Abstract
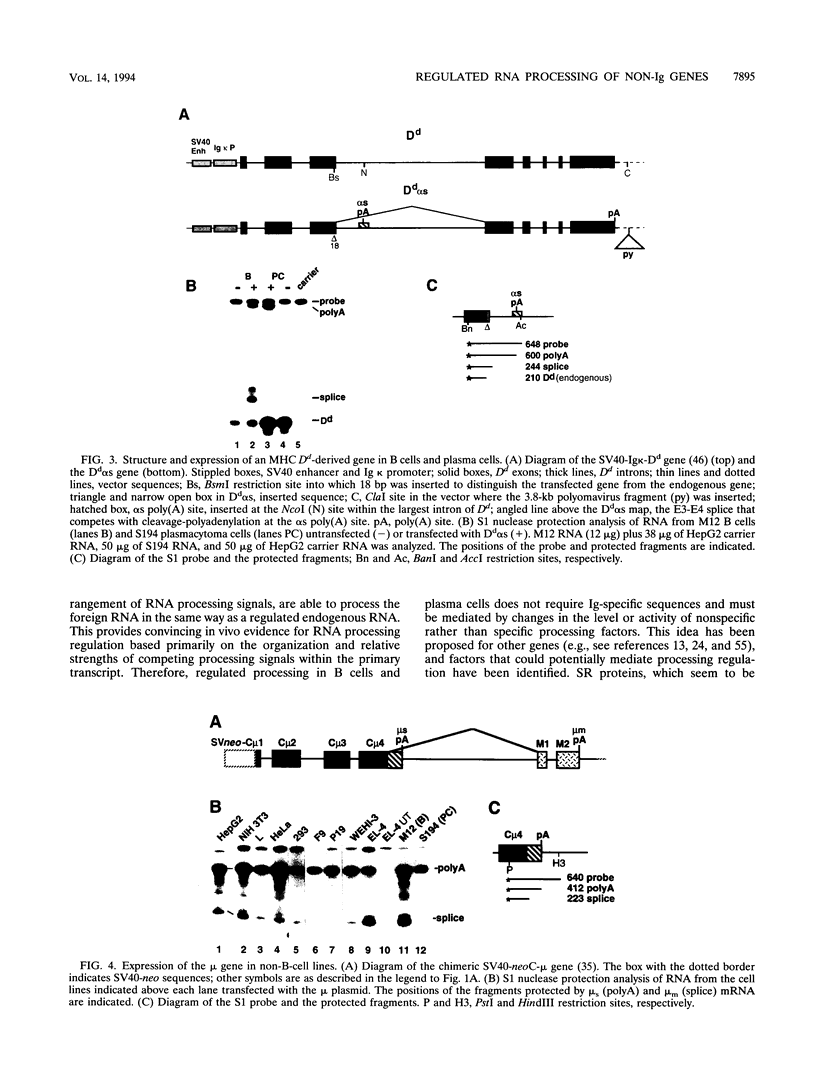
Alternative RNA processing of the heavy-chain immunoglobulin mu gene is regulated during B-cell maturation and requires competition between splice and cleavage-polyadenylation reactions that have balanced efficiencies. Studies with modified mu genes have failed to identify gene-specific sequences required for regulation. Thus, the only important feature for regulation may be the balanced competing splice and cleavage-polyadenylation reactions themselves. If this is so, then alternative RNA processing from any gene with similar competitive RNA processing pathways should also be regulated when expression is compared between B cells and plasma cells. To test this prediction, two nonimmunoglobulin genes engineered to have competing splice and cleavage-polyadenylation reactions were expressed in B cells and plasma cells. The ratios of alternative RNAs produced from both genes are different in the two cell types; like the mu gene, relatively more spliced RNA is produced in B cells than in plasma cells. Also, in a survey of mu gene expression in nine non-B-cell lines, only a T-cell line had an expression pattern similar to that of B cells; the expression patterns of all other lines resembled that of the plasma cells. Therefore, regulated mu RNA processing must be mediated by changes in general processing factors whose activity or abundance is regulated, most likely, in B cells.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Benech P., Merlin G., Revel M., Chebath J. 3' end structure of the human (2'-5') oligo A synthetase gene: prediction of two distinct proteins with cell type-specific expression. Nucleic Acids Res. 1985 Feb 25;13(4):1267–1281. doi: 10.1093/nar/13.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Tucker P. W. The molecular biology of immunoglobulin D. Nature. 1984 Feb 2;307(5950):417–422. doi: 10.1038/307417a0. [DOI] [PubMed] [Google Scholar]
- Brown S. L., Morrison S. L. Developmental regulation of membrane and secretory Ig gamma 2b mRNA. J Immunol. 1989 Mar 15;142(6):2072–2080. [PubMed] [Google Scholar]
- Buchman A. R., Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988 Oct;8(10):4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Chung S., Perry R. P. Importance of introns for expression of mouse ribosomal protein gene rpL32. Mol Cell Biol. 1989 May;9(5):2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris P., Levitt N., Briggs D., Proudfoot N. J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991 Jul;10(7):1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshhar Z., Blatt C., Bergman Y., Heimovich J. Induction of secretion of IgM from cells of the B cell line 38c-13 by somatic cell hybridization. J Immunol. 1979 Jun;122(6):2430–2434. [PubMed] [Google Scholar]
