, 46, 5097–5116. [DOI] [PubMed] [Google Scholar]
- Paris M.; Porcelloni M.; Binaschi M.; Fattori D. Histone deacetylase inhibitors: From bench to clinic. J. Med. Chem. 2008, 51, 1505–1529. [DOI] [PubMed] [Google Scholar]
- DeSimone R. W.; Currie K. S.; Mitchell S. A.; Darrow J. W.; Pippin D. A. Privileged structures: Applications in drug discovery. Comb. Chem. High Throughput Screening 2004, 7, 473–493. [DOI] [PubMed] [Google Scholar]
- Costantino L.; Barlocco D. Privileged structures as leads in medicinal chemistry. Curr. Med. Chem. 2006, 13, 65–85. [PubMed] [Google Scholar]
- Kamal A.; Srikanth Y. V.; Ramaiah M. J.; Khan M. N.; Kashi Reddy M.; Ashraf M.; Lavanya A.; Pushpavalli S. N.; Pal-Bhadra M. Synthesis, anticancer activity and apoptosis inducing ability of bisindole linked pyrrolo[2,1-c][1,4]benzodiazepine conjugates. Bioorg. Med. Chem. Lett. 2012, 22, 571–578. [DOI] [PubMed] [Google Scholar]
- Olsen C. A.; Montero A.; Leman L. J.; Ghadiri M. R. Macrocyclic Peptoid–Peptide Hybrids as Inhibitors of Class I Histone Deacetylases. ACS Med. Chem. Lett. 2012, 3, 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini G.; Marzi M.; Di Marzo M.; Battistuzzi G.; Pezzi R.; Brunetti T.; Cabri W.; Vesci L.; Pisano C. Exploring bis-(indolyl)methane moiety as an alternative and innovative CAP group in the design of histone deacetylase (HDAC) inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 2840–2843. [DOI] [PubMed] [Google Scholar]
- Attenni B.; Ontoria J. M.; Cruz J. C.; Rowley M.; Schultz-Fademrecht C.; Steinkuhler C.; Jones P. Histone deacetylase inhibitors with a primary amide zinc binding group display antitumor activity in xenograft model. Bioorg. Med. Chem. Lett. 2009, 19, 3081–3084. [DOI] [PubMed] [Google Scholar]
- Lai M.-J.; Huang H.-L.; Pan S.-L.; Liu Y.-M.; Peng C.-Y.; Lee H.-Y.; Yeh T.-K.; Huang P.-H.; Teng C.-M.; Chen C.-S.; Chuang H.-Y.; Liou J.-P. Synthesis and Biological Evaluation of 1-Arylsulfonyl-5-(N-hydroxyacrylamide)indoles as Potent Histone Deacetylase Inhibitors with Antitumor Activity in Vivo. J. Med. Chem. 2012, 55, 3777–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontiki E.; Hadjipavlou-Litina D. Histone deacetylase inhibitors (HDACIs). Structure–activity relationships: History and new QSAR perspectives. Med. Res. Rev. 2012, 32, 1–165. [DOI] [PubMed] [Google Scholar]
- Inks E. S.; Josey B. J.; Jesinkey S. R.; Chou C. J. A novel class of small molecule inhibitors of HDAC6. ACS Chem. Biol. 2012, 7, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner J. E.; West N.; Grachan M. L.; Greenberg E. F.; Haggarty S. J.; Warnow T.; Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010, 6, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass D. M.; Shah R.; Ghosh B.; Hennig K.; Norton S.; Zhao W. N.; Reis S. A.; Klein P. S.; Mazitschek R.; Maglathlin R. L.; Lewis T. A.; Haggarty S. J. Short-Chain HDAC Inhibitors Differentially Affect Vertebrate Development and Neuronal Chromatin. ACS Med. Chem. Lett. 2011, 2, 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M. F.; Ballestar E.; Villar-Garea A.; Boix-Chornet M.; Espada J.; Schotta G.; Bonaldi T.; Haydon C.; Ropero S.; Petrie K.; Iyer N. G.; Perez-Rosado A.; Calvo E.; Lopez J. A.; Cano A.; Calasanz M. J.; Colomer D.; Piris M. A.; Ahn N.; Imhof A.; Caldas C.; Jenuwein T.; Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.