Abstract
A vertex-differentiated icosahedral closo-B122− core was utilized to construct a αvβ3 integrin receptor-targeted (via cRGD peptide) high payload MRI contrast agent (CA-12) carrying 11 copies of Gd3+-DOTA chelates attached to the closo-B122− surface via suitable linkers. The resulting polyfunctional MRI contrast agent possessed a higher relaxivity value per-Gd compared to Omniscan, a small molecular contrast agent commonly used in clinical settings. The αvβ3 integrin receptor specificity of CA-12 was confirmed via in vitro cellular binding experiments and in vivo MRI of mice bearing human PC-3 prostate cancer xenografts. Integrin αvβ3-positive MDA-MB-231 cells exhibited 300% higher uptake of CA-12 than αvβ3-negative T47D cells. Serial T1-weighted MRI showed superior contrast enhancement of tumors by CA-12 compared to both a non-targeted 12-fold Gd3+-DOTA closomer control (CA-7) and Omniscan. Contrast enhancement by CA-12 persisted for 4 h post-injection, and subsequent enhancement of kidney tissue indicated a renal elimination route similar to Omniscan. No toxic effects of CA-12 were apparent in any mice for up to 24 h post-injection. Post-mortem ICP-OES analysis at 24 hours detected no residual Gd in any of the tissue samples analyzed.
INTRODUCTION
Magnetic resonance imaging (MRI) is one of the most powerful diagnostic modalities in clinical medicine and biomedical research. The resolution and sensitivity of MRI is further enhanced through the administration of a contrast agent (CA). Due to their positive contrast enhancement properties, paramagnetic gadolinium-based small molecular CAs have been used most widely in clinical settings.1 A significant drawback of these CAs is their lack of specificity and failure to provide sufficient contrast at low concentrations. With regard to cancer detection, targeting of MRI CA to cellular markers specifically expressed by tumor cells would greatly facilitate diagnosis, monitoring of disease progression, and assessment of treatment success. Research toward the development of targeted CAs has been ongoing, with most efforts directed toward conjugation of a CA to specific antibodies or a peptide.2,3
Efforts have also been directed at producing tissue specific MRI CAs.4 In particular, the αvβ3 integrin receptor has been extensively investigated as a molecular target due to its involvement in angiogenesis, proliferation, and metastasis of several tumor types.5 Integrin αvβ3 has high expression on proliferating endothelial cells during tumor angiogenesis and metastasis in contrast to resting cells.6,7 Since the αvβ3 integrin receptors binds to certain extracellular matrix proteins via the peptide sequence Arg-Gly-Asp (RGD), diverse RGD peptide ligands have been investigated as a targeting agent for αvβ3 integrin.8 In particular, the cyclic RGD peptide (cRGD) has a relatively high and specific affinity for αvβ3 integrin receptors. The synthesis of several small molecular MRI CAs possessing a cRGD peptide conjugated to a Gd3+-chelate have been reported in the literature.5
However, small molecular CAs are in practice unsuitable for targeted imaging applications because of the high tissue concentrations of Gd3+ ions necessary to obtain sufficient contrast. Target receptors associated with particular tumor types or tissue pathologies are typically expressed at very low levels. In the case of αvβ3 integrin, the αv and β3 subunits number only 3×103 to 1.4×104 and 5.3×102 to 1.1×104 copies per cell, respectively.9 In order to obtain sufficient contrast enhancement, the number of Gd3+ ions bound per target must be substantially increased.
The problem of insufficient Gd concentration in targeted imaging has been addressed by the use of macromolecules (dendrimers, liposomes, micelles, nanoparticles) to which large numbers of Gd3+-chelates are attached along with targeting moieties.10 Conjugation of Gd3+-chelates to macromolecules offers the additional advantage of increasing the rotational correlation time, resulting in improved relaxivity (r1) and therefore increased contrast enhancement.11 However, despite their satisfactory in vivo biodistribution profiles and high r1 values, these macromolecular CAs suffer from the problem of polydispersity. Polydispersity has impact on the stability, solubility, appearance and texture of the macromolecule and thus can affect the overall efficacy of the drug or imaging agent.