- Fu X. D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993 Sep 2;365(6441):82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise J. W., McDevitt M. A., Tucker P. W., Nevins J. R. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted mu-chain expression during B-cell development. Genes Dev. 1987 Jul;1(5):471–481. doi: 10.1101/gad.1.5.471. [DOI] [PubMed] [Google Scholar]
- Ge H., Manley J. L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990 Jul 13;62(1):25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985 Jul;41(3):885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- Huang M. T., Gorman C. M. Intervening sequences increase efficiency of RNA 3' processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990 Feb 25;18(4):937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp D. J., Harris A. W., Adams J. M. Transcripts of the immunoglobulin C mu gene vary in structure and splicing during lymphoid development. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7400–7404. doi: 10.1073/pnas.77.12.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Kraus M. E., Lis J. T. The concentration of B52, an essential splicing factor and regulator of splice site choice in vitro, is critical for Drosophila development. Mol Cell Biol. 1994 Aug;14(8):5360–5370. doi: 10.1128/mcb.14.8.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskov R., Kim K. J., Asofsky R. Induction of amplified synthesis and secretion of IgM by fusion of murine 'b lymphoma with myeloma cells. Proc Natl Acad Sci U S A. 1979 Feb;76(2):915–919. doi: 10.1073/pnas.76.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassman C. R., Matis S., Hall B. L., Toppmeyer D. L., Milcarek C. Plasma cell-regulated polyadenylation at the Ig gamma 2b secretion-specific poly(A) site. J Immunol. 1992 Feb 15;148(4):1251–1260. [PubMed] [Google Scholar]
- Lassman C. R., Milcarek C. Regulated expression of the mouse gamma 2b Ig H chain gene is influenced by polyA site order and strength. J Immunol. 1992 Apr 15;148(8):2578–2585. [PubMed] [Google Scholar]
- Levitt N., Briggs D., Gil A., Proudfoot N. J. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989 Jul;3(7):1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- Mann K. P., Weiss E. A., Nevins J. R. Alternative poly(A) site utilization during adenovirus infection coincides with a decrease in the activity of a poly(A) site processing factor. Mol Cell Biol. 1993 Apr;13(4):2411–2419. doi: 10.1128/mcb.13.4.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992 Jan 24;68(2):365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi T., Tennyson G. E., Nikodem V. M. Alternative splicing generates messages encoding rat c-erbA proteins that do not bind thyroid hormone. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5804–5808. doi: 10.1073/pnas.85.16.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., Vuocolo G. A. Synthesis of two mRNAs by utilization of alternate polyadenylation sites: expression of SV40-mouse immunoglobulin mu chain gene recombinants in Cos monkey cells. EMBO J. 1984 Apr;3(4):689–699. doi: 10.1002/j.1460-2075.1984.tb01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M., MacDonald C. C., Berget S. M. Are vertebrate exons scanned during splice-site selection? Nature. 1992 Nov 19;360(6401):277–280. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- Peterson M. L. Balanced efficiencies of splicing and cleavage-polyadenylation are required for mu-s and mu-m mRNA regulation. Gene Expr. 1992;2(4):319–327. [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Bryman M. B., Peiter M., Cowan C. Exon size affects competition between splicing and cleavage-polyadenylation in the immunoglobulin mu gene. Mol Cell Biol. 1994 Jan;14(1):77–86. doi: 10.1128/mcb.14.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Gimmi E. R., Perry R. P. The developmentally regulated shift from membrane to secreted mu mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol Cell Biol. 1991 Apr;11(4):2324–2327. doi: 10.1128/mcb.11.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989 Feb;9(2):726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D. J., Hawley J., Dang T., Yuan D. Role of differential mRNA stability in the regulated expression of IgM and IgD. J Immunol. 1994 Jun 1;152(11):5330–5336. [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruether J. E., Maderious A., Lavery D., Logan J., Fu S. M., Chen-Kiang S. Cell-type-specific synthesis of murine immunoglobulin mu RNA from an adenovirus vector. Mol Cell Biol. 1986 Jan;6(1):123–133. doi: 10.1128/mcb.6.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Marcu K. B., Perry R. P. The synthesis and processing of the messenger RNAs specifying heavy and light chain immunoglobulins in MPC-11 cells. Cell. 1978 Dec;15(4):1495–1509. doi: 10.1016/0092-8674(78)90072-7. [DOI] [PubMed] [Google Scholar]
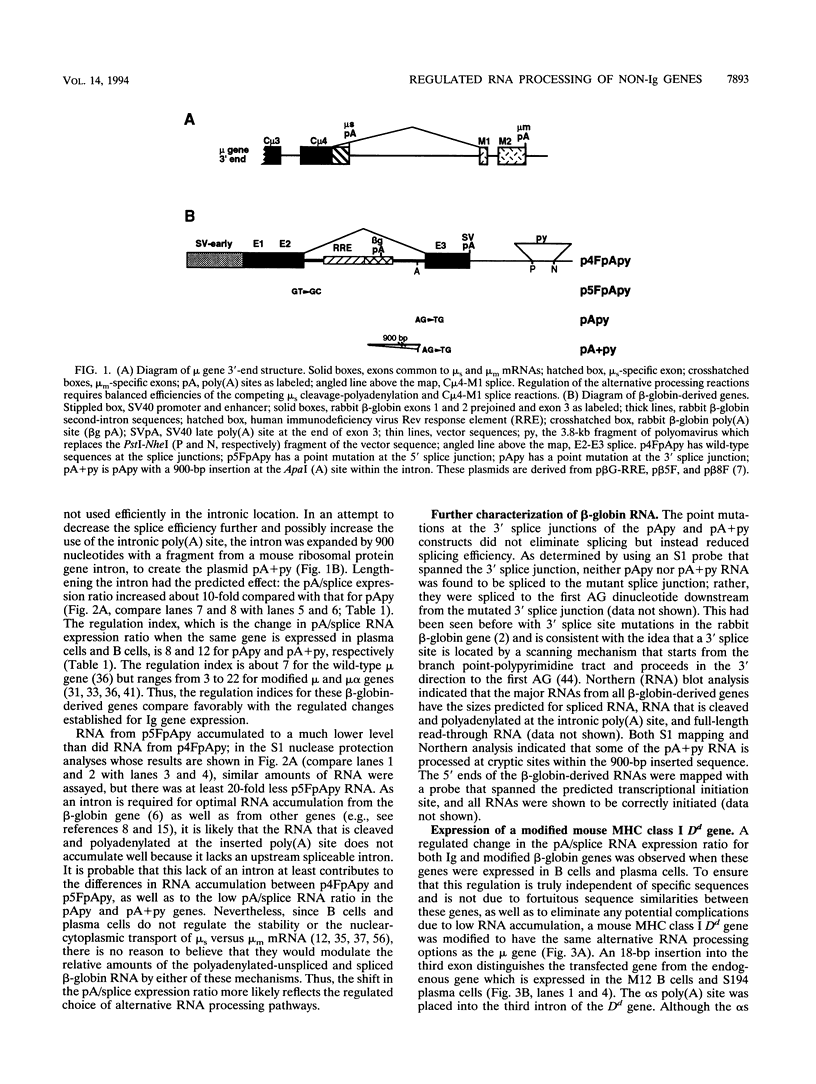
- Sher B. T., Nairn R., Coligan J. E., Hood L. E. DNA sequence of the mouse H-2Dd transplantation antigen gene. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1175–1179. doi: 10.1073/pnas.82.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Porro E. B., Patton J. G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3' splice site of mammalian introns. Nature. 1989 Nov 16;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Spear B. T., Tilghman S. M. Role of alpha-fetoprotein regulatory elements in transcriptional activation in transient heterokaryons. Mol Cell Biol. 1990 Oct;10(10):5047–5054. doi: 10.1128/mcb.10.10.