Abstract
HOXC13 is a homeobox containing gene that plays crucial roles in hair development and origin of replication. Herein, we investigated the biochemical functions of HOXC13 and explored its potential roles in tumor cell viability. We have designed a phosphorothioate based antisense-oligonucleotide that specifically knockdown HOXC13 in cultured cells. Cell viability and cytotoxicity assays demonstrated that HOXC13 is essential for cell growth and viability. Antisense-mediated knockdown of HOXC13 affected the cell viability and induced apoptosis in cultured tumor cells. HOXC13 regulates the expression of cyclins and antisense-mediated knockdown of HOXC13 resulted in cell cycle arrest and apoptosis in colon cancer cells. Finally over expression of HOXC13 resulted in 3D-colony formation in soft-agar assay indicating its potential roles in cell proliferation and tumorigenesis.
Introduction
Antisense-oligonucleotide mediated gene targeting has been a promising strategy for down regulation of various genes for understanding their biochemical functions and in developing potential therapy (1, 2). The sequence specific binding of an antisense-oligonucleotide to a target mRNA results in degradation of the target mRNA and prevent gene translation. Various genes associated with apoptosis, cell growth, metastasis, and angiogenesis are potential targets for antisense therapy (3–5). For example, antisense against Bcl-2, which is an anti-apoptotic gene, is a target for cancer therapy and in clinical trial (6). Herein, we have developed an antisense oligonucleotide against a homeobox containing gene, HOXC13 and examined the roles of HOXC13 in growth and viability of tumor cells and explored its potential in novel cancer therapy.
Homeobox containing genes (HOX genes) are evolutionary conserved family of genes that play crucial roles during embryogenesis and development (7, 8). There are 39 different HOX genes in human that are clustered in four different groups, HOXA-D. HOX genes act as transcription factors and they bind to their target gene promoters regulating target gene expression (9, 10). Colinear expression of different HOX genes are crucial for proper cell differentiation and development (11, 12). In addition to their crucial roles in embryonic development, HOX gene appeared to play important roles in postnatal development (13). Various HOX genes such as HOXA5, HOXA7, HOXA10 etc are shown to be important for cell cycle regulation (14). Studies from our laboratory and others demonstrated that HOX genes are misregulated in various cancer including breast and prostate cancer (15, 16). For example, HOXC6, a gene which is important for mammary gland development, is closely associated with prostate cancer and prostate branching morphogenesis (17, 18). HOXB9, that is crucial for mammary gland development, is important for angiogenesis and breast cancer (19). Similarly, HOXC10, that is critical for spinal cord development, is transcriptionally regulated by estradiol and is over expressed in breast cancer tissue (16). Thus increasing amount of studies indicate that HOX genes are misregulated in various human diseases including cancer and are potential targets for novel cancer therapy.
Herein, we have investigated the function of HOXC13 in cell cycle progression, growth and tumorigenesis and explored its therapeutic potential. It is well recognized that HOXC13 is an important regulator of hair keratin gene cluster (20–22). Over expression of HoxC13 in transgenic mice develops alopecia, accompanied by a progressive pathological skin condition (21, 22). HoxC13 knockout mice lack external-hair indicating its important roles in hair development (22). In addition to its developmental function, recent study demonstrated that HOXC13 is a member of human DNA replication complex, interacts with ORC2 and binds to the origin of replication and is involved in DNA replication (23–25). HOXC13 is also shown to be fused with NUP98 and associated with acute myeloid leukemia (26). Recently, we demonstrated that HOXC13 is transcriptionally regulated by estradiol in placental choriocarcinoma cells (JAR) and mixed lineage leukemia (MLL) family of histone methylases, that are key players in gene activation, coordinate with estrogen receptors (ERs), and regulate estradiol-induced activation of HOXC13 (27). In this study, we have shown that HOXC13 is a critical player in cell viability, cell cycle progression and its over expression induces three-dimensional (3D) colony formation in soft agar media indicating its potential role in tumorigenesis.
Experimental procedures
Cell culture and antisense transfection
Human colorectal adenocarcinoma (SW480), human breast adenocarcinoma (MCF7), human lung metastasized prostate carcinoma (PC3ML), human cervical cancer (HeLa), non-malignant colon fibroblast (CCD-18Co), human bronchioalveolar carcinoma (H358) and human embryonic kidney (HEK293) cell lines were obtained from American Type Cell Culture Collection (ATCC). Cells were grown and maintained in DMEM (Dulbecco’s modified Eagle’s media; Sigma) supplemented with 10 % heat inactivated fetal bovine serum, 1 % L-glutamine, 100 units/ml penicillin and 0.1 mg streptomycin/ml. All cell lines were maintained in a humidified incubator with 5 % CO2 and 95 % air at 37 °C (28).
For antisense transfection, cells were grown up to 60% confluency (about 0.3 million cells) in 60 mm culture plates and then transfected with HOXC13 antisense oligonucleotide (Anti-HOXC13: 5'-GCTGACCACCTTCTTCTCTT-3') using iFect transfection reagent (K.D. Medical Inc) as previously described (28). Prior to trasnfection, a cocktail of transfection reagent and antisense oligonucleotide was made as follows. Initially, 12 µL (3 µg/mL final concentration, stock is 1 mg/mL) of iFect reagent was mixed with 200 µL DMEM (without FBS and antibiotics) in an eppendorf tube. In a separate eppendorf tube, antisense oligonucleotide (varying concentrations, 0.75 – 1.75 µg/mL (equivalent to 120–280 nM final concentration) was mixed with 100 µL DMEM (without supplements). Then the diluted antisense solution was mixed with diluted iFect reagent and allowed to stand for 45 min in dark. In the meantime, cells were washed twice with supplement free DMEM and then 1.7 mL of supplement free DMEM was added to each cell culture plate. Finally, antisense-transfection reagents cocktail was applied to the cell plates and incubated for 24 h. Then 2 mL of DMEM with all supplements including 20% FBS was added to all cell plates. The cells were incubated for additional 24 h then harvested for RNA/protein extraction or fixed for microscopic and other analysis.
RNA and protein extraction
For the RNA and protein extraction, we followed similar protocols as described by us earlier (28). In brief, for the RNA extraction, cells were harvested and centrifuged at 500g for 5 min at 4 °C, then resuspended in diethyl pyrocarbonate (DEPC) treated buffer A (20 mM Tris-HCl, pH 7.9; 1.5 mM MgCl2; 10 mM KCl,; 0.5 mM dithiothreitol (DTT) and 0.2 mM phenylmethanesulfonyl fluoride (PMSF)), for 10 min on ice and then centrifuged at 3500g for 5 min at 4 °C. Supernatant was subjected to phenol-chloroform (1:1 phenol and chloroform saturated with 1×TE) extraction. The aqueous layer was mixed with 3 volume ethanol, incubated at −80 °C for 4 h and then centrifuged at 13000g for 30 min at 4 °C. The RNA pellets were air dried, dissolved in DEPC treated water containing 0.5 mM EDTA and quantified using nanodrop spectrophotometer.