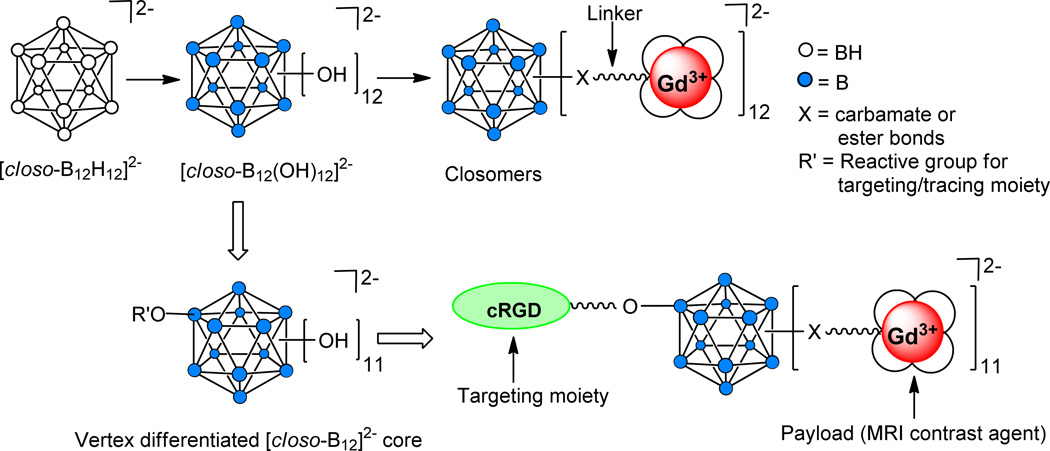
Polyhedral boranes and carboranes have generated considerable interest in biomedical research because of their potential as monodispersed nanocarriers of diagnostic and therapeutic agents.12 Hydroxylation of all 12 B-H vertices13 of icosahedral [closo-B12H12]2− to [closo-B12(OH)12]2− permits the attachment of up to 12 copies of a desired functionality through suitable linkers even at zero generation.14 The 12 linker arms are in close proximity to one another, a configuration that makes these compounds ideally suited for the construction of monodispersed nanomolecular MRI CAs. In a companion paper, we reported the synthesis of the first such contrast agent, an icosahedral closo-B122 scaffold possessing 12 Gd+3-chelates (Figure 1).15
Figure 1.
Schematic representation of a vertex-differentiated [closo-B12]2− core supporting a targeting moiety (cRGD) and 11 copies of a MRI contrast agent.
Synthesis of a targeted nanomolecular CA requires the presence of heterobifunctionalized linker arms on the same scaffold to permit attachment of both the targeting moiety as well as multiple copies of Gd3+-chelates. We recently developed a synthetic methodology to differentiate a single B-OH vertex of [closo-B12(OH)12]2− to provide [closo-B12(OR)(OH)11]2−. This methodology permitted the preparation of several vertex-differentiated closomers [closo-B12(OR)(OR’)11]2− utilizing orthogonal chemical reactions.16
As proof of principle, we herein report the first application of this unique approach to synthesize a tumor-targeting nanomolecular MRI CA derived from a vertex-differentiated icosahedral closo-B122− core. This closomer supports an αvβ3 integrin-targeting peptide (cRGD) covalently linked to one vertex, with the remaining 11 vertices carrying a high payload of Gd3+-chelates (Figure 1). Using a cell binding assay, we demonstrate the specificity of this targeted CA for αvβ3 integrin receptors. Also presented is a comparative analysis of the relaxivity and contrast enhancement kinetics of the targeted CA to a non-targeted 12-fold closomer CA and to the commercially available small molecular CA Omniscan.
RESULTS AND DISCUSSION
Abbreviations. N,N’-dimethylformamide (DMF), triethylamine (Et3N), sodium sulfate (Na2SO4), sodium hydride (NaH), hexaethylene glycol (HEG), 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA), 1,4,7,10-tetraazacyclododecane-1,4,7-tris(t-butyl acetate) (DO3A-t-Buester), potassium hydrogen carbonate (KHCO3) tetrahydrofuran (THF), dichloromethane (DCM), ethyl acetate (EtOAc ), methanesulfonyl chloride (MsCl) acetonitrile (ACN), ammonium chloride (NH4Cl), methanol (MeOH), hydrochloric acid (HCl), N,N-diisopropylethylamine (DIPEA), room temperature (RT), diethylenetriaminepentaacetic acid (DTPA), diethylenetriaminetetraacetic acid (DTTA), trifluoroacetic acid (TFA), tetrabutyl ammonium (TBA), phosphate-buffered saline (PBS), molecular weight cut-off (MWCO), inductively coupled plasma optical emission spectroscopy (ICP-OES), and hours (h).