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Mayeda A., Hampson R. K., Krainer A. R., Rottman F. M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993 Dec;7(12B):2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- Tsurushita N., Avdalovic N. M., Korn L. J. Regulation of differential processing of mouse immunoglobulin mu heavy-chain mRNA. Nucleic Acids Res. 1987 Jun 11;15(11):4603–4615. doi: 10.1093/nar/15.11.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurushita N., Korn L. J. Effects of intron length on differential processing of mouse mu heavy-chain mRNA. Mol Cell Biol. 1987 Jul;7(7):2602–2605. doi: 10.1128/mcb.7.7.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P. W., Slightom J. L., Blattner F. R. Mouse IgA heavy chain gene sequence: implications for evolution of immunoglobulin hinge axons. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7684–7688. doi: 10.1073/pnas.78.12.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Perry R. P. Characterization of the multigene family encoding the mouse S16 ribosomal protein: strategy for distinguishing an expressed gene from its processed pseudogene counterparts by an analysis of total genomic DNA. Mol Cell Biol. 1985 Dec;5(12):3560–3576. doi: 10.1128/mcb.5.12.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Sakamoto H., Shimura Y. Repositioning of an alternative exon sequence of mouse IgM pre-mRNA activates splicing of the preceding intron. Gene Expr. 1991;1(3):175–184. [PMC free article] [PubMed] [Google Scholar]
- Winkelmann J. C., Costa F. F., Linzie B. L., Forget B. G. Beta spectrin in human skeletal muscle. Tissue-specific differential processing of 3' beta spectrin pre-mRNA generates a beta spectrin isoform with a unique carboxyl terminus. J Biol Chem. 1990 Nov 25;265(33):20449–20454. [PubMed] [Google Scholar]
- Yang X., Bani M. R., Lu S. J., Rowan S., Ben-David Y., Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5' splice site selection in vivo. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley J. M., Hedjran F., Morfin J. P., Merillat N., Rosenfeld M. G., Emeson R. B. Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol Cell Biol. 1993 Oct;13(10):5999–6011. doi: 10.1128/mcb.13.10.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D., Dang T. Regulation of microM vs microS mRNA expression in an inducible B cell line. Mol Immunol. 1989 Nov;26(11):1059–1067. doi: 10.1016/0161-5890(89)90070-9. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Lane W. S., Stolk J. A., Roth M. B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992 May;6(5):837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Neugebauer K. M., Lane W. S., Roth M. B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993 Apr 9;260(5105):219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]