For preparation of protein extracts, antisense-treated and control cells were resuspended in whole cell extraction buffer (50 mm Tris/HCI, pH 8.0; 150 mM NaCl; 1 mM EDTA; 0.05% NP-40; 0.2 mM PMSF; 0.5 mM DTT and 1X protease inhibitors cocktail) for 25 min on ice followed by centrifugation at 10000 g for 20 min at 4 °C. The supernatant contained the cell nuclear protein extract and stored at − 80 °C prior to Western blot analysis.
RT-PCR and Western blot analysis
For cDNA synthesis, 2.4 µM of oligo dT (Promega) was mixed with 500 ng of the RNA in a 10 µL total volume and incubated at 70°C for 10 min. This mixture was added to a cocktail of 100 units of MMLV reverse transcriptase, 1X first strand buffer (Promega), 100 µM each dNTPs (dATP, dGTP, dCTP and dTTP, Promega), 1 mM DTT and 20 units of RNaseOut (Invitrogen) and the volume was made up to 25 µL. This mixture was incubated at 37 °C for 1 h for the reverse transcription. Each cDNA product was diluted to 100 µL, and 5 µL of the diluted cDNA was subjected to PCR amplification using specific primer pairs described in Table 1.
Table 1.
Primers used for RT-PCR, cloning and antisense
| Primers | Forward primer (5′- 3′) | Reverse primer (5′ - 3′) |
|---|---|---|
| GAPDH | CAATGACCCCTTCATTGACC | GACAAGCTTCCCGTTCTCAG |
| HOXC13 | GGAAGTCTCCCTTCCCAGAC | CGATTTGCTGACCACCTTCT |
| Cyclin A | AAGAAGCAGCCAGACATCACGGAA | AGCTGCAGTTTCCCTCTCAGAACA |
| Cyclin B | TTGATACTGCCTCTCCAAGCCCAA | TTGGTCTGACTGCTTGCTCTTCCT |
| Cyclin D | AGAAGCTGTGCATCTACACCGACA | AGAAGCTGTGCATCTACACCGACA |
| Cyclin E | TTTCAGGGTATCAGTGGTGCGACA | ACAACATGGCTTTCTTTGCTCGGG |
| HOXC13 cloning primer | AAGCTTACGACTTCGCTGCTCCTGC | GGATCCTCAGGTGGAGTGGAGATGAGGC |
| HOXC13 antisense | GCTGACCACCTTCTTCTCTTa | |
| Scramble antisense | CGTTTGTCCCTCCAGCATCTa |
Phosphorothioate antisense oligonucleotide
Regular PCR reactions were carried out for 31 cycles (30 s at 94 °C for denaturation, 30 s at 60 °C for annealing, 45 s at 72 °C for elongation) and finally PCR products were analyzed in 1.5 % agarose gel electrophoresis. For the qPCR reactions, 5 µL of diluted cDNA were mixed with 5 µL Sso EvaGreen supermix (Bio-Rad) and 2 µM each primer and final volume was made up to 12 µL. PCR reactions were carried out in CFX96 real-time detection system (Bio-Rad) for 40 cycles (5s at 95 °C for denaturation and 10s at 60 °C for both annealing and elongation). Data analysis was performed using CFX manager software (Bio-Rad). Each experiment was repeated three times with three replicates each time.
For Western blot analysis, 30µg protein extract from each sample were electrophoresed in SDS-PAGE, transferred to nitrocellulose membrane and then subjected to Western blotting with anti-HOXC13 (Abcam) and anti-actin (Sigma) antibodies. Western blot was developed using alkaline phosphatase method.
Cytotoxicity (MTT assay) and growth rate measurement
MTT assay was performed as described before (29, 30). Briefly, about 10000 SW480 cells (in 200 µL DMEM with all the supplements) were seeded in each well of 96 micro titer plates and incubated overnight inside the tissue culture incubator with 5 % CO2 and 95 % air at 37 °C to reach about 60% confluency. Then prior to transfection, supernatant media from the cells were discarded, cells were washed twice with supplement-free DMEM and then 85 µL supplement-free DMEM was added in each well. Cells were then transfected with HOXC13 antisense by adding a cocktail (15 µL) of HOXC13 antisense (1.75 µg/mL final concentration) and iFect reagent (3µg/mL), incubated inside the tissue culture incubator for 24 h followed by addition of another 100 µL of DMEM supplemented with 20 % FBS and other antibiotics supplements. Cells were incubated for additional 24 h and then 30 µL of MTT (5 mg/mL stock in PBS) solution was added to each well and all samples were incubated in standard cell culture condition for 4 h. Supernatant in each well was removed and replaced by 100 µL of DMSO, incubated at room temperature on a shaker for 1 h. All plates were analyzed by micro plate reader (Fluostar-omega, BMG Labtech) at 560 nm. Each experiment was performed in 4 replicates and experiments were repeated at least two times. For growth rate measurement, SW480 cells were grown in 60 mm cell culture plates and transfected with HOXC13 and scramble antisenses separately, and incubated for varying time periods. Cells were harvested, stained with trypan blue and counted using hemocytometer and plotted. Each experiment was performed in 3 replicates and experiments were repeated at least two times.
Flow cytometry analysis
SW480 cells were transfected with varying concentrations of HOXC13 (0.75–1.75 µg/mL) and scramble antisense (1.25 µg/mL) for 48 h. Supernatant media (containing floating cells) was transferred to a falcon tube and attached cells (on the plate) were harvested by trypsinization. Both supernatant media and trypsinized cells were mixed together, centrifuged (1000 g for 5 min at 4 °C), washed twice with cold PBS and then fixed by 70% ethanol/PBS for 1 h in 4 °C then stored in −20 °C overnight. Fixed cells were washed with cold PBS and resuspended with propidium iodide (PI) solution (0.5 µg/mL) for 2 h. Stained cells were subjected to FACS analysis (Beckman Coulter, Fullerton, CA, USA) (31).
Cytochrome c immunostaining
SW480 cells were grown on cover slips (placed inside tissue culture plate), transfected with HOXC13 (1.75 µg/mL) or scramble antisenses (1.75 µg/mL) for 48 h and subjected to immunostaining with anti-cytochrome c antibody (32). In brief, cells were fixed by 4% formaldehyde, washed with cold PBS twice, permeabilized by 0.2 % Triton X-100/PBS for 15 min, blocked with goat serum and then incubated with cytochrome c antibody (Upstate) for at least 2 h. Samples were washed with 0.2% tween 20/PBS twice and incubated with FITC conjugated secondary antibody for 1 h. DAPI staining of the cells were done to visualize nuclear integrity. Samples were mounted on the microscope slides and subjected to fluorescence microscopy (Nikon Eclipse TE2000-U, Japan).