Synthesis of DOTA ligands
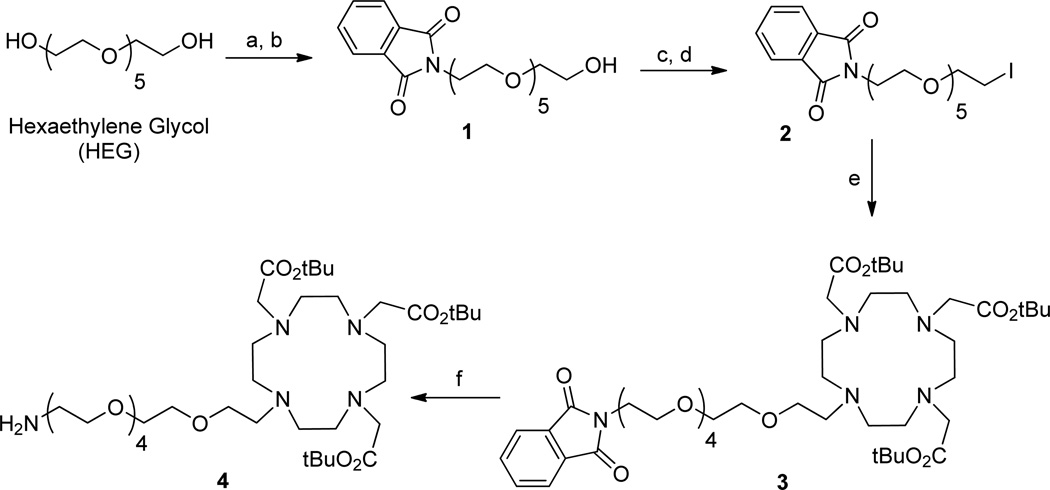
For this study, DOTA was selected as the Gd3+-chelating ligand because of the higher thermodynamic stability of Gd3+-DOTA (log KGdL~23) relative to Gd3+-DTPA (log KGdL~22) or Gd3+-DTTA (log KGdL~17–19) chelates.1c An amino-terminated DOTA ligand (4) was prepared via a six-step synthesis starting from hexaethylene glycol (HEG) (1) (Scheme 1). First, a phthalimide-functionalized HEG linker (1) was synthesized from commercially available HEG via a two-step process that converted one of the two hydroxyl groups to a phthalimide group in 44% yield. The remaining hydroxyl group in 1 was converted to a mesylate (−OMs) group, which upon reaction with sodium iodide, gave iodo-terminated HEG linker 2 in quantitative yield. The reaction of 2 with DO3A-t-Bu-ester produced DOTA derivative 3 in 96% yield. Deprotection of the phthalimide group on 3 using hydrazine gave the desired amino-terminated HEG-linked DOTA derivative 4 in 96% yield. The structures of compounds 1–4 were characterized by IR, NMR, and HRMS spectroscopy.
Scheme 1.
Synthesis of amino-terminated hexethylene glycol (HEG) linked DOTA ligand 4.
Reagents and conditions: (a) 1.0 eq. MsCl, Et3N, DCM, 0 °C-RT, 3 h; (b) Potassium phthalimide, DMF, 110 °C, 15 h, 44% in two steps; (c) 1.5 eq. MsCl, Et3N, DCM, 0 °C-RT, 3 h; (d) NaI, acetone, 65 °C, 15 h, 95% in two steps; (e) DO3A-t-Bu-ester, KHCO3, DMF, 12 h, RT, 96%; (f) Hydrazine, ethanol, 65 °C, 4 h, 96%.