TUNEL assay
The TUNEL (terminal dUTP nicked end labeling) assay was performed using ApoAlert DNA fragmentation assay kit (Clontech) (31, 33). Briefly, SW480 cells were grown up to 60% confluency in 60 mm culture plates and then transfected with HOXC13 (1.75 µg/mL) and scramble-antisense (1.75 µg/mL) oligonucleotides separately for 48 h. Cells were fixed by 4% formaldehyde for 25 min at room temperature. The cells from the supernatant as well as adhered on plate were harvested and washed twice with PBS. All samples were permeabilized with 0.2% Triton X-100/PBS for 15 min, washed twice with PBS, equilibrated each sample in 100 µL equilibration buffer for 15 min and then centrifuged (500 g). Each sample was resuspended with 50 µL of reaction mix containing nucleotides and TdT enzyme for 60 min at 37 °C. To stop the reaction, each sample was mixed with 1 mL of 2X SSC buffer for 15 min, washed twice with PBS and then stained with DAPI (5 mg/mL) for 5 min, then briefly washed with PBS, and resuspended in 20 µL PBS and spread on the microscope slide, mounted with coverslip and analyzed under fluorescence microscope.
Generation of HOXC13 stable cell line and soft agar based colony formation assay
HOXC13 gene was cloned into pFlag-CMV4 human expression construct. In brief HOXC13 gene was PCR-amplified from HeLa cell cDNA library using primers specific to HOXC13 open-reading frame (ORF) (Table 1) and initially cloned in a TA-cloning vector (pGEM-T, Promega). Notably the HOXC13 cloning primers are flanked by HindIII (forward primer: 5’-AAGCTTACGACTTCGCTGCTCCTGC-3’) and BamH1 (reverse primer: 5’-GGATCCTCAGGTGGAGTGGAGATGAGGC-3’) restriction site in frame for cloning into pFlag-CMV4 construct and expression in mammalian cells. The insert of HOXC13 was verified by sequencing and the HOXC13 insert was digested out from the pGEM-T vector using BamH1 and HindIII endonucleases and subcloned into the pFlag-CMV4 human expression construct.
For the generation of HOXC13 overexpressing stable cell, pFlag-CMV4-HOXC13 construct was transfected into HEK293 cells using lipofectamine (Invitrogen) transfection reagent for 48 h and stable transformant cells expressing Flag-HOXC13 were selected using G418 antibiotic (Sigma) selection procedure (34). In brief, the transfected cells (after 48 h of transfection) were splited into fresh DMEM containing 1 mg/mL G418 antibiotic and continued to grow in the G148 containing media. After about 2 weeks, most of the untransfected cells were dead and the stably transfected cells expressing Flag-HOXC13 were grown into distinctly visible individual colony. Individual colonies were picked up and cultured separately in G418 containing media and stably transfected cell colonies expressing flag-HOXC13 were confirmed by Western blot analysis using anti-Flag antibody (Sigma).
Colony forming ability of HEK293 and Flag-HOXC13 stable cell lines were assessed using soft agar method as described previously (35, 36). Briefly, 1% agar (in PBS) was autoclaved, cooled down (to approximately 40°C) and mixed with equal volume of DMEM containing 20 % FBS, 4 mM L-glutamine and 2% Penicillin/Streptomycin (200 unit and 0.2 mg/mL respectively). The mixture was plated on 6 well culture plates and the plates were allowed to cool down to solidify. Then about 5000 HEK293 and Flag-HOXC13 stable cells were added to a mixture of 0.5 mL 2× DMEM and 0.5 ml of 0.7% agar (in PBS) and then plated on top of the base agar layer. The dishes were cooled to room temperature (15 min) and then incubated in the tissue culture incubator. The cells were fed with 0.5 mL of normal growth media at 2 days interval for 4–5 weeks, until colonies were visible. Colonies were stained with 0.005 % crystal violet for 2 h and then examined under light microscope.
Results and Discussion
Design and characterization of knockdown efficacy of HOXC13 antisense-oligonucleotide
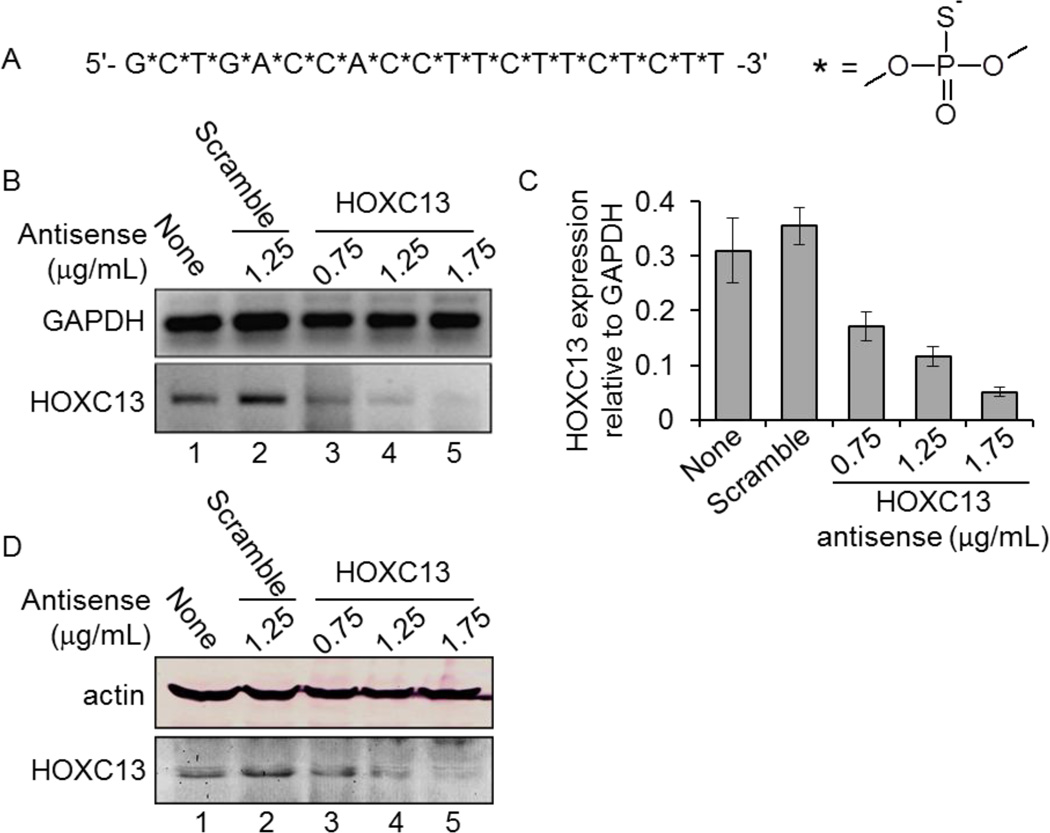
To understand the biochemical function of HOXC13, we knocked it down in cultured human cells using an antisense oligonucleotide. HOXC13 antisense oligonucleotide, (5'-GCTGACCACCTTCTTCTCTT-3') was designed to bind at 940–960 nt of HOXC13 mRNA (Figure 1A, Table 1). Importantly, to enhance the in vivo stability of the HOXC13 antisense, we introduced phosphorothioate linkages instead of regular phosphodiester bonds throughout the antisense oligonucleotide (Figure 1A).
Figure 1.
HOXC13 antisense oligonucleotide design and characterization. (A) Oligonucleotide sequences of HOXC13 antisense. Nucleotides are linked by phosphorothioate linkages instead of phosphodiester bonds. (B–D) Antisense–mediated of knockdown of HOXC13. SW480 cells were transfected with different concentration of HOXC13 (0.75 – 1.75 µg/mL) and scramble antisense (1.25 µg/mL) for 48 h. RNA was extracted, reverse-transcribed and subjected to PCR with primers specific to HOXC13 and actin (control) (in panel B). Real-time PCR for quantification of HOXC13 knockdown in control and antisense treated cells are shown in panel C (GAPDH was used as control). Bars are indicating standard error (n=6, p<0.05). (D) Protein extracts from the control and antisense-treated cells were analyzed by Western blotting using actin (control) and HOXC13 antibodies.
To assess the efficiency of the antisense, we transfected human colon cancer cells (SW480) with varying concentrations (0.75– 1.75 µg/mL, equivalent to 120–280 nM final concentration) of HOXC13 antisense using iFect transfection reagent and incubated for 48 h. A scramble antisense (with no homology to HOXC13, 1.25 µg/mL) was also transfected in parallel as a negative control (Table 1). RNA was isolated from the control and antisense-treated cells, reverse-transcribed into cDNA and subjected to PCR-amplification using real-time PCR and as well as regular PCR. Our analysis demonstrated that transfection with HOCX13 antisense efficiently knocked down HOXC13 both at mRNA (Figure 1B, real-time quantification is in figure 1C) and protein level (Figure 1D). HOXC13 knockdown was highest at 1.75 µg/mL antisense concentration (Figure 1B–D). GAPDH level remained mostly unaffected upon HOXC13 antisense transfection (Figures 1B–C). Scramble antisense had no significant impact on HOXC13 expression (Figures 1B–D). These analyses demonstrated that HOXC13 antisense effectively and specifically knocked down HOXC13 in SW480 cells.
Antisense-mediated knockdown of HOXC13 affects cell growth and viability
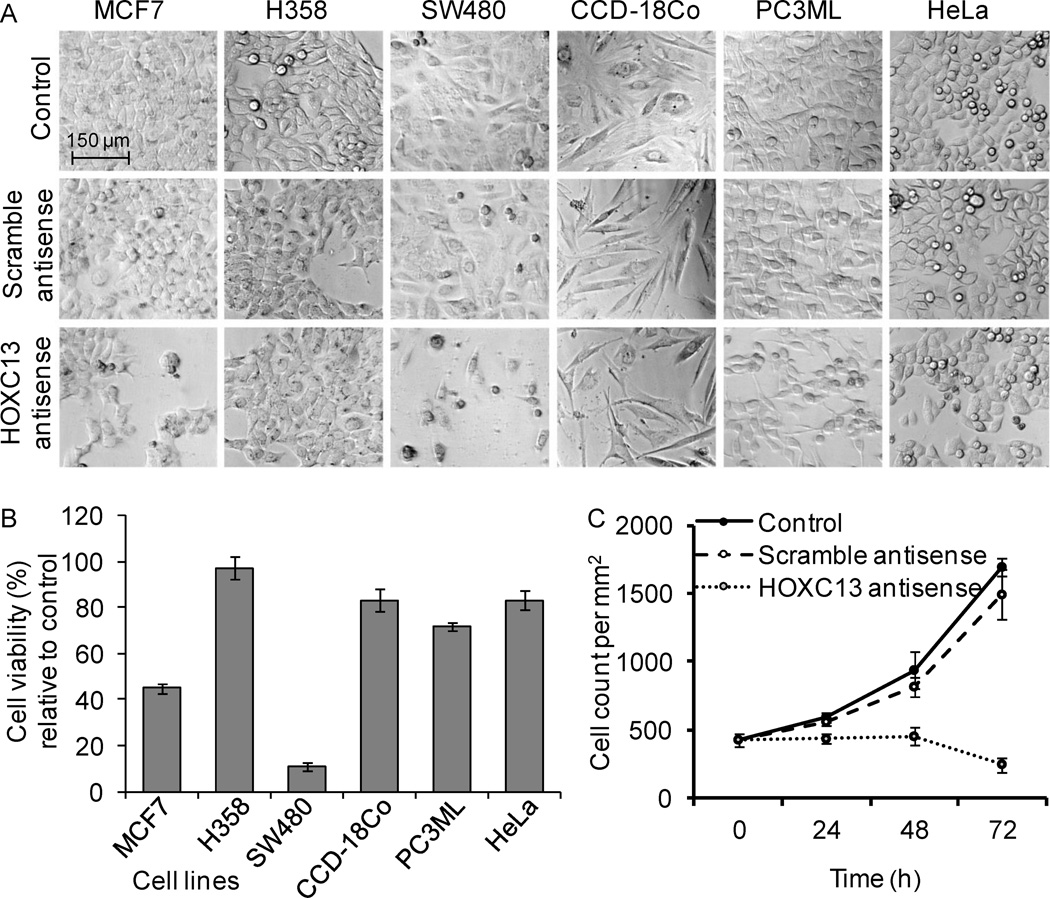
To examine the impact of HOXC13 knockdown on cell viability, we transfected different types of cancer and normal cells with HOXC13 antisense for 48 h and then examined its impact on cell viability using microscopic analysis and MTT assay. The cell lines include MCF7 (human breast adenocarcinoma), H358 (human bronchioalveolar carcinoma), SW480 (human colorectal adenocarcinoma), CCD-18Co (non-malignant colon fibroblast), PC3ML (human lung metastasized prostate carcinoma), and HeLa (human cervical cancer). Microscopic analysis showed that transfection with scramble antisense had little or no significant impact on cellular morphologies and appearance (Figure 2A). However, upon transfection with HOXC13 antisense, growth of most cells was impaired though to a different extent. SW480 (colon cancer) and MCF7 cells (human breast cancer) were more sensitive to HOXC13 knockdown compared to other cancer cells (PC3ML, HeLa, and H358) (Figure 2A). Non-malignant colon epithelial cells (CCD-18Co) appeared to be not significantly affected upon HOXC13 knockdown indicating preferential sensitivity of colon tumor cells over normal cells. MTT assay also demonstrated that HOXC13 knockdown affected significantly the viability of colon cancer (~ 90% cell death) and breast cancer cell (>55 % cell death) (Figure 2B). These results demonstrated that HOXC13 is critical for viability of most cell types and its knockdown induced death in cultured tumor cells.