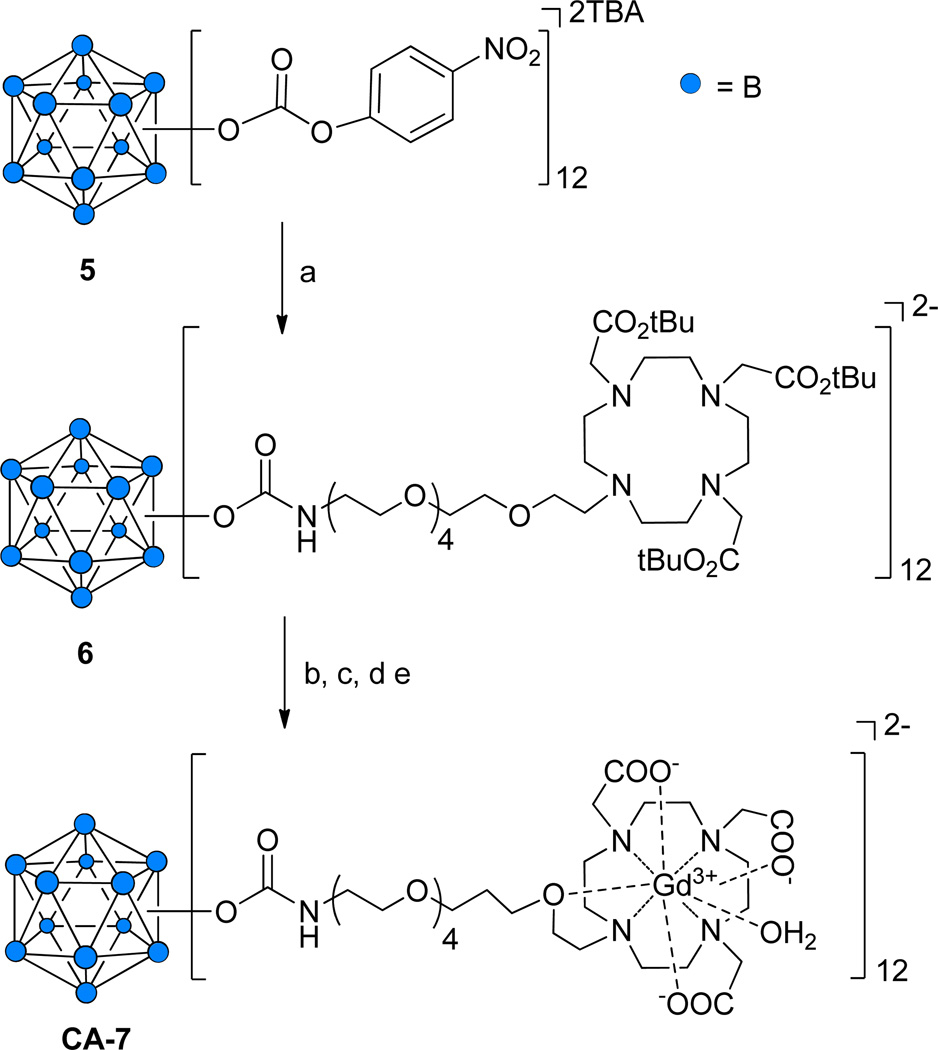
Synthesis of 12-fold Gd3+-DOTA-conjugated closomer CA-7
The preparation of a closomer conjugated to 12 Gd+3-DOTA chelates was accomplished in a stepwise procedure that relied upon our recently reported synthesis of 12-fold carbonate closomers capable of reacting with primary amines to yield 12-fold carbamate closomers.14a The carbonate closomer 5 was reacted with the amino-terminated DOTA ligand 4 to produce the 12-fold DOTA conjugate 6 in 80% yield (Scheme 2). Closomer 6 was purified by size-exclusion column chromatography on Lipophilic Sephadex LH-20 using MeOH as the eluent and characterized by IR, NMR, and HRMS spectroscopy. To remove the tert-butyl ester groups present on the DOTA ligands, closomer 6 was treated with 80% TFA in DCM. Complete deprotection was confirmed by the absence of a large singlet at δ 1.47 ppm from the 1H NMR spectrum of 6. Gd3+ chelation was achieved by reacting deprotected closomer 6 with GdCl3·6H2O in citrate buffer at pH ~7. Following extensive dialysis and lyophilization, the 12-fold Gd3+-DOTA-conjugated closomer CA-7 was obtained in 71% yield.
Scheme 2.
Synthesis of 12-fold Gd3+-DOTA closomer conjugate CA-7.
Reagents and conditions: (a) 4, ACN, 7 days, RT, 76%; (b) 80% TFA-DCM, 6 h, RT; (c) GdCl3·6H2O, citrate buffer, pH 7, 24 h, RT, sonication; (d) Dialysis, 1,000 MWCO, 48 h; (e) Lyophilization, 82%.