Figure 2.
Effect of HOXC13 knockdown on cell viability. (A) Miscroscopic analysis: Different cell lines were transfected with HOXC13 (1.75 µg/mL) and scramble (1.75 µg/mL) antisenses separately for 48 h and visualized under a microscope. (B) Cytotoxicity analysis: Different cell lines were transfected with 1.75 µg/mL of HOXC13 and scramble antisenses (1.75 µg/mL) for 48 h and then subjected to MTT assay. The percent viability of HOXC13 antisense versus scramble antisense samples were plotted for different cell types. Bars indicate standard errors (n=8, p<0.05). (C) Effect of HOXC13 knockdown on cell growth: SW480 cells were transfected with 1.75 µg/mL HOXC13 and scramble antisenses (1.75 µg/mL) and harvested at different times (24, 48 and 72 h), stained with trypan blue and counted under microscope and plotted. Three replicates of each experiment were repeated for at least two times. Bars indicate standard errors.
To examine further the impact of HOXC13 knockdown on cellular growth, we transfected SW480 cells with HOXC13 antisense (1.75 µg/mL) and a scramble antisense (1.75 µg/mL) for varying time periods and analyzed its impact on cell growth. In brief, control and antisense treated cells were harvested at different time points and counted under a microscope. Cell numbers were plotted against time. This analysis showed that control grew exponentially with similar fashion and almost tripled in 72 h (Figure 2C). The scramble antisense-treated cells also grew exponentially in a similar fashion as the control cells, however their growth rate was slightly slowed down under scramble antisense-treatment condition (Figure 2C). This slightly slower growth could be due to the toxic effects of the liposomes used for transfection or potential off target effects of scramble antisense. Interestingly, under HOXC13 antisense treatment condition, cell growth was significantly perturbed. Cells did not grow further (up to 48 h) and upon longer time (72 h) incubation, cells populations decayed likely because of death (Figure 2C). These results demonstrated that HOXC13 expression is critical for cell growth and viability and knockdown of HOXC13 induced cell death in colon cancer cells.
HOXC13 knockdown affects cell cycle progression
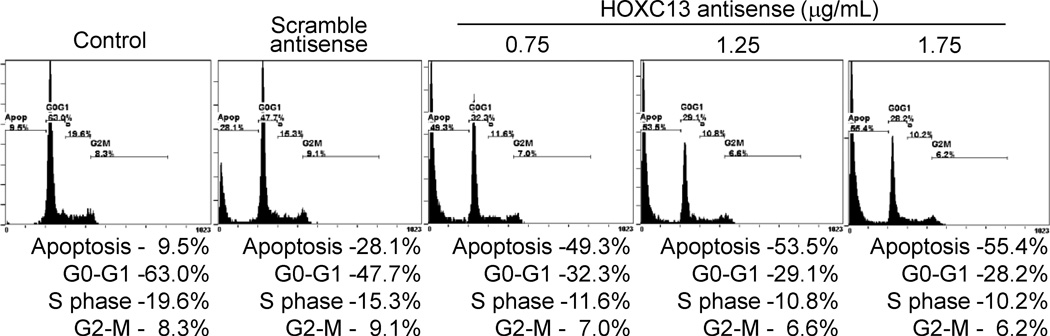
To examine further the impact of HOXC13 knockdown on cellular function, we examined the impact of HOXC13 knockdown on cell cycle progression. We transfected SW480 cells with varying concentrations (0.75–1.75 µg/mL) of HOXC13 antisense for 48 h, stained with propidium iodide and analyzed by flow cytometry. Transfection with scramble antisense (1.25 µg/mL) was also performed in parallel as a negative control. Our analysis demonstrated that transfection with the scramble-antisense increased the amount of apoptotic cell population (from 9.5 % (control) to 28 % (scramble) and also affected cell populations at G0/G1 phase (decreased in comparison to control) and these changes are likely associated with the toxic effect of the transfection reagent used (Figure 3). Interestingly however, transfection with HOXC13 antisense, drastically decreased the cell population at G0/G1 and S phase, while apoptotic cells were significantly increased, in comparison to the control or scramble antisense treated cells (Figure 3). Upon transfection with 0.75 µg/mL of HOXC13 antisense, about 50% cells underwent apoptosis, while G0/G1 population dropped from 63 % to 32.3 %, S-phase population dropped from 19.6 % to 11.6 % (Figure 3). G2/M-phase population was not significantly affected (Figure 3). Increased HOXC13 antisense concentration (1.25 and 1.75 µg/mL) further increased the percent apoptotic cell death (Figure 3). These results demonstrated that HOXC13 is a key player in cell cycle regulation and growth and knockdown of HOXC13 affected the cell cycle progression and ultimately induced apoptosis in SW480 cells.
Figure 3.
HOXC13 knockdown affects cell cycle progression. SW480 cells were transfected with varying concentrations (0.75 – 1.75 µg/mL) of HOXC13 and scramble antisenses (1.25 µg/mL) for 48 h. Cells were harvested, fixed with 70% ethanol/PBS, stained with PI and subjected to flow cytometry analysis. Cell population percentage at each stages of the cell cycle are shown in the bottom panel.
HOXC13 knockdown induced nuclear fragmentation and cytochrome c release and misregulated cyclin expression resulting in apoptosis
Apoptosis is a programmed cell death mechanism by which body eliminates unwanted cells and can be induced by various external or internal stimuli. Nuclear fragmentation, changes in mitochondrial membrane potential, caspase activation etc are different signatures of apoptotic cells (37–39). Herein, to examine the nature of cell death under HOXC13 knockdown environment, we transfected SW480 cells with HOXC13 antisense and subjected to TUNEL assay, DAPI (nuclear staining dye) staining, and cytochrome-c immunostaining analysis.
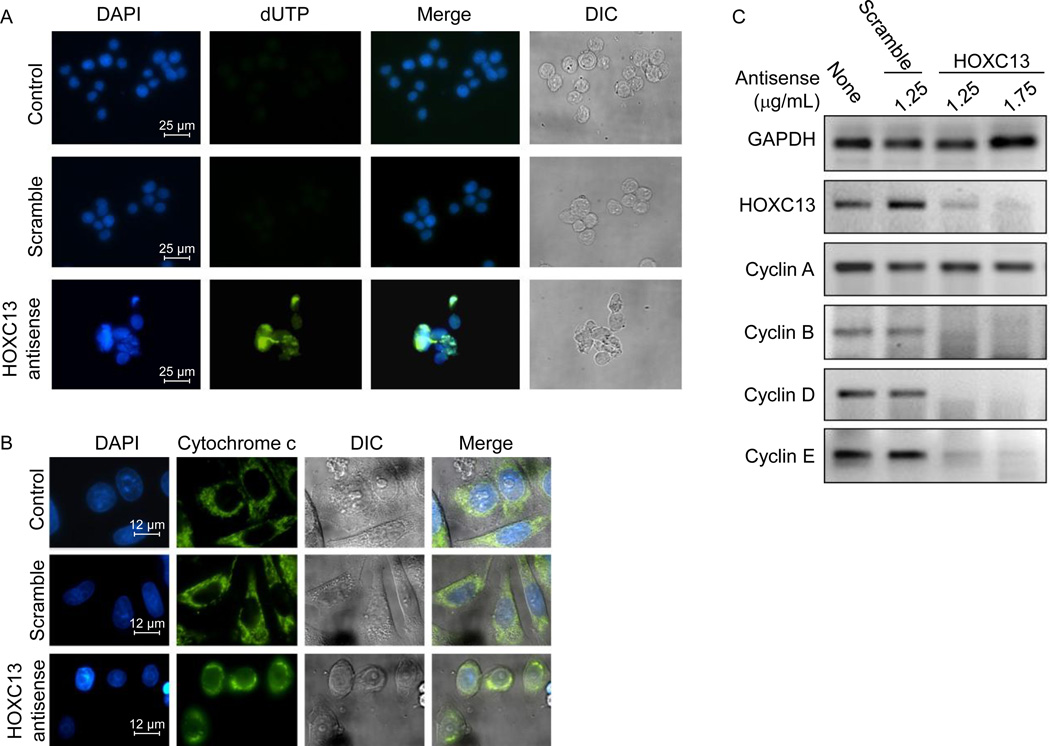
DAPI staining showed that knockdown of HOXC13 induced nuclear fragmentation and condensation (intense DAPI staining) in SW480 cells (Figure 4A, bottom panel). TUNEL assay demonstrated that transfection with HOXC13 antisense resulted in DNA fragmentation (dUTP labeling at the nicked DNA sites, shown by green colored nuclei, Figure 4A). Scramble antisense had no significant impact on nuclear staining and morphologies and most cells remained healthy (Figure 4A). Light microscopic images (differential interference contrast or DIC images) also showed that under HOXC13 antisense-treated environment, cells appeared very sick or dead or converted into cell debris, in comparison to the control cells (Figure 4A, right panels). Notably, cell populations in the HOXC13 antisense-treated cell were low because the HOXC13 knockdown affected the cell growth (Figure 4A).
Figure 4.
HOXC13 knockdown affects cell viability, nuclear integrity and cell cycle regulatory gene expressions. (A)TUNEL assay: SW480 cells were transfected with 1.75 µg/mL of HOXC13 and scramble antisense (1.75 µg/mL) separatally for 48 h, fixed with 4% formaldehyde and then subjected to terminal dUTP nicked end labeling and stained with DAPI. The green speckles are showing apoptotic cells containing fragmented DNA. (B) Cytochrome-c release assay: HOXC13 knocked down and control cells were immuno-stained with cytochrome-c antibody and then FITC-labeled secondary antibody and visualized under fluorescence microscope. Nuclear staining was performed with DAPI. (C) Effect of HOXC13 knockdown on cyclin expression. SW480 cells were treated with 1.25 and 1.75 µg/mL of HOXC13 and scramble antisense (1.25 µg/mL) for 48 h. RNA from control and antisense treated cells were harvested, reverse-transcribed and subjected to PCR using specific primers.
Mitochondrial membrane potential is known to be perturbed upon apoptotic cell death and these results in release of cytochrome-c from the mitochondria to cytosol. To examine the impacts of HOXC13 knockdown on mitochondrial membrane potential and integrity, we have immunostained HOXC13 antisense treated cells with cytochrome-c antibody and subjected to immunofluorescence analysis. DAPI staining was performed to stain the nuclei. This analysis demonstrated that cytochrome-c is located inside the mitochondria (green speckles outside the nucleus) in both control and scramble antisense-treated cells (Figure 4B). However upon transfection with HOXC13 antisense, cytochrome-c was released from mitochondria to cytosol in most cells indicating perturbation in mitochondrial membrane potential and apoptotic cell death induced by HOXC13 knockdown (Figure 4B).
To understand the potential mechanism of cell cycle arrest and cell death induced by HOXC13 knockdown, we examined the expression of various cell cycle regulatory genes such as cyclins in the absence and presence of HOXC13 knockdown environment. Notably, the changes in expression levels of cyclins are associated with normal cell cycle progression (40–42). For example, cyclin D and E are required for progression through G1 phase, cyclin A is produced in late G1 and it accumulates during S and G2 phase, while cyclin B is typically expressed during the G2 to M phase transition (40, 41). Our results showed that HOXC13 knockdown down-regulated the expression of cyclin B, D and E, while cyclin A was mostly unaffected (Figure 4C). The misregulation of cyclin expression is likely the cause of observed cell cycle arrest and apoptosis induced upon knockdown of HOXC13 in SW480 cells.
HOXC13 over expression induced 3D-tumor colony formation in soft-agar assay
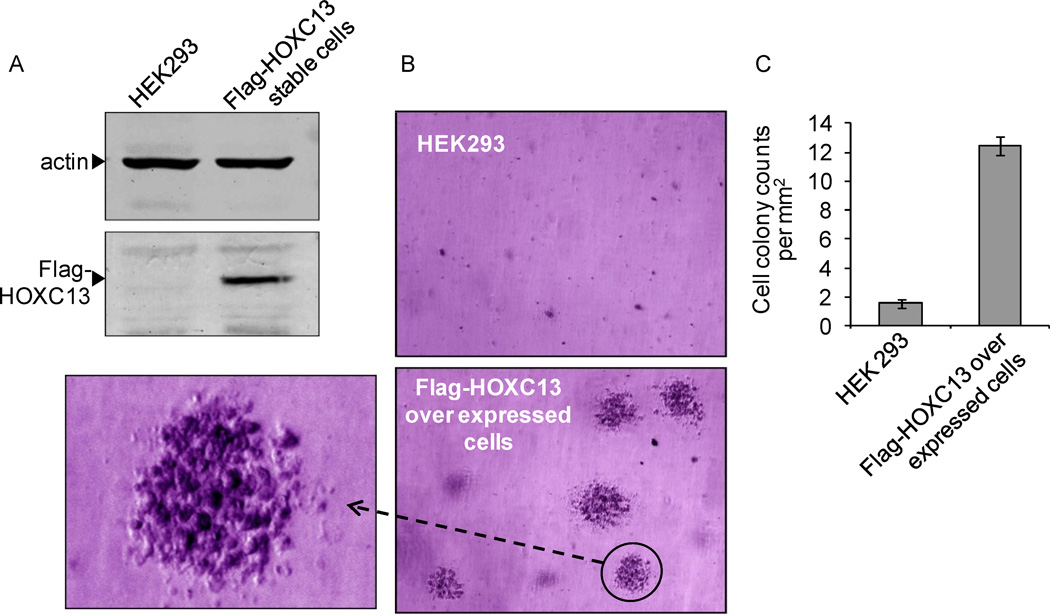
As our studies showed that HOXC13 is critical for growth and viability of tumor cells and its knockdown induces apoptosis, we examined the impact of HOXC13 over expression on tumor cell proliferation and growth using colony formation assay. In brief, initially we generated a stable transfected cell line over expressing HOXC13 in HEK293 cells (that is originated from a healthy embryonic tissue). Human HOXC13 gene was cloned in Flag-CMV4 human expression construct, transfected into HEK293 cells and stable transformants were selected using G418 antibiotic selection procedure (34, 36). Western blotting analysis demonstrated that Flag-HOXC13 is overexpressed in Flag-HOXC13 stable transfected cell line (Figure 5A).
Figure 5.
HOXC13 over expression induces tumorigenesis. (A) Western blotting for HOXC13 overexpression analysis. Proteins were isloated from HEK293 cells and stable cells expressing Flag-HOXC13 and subjected to Westen blotting using actin(control) and flag antibodies. (B) 3D-colony formation ability of HOXC13 over expressed cells. HEK293 and Flag-HOXC13 stable cells were incubated in soft agar for 4–5 weeks stained with 0.005% crystal violet and analyzed under microscope. Magnified view of one HOXC13 over expressed colony is shown on the left side. (C) Number of colonies grown in soft agar cultures were counted under light microscope and plotted. Bars indicate standard errors.
In order to examine the impact of HOXC13 in tumorigenesis, we performed 3D-colony formation assay using Flag-HOXC13 overexpressed stable cell line and control (non-transfected) HEK293 cells in soft agar (36). HEK293 cells and Flag-HOXC13 stable cells were separately plated in soft agar and were grown for 4–5 weeks, until distinct colonies were visible. Our analysis showed that while very few small colonies were observed in non-transfected HEK293 cells, HOXC13 over-expressing cells (Flag-HOXC13 stable cell line) formed distinctly visible large three dimensional colonies embedded in different layers of soft agar (Figure 5B, expanded view of colonies is shown in the side). The number of colonies were about 6–7 fold more in the HOXC13 over expressed cell line in comparison to the non-transfected HEK293 cells. Colonies in the HOXC13 over expressed cells appeared three dimensional and dense in cell population. These observations indicated that HOXC13 over expression enhanced the cell proliferation and formation of large 3D-growth of cell colonies indicating its potential role in tumorigenesis.
Conclusion
In this study we have developed a phosphorothioate based antisense-oligonucleotide for HOXC13 that specifically and efficiently knocked down HOXC13 in cultured human cells. Our studies demonstrated that HOXC13 is a key player in cell growth and viability. Antisense-mediated knockdown of HOXC13 affects the cell viability and apoptosis in cultured tumor cells. HOXC13 regulates the expression of cyclins that play crucial role in cell cycle progression. Antisense-mediated knockdown of HOXC13 resulted in cell cycle arrest and ultimately led to apoptosis in colon cancer cells. Finally over expression of HOXC13 resulted in 3D-colony formation in soft-agar assay indicating potential roles of HOXC13 in cell proliferation and tumorigenesis. Overall, we showed that HOXC13 is a critical player in tumor cell viability, proliferation and tumorigenesis and can be targeted for developing novel cancer therapy.
Acknowledgement
We thank Mandal lab members for helpful discussions. Research in Mandal laboratory is supported in parts by grants from National Institute of Health (1R15 ES019129-01, 2R15 CA113747-02) and National Science Foundation (1148541).
References
- 1.Gleave ME, Monia BP. Antisense therapy for cancer. Nat Rev Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 2.Kurreck J. Therapeutic oligonucleotides. Cambridge: RSC Pub.; 2008. [Google Scholar]
- 3.Rayburn ER, Zhang R. Antisense, RNAi, and gene silencing strategies for therapy: Mission possible or impossible? Drug Discovery Today. 2008;13:513–521. doi: 10.1016/j.drudis.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Nigris F, Balestrieri ML, Napoli C. Targeting c-Myc, Ras and IGF cascade to treat cancer and vascular disorders. Cell cycle (Georgetown, Tex) 2006;5:1621–1628. doi: 10.4161/cc.5.15.3138. [DOI] [PubMed] [Google Scholar]
- 5.Crooke ST. Potential roles of antisense technology in cancer chemotherapy. Oncogene. 2000;19:6651–6659. doi: 10.1038/sj.onc.1204093. [DOI] [PubMed] [Google Scholar]
- 6.Smith S. Anti-Bcl2 therapy in chronic myelogenous leukemia. Leukemia & lymphoma. 2008;49:1232–1233. doi: 10.1080/10428190802105090. [DOI] [PubMed] [Google Scholar]
- 7.Lappin TR, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. Ulster Med J. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence HJ, Sauvageau G, Humphries RK, Largman C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells. 1996;14:281–291. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- 9.Rubin MR, King W, Toth LE, Sawczuk IS, Levine MS, D'Eustachio P, Nguyen-Huu MC. Murine Hox-1.7 homeo-box gene: cloning, chromosomal location, and expression. Mol Cell Biol. 1987;7:3836–3841. doi: 10.1128/mcb.7.10.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen WF, Chang CP, Rozenfeld S, Sauvageau G, Humphries RK, Lu M, Lawrence HJ, Cleary ML, Largman C. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science (New York, N Y) 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 12.Reid AI, Gaunt SJ. Colinearity and non-colinearity in the expression of Hox genes in developing chick skin. Int J Dev Biol. 2002;46:209–215. doi: 10.1387/ijdb.011495. [DOI] [PubMed] [Google Scholar]
- 13.Neville SE, Baigent SM, Bicknell AB, Lowry PJ, Gladwell RT. Hox gene expression in adult tissues with particular reference to the adrenal gland. Endocrine Res. 2002;28:669–673. doi: 10.1081/erc-120016984. [DOI] [PubMed] [Google Scholar]
- 14.Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. FEBS J. 2009;276:1629–1640. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Sukumar S. HOX genes: emerging stars in cancer. Cancer Biol Ther. 2003;2:524–525. doi: 10.4161/cbt.2.5.525. [DOI] [PubMed] [Google Scholar]
- 16.Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. J Mol Endocrinol. 2012;48:61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- 17.McCabe CD, Spyropoulos DD, Martin D, Moreno CS. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68:1988–1996. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, Lucia MS, Nordeen SK. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 19.Hayashida T, Takahashi F, Chiba N, Brachtel E, Takahashi M, Godin-Heymann N, Gross KW, Vivanco MM, Wijendran V, Shioda T, Sgroi D, Donahoe PK, Maheswaran S. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci U S A. 2010;107:1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jave-Suarez LF, Schweizer J. The HOXC13-controlled expression of early hair keratin genes in the human hair follicle does not involve TALE proteins MEIS and PREP as cofactors. Arch Dermatol Res. 2006;297:372–376. doi: 10.1007/s00403-005-0623-3. [DOI] [PubMed] [Google Scholar]
- 21.Awgulewitsch A. Hox in hair growth and development. Naturwissenschaften. 2003;90:193–211. doi: 10.1007/s00114-003-0417-4. [DOI] [PubMed] [Google Scholar]
- 22.Godwin AR, Capecchi MR. Hair defects in Hoxc13 mutant mice. J Investig Dermatol Symp Proc. 1999;4:244–247. doi: 10.1038/sj.jidsp.5640221. [DOI] [PubMed] [Google Scholar]
- 23.Marchetti L, Comelli L, D'Innocenzo B, Puzzi L, Luin S, Arosio D, Calvello M, Mendoza-Maldonado R, Peverali F, Trovato F, Riva S, Biamonti G, Abdurashidova G, Beltram F, Falaschi A. Homeotic proteins participate in the function of human-DNA replication origins. Nucleic Acids Res. 2010;38:8105–8119. doi: 10.1093/nar/gkq688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falaschi A, Abdurashidova G, Biamonti G. DNA replication, development and cancer: a homeotic connection? Critical Rev Biochem Mol Biol. 2010;45:14–22. doi: 10.3109/10409230903365608. [DOI] [PubMed] [Google Scholar]
- 25.Comelli L, Marchetti L, Arosio D, Riva S, Abdurashidova G, Beltram F, Falaschi A. The homeotic protein HOXC13 is a member of human DNA replication complexes. Cell cycle (Georgetown, Tex) 2009;8:454–459. doi: 10.4161/cc.8.3.7649. [DOI] [PubMed] [Google Scholar]
- 26.Panagopoulos I, Isaksson M, Billstrom R, Strombeck B, Mitelman F, Johansson B. Fusion of the NUP98 gene and the homeobox gene HOXC13 in acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36:107–112. doi: 10.1002/gcc.10139. [DOI] [PubMed] [Google Scholar]
- 27.Ansari KI, Kasiri S, Hussain I, Mandal SS. Mixed lineage leukemia histone methylases play critical roles in estrogen-mediated regulation of HOXC13. FEBS J. 2009;276:7400–7411. doi: 10.1111/j.1742-4658.2009.07453.x. [DOI] [PubMed] [Google Scholar]
- 28.Ansari KI, Hussain I, Shrestha B, Kasiri S, Mandal SS. HOXC6 Is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. J Mol Biol. 2011;411:334–349. doi: 10.1016/j.jmb.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ansari KI, Grant JD, Woldemariam GA, Kasiri S, Mandal SS. Iron(III)-salen complexes with less DNA cleavage activity exhibit more efficient apoptosis in MCF7 cells. Org Biomol Chem. 2009;7:926–932. doi: 10.1039/b816858j. [DOI] [PubMed] [Google Scholar]
- 30.Ansari KI, Grant JD, Kasiri S, Woldemariam G, Shrestha B, Mandal SS. Manganese(III)-salens induce tumor selective apoptosis in human cells. J Inorg Biochem. 2009;103:818–826. doi: 10.1016/j.jinorgbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Ansari KI, Kasiri S, Mishra BP, Mandal SS. Mixed lineage leukaemia-4 regulates cell-cycle progression and cell viability and its depletion suppresses growth of xenografted tumour in vivo. Bri J Cancer. 2012;107:315–324. doi: 10.1038/bjc.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ansari KI, Kasiri S, Grant JD, Mandal SS. Fe(III)-Salen and Salphen Complexes Induce Caspase Activation and Apoptosis in Human Cells. J Biomol Screen. 2011;16:26–35. doi: 10.1177/1087057110385227. [DOI] [PubMed] [Google Scholar]
- 33.Ansari KI, Kasiri S, Mandal SS. Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene. 2012 doi: 10.1038/onc.2012.352. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ansari KI, Mishra BP, Mandal SS. Human CpG binding protein interacts with MLL1, MLL2 and hSet1 and regulates Hox gene expression. Biochim Biophys Acta. 2008;1779:66–73. doi: 10.1016/j.bbagrm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Park SI, Artime MC, Summy JM, Shah AN, Bomser JA, Dorfleutner A, Flynn DC, Gallick GE. AFAP-110 is overexpressed in prostate cancer and contributes to tumorigenic growth by regulating focal contacts. J Clin Invest. 2007;117:2962–2973. doi: 10.1172/JCI30710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha B, Ansari KI, Bhan A, Kasiri S, Hussain I, Mandal SS. Homeodomain-containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth invitro and is overexpressed in breast cancer tissue. FEBS J. 2012;279:3715–3726. doi: 10.1111/j.1742-4658.2012.08733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park MS, De Leon M, Devarajan P. Cisplatin induces apoptosis in LLC-PK1 cells via activation of mitochondrial pathways. J Am Soc Nephrol. 2002;13:858–865. doi: 10.1681/ASN.V134858. [DOI] [PubMed] [Google Scholar]
- 38.Orrenius S, Gogvadze A, Zhivotovsky B. Mitochondrial oxidative stress: Implications for cell death. Ann Rev Pharmacol Toxicol. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 39.Woldemariam GA, Mandal SS. Iron(III)-salen damages DNA and induces apoptosis in human cell via mitochondrial pathway. J Inorg Biochem. 2008;102:740–747. doi: 10.1016/j.jinorgbio.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Obaya AJ, Sedivy JM. Regulation of cyclin-Cdk activity in mammalian cells. Cell Mol Life Sci. 2002;59:126–142. doi: 10.1007/s00018-002-8410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 42.Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Front Biosci. 2009;14:3483–3495. doi: 10.2741/3466. [DOI] [PubMed] [Google Scholar]