CA-7 was characterized by IR spectroscopy and ICP-OES analysis. The IR spectrum of CA-7 exhibited the characteristic shift of the carbonyl stretch from 1725 cm−1 to 1605 cm−1 indicative of complexation of Gd3+ with the DOTA ligands (Supporting Information). The gadolinium/boron content of CA-7 was determined using ICP-OES analysis. CA-7 contained an average of 11.4 Gd3+ ions per closomer, indicating essentially fully loaded DOTA ligands. Mass spectrometry analysis of CA-7 was unsuccessful; we could not obtain the desired mass peaks possibly due to the production of multiple charged species during the ionization step. The presence of free Gd3+ ions in the solution of closomer CA-7 was determined by a spectrophotometric method using xylenol orange.17 No free Gd3+ ions were detected. Dynamic light-scattering (DLS) analysis of CA-7 in PBS and 2% Tween-80/PBS solution gave an average particle size of 10 nm (Supporting Information).
Synthesis of targeted 11-fold Gd3+-DOTA conjugated closomer CA-12
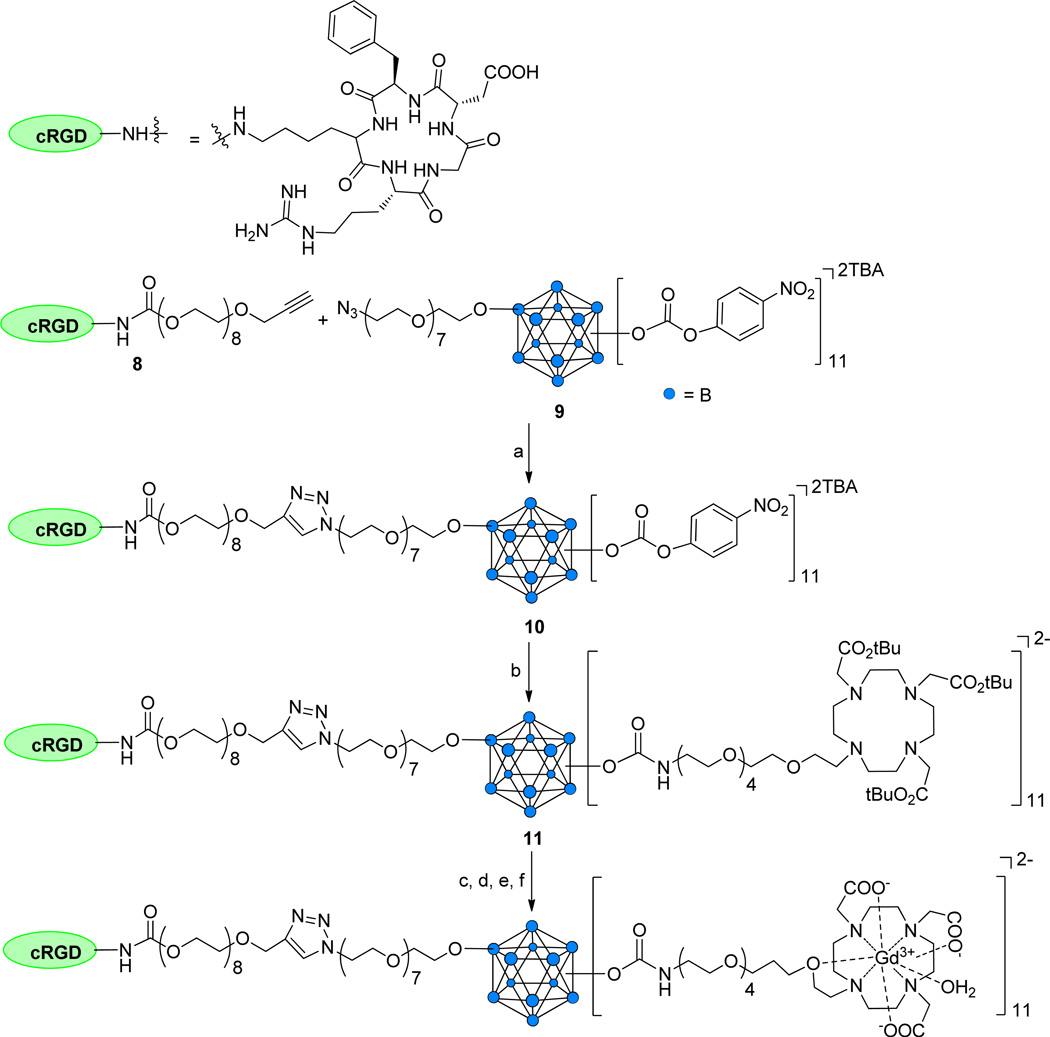
The synthesis of a targeted MRI CA assembled on an icosahedral closo-B122− core is based on our recently communicated work regarding preparation of vertex-differentiated closomers.16 Attachment of the targeting peptide cRGD was accomplished by an azide-alkyne click reaction of alkyne-terminated cRGD peptide derivative 818 with the 11-fold carbonate closomer 9,16 which possesses an azido-termined linker at a single vertex (Scheme 3). The resulting cRGD-conjugated closomer 10 was purified by size-exclusion column chromatography on Lipophilic Sephadex LH-20 using MeOH as the eluent. Purified closomer 10 was characterized by NMR and HRMS spectroscopy. The 1H NMR spectral analysis of 10 showed a singlet at δ 7.94 ppm attributed to the single proton of the 1,4-triazole group, confirming attachment of the cRGD peptide.
Scheme 3.
Synthesis of 11-fold Gd+3-DOTA-cRGD closomer conjugate CA-12.
Reagents and conditions: (a) CuI, DIPEA, THF-ACN, 15 h, RT, 87%; (b) 4, ACN, 7 days, RT, 83%; (c) 80% TFA-DCM, 6 h, RT; (d) GdCl3·6H2O, citrate buffer, pH 7, 24 h, RT, sonication; (e) Dialysis, 1,000 MWCO, 48 h; (f) Lyophilization, 82%.
Addition of DOTA ligands to the remaining 11 vertices of cRGD-conjugated closomer 10 was accomplished by reaction with amino-terminated DOTA derivative 4 (5 equiv per vertex) in ACN for seven days at RT to obtain the 11-fold carbamate closomer 11 in 83% yield (Scheme 3). Closomer 11 was purified by size-exclusion column chromatography on Lipophilic Sephadex LH-20 using MeOH as the eluent and its structure verified by IR, NMR, and HRMS spectroscopy. Removal of the tert-butyl ester groups from 11 was achieved by treatment with 80% TFA in DCM. Complete deprotection of tert-butyl ester groups from 11 was confirmed by the absence of the large singlet at δ 1.50 ppm from its 1H NMR spectrum.
Deprotected closomer 11 was reacted with GdCl3·6H2O (10 equiv per vertex) in citrate buffer at pH ~7 to give closomer CA-12 in 82% yield. CA-12 was purified by exhaustive dialysis in ultrapure water and was characterized by IR spectroscopy and ICP-OES analysis. IR spectral analysis showed a characteristic shift of the carbonyl stretch from 1730 cm−1 to 1595 cm−1, confirming complexation of Gd3+ with the DOTA ligands. The mass spectrometry analysis of CA-12 was unsuccessful; however, the ICP-OES analysis of CA-12 showed an average of 10.6 Gd3+ ions per closomer, indicating the formation of essentially fully loaded complex. The presence of any free Gd3+ ions in solution of targeted closomer CA-12 was determined by a spectrophotometric method using xylenol orange.17 No free Gd3+ ions were detected in the solution. DLS analysis of CA-12 in PBS and 2% Tween-80/PBS solution gave an average particle size of 12–13 nm (Supporting Information).
Relaxivity Studies
The per-Gd relaxivity (r1) of CA-7, CA-12 and Omniscan at 25 °C and 7 Tesla were compared in three different formulations: PBS, 2% Tween/PBS, and bovine calf serum (Figure 2). The relaxivity of each contrast agent was relatively unaffected by formulation type, but the r1 values of CA-7 and CA-12 were slightly higher than Omniscan (4.2 mM−1s−1 in PBS) across all formulations. Nonetheless, the r1 values for CA-7 and CA-12 (e.g., 4.8 mM−1s−1 per Gd and 5.9 mM−1s−1 per Gd in PBS) fall well within the range of other macromolecular MRI CAs carrying multiple Gd3+ chelates that have one inner sphere water-exchange site.11 Macromolecular CAs are known to exhibit higher r1 values due to their larger size and the confinement of the Gd3+ ions in a sterically constrained space. However, the slightly higher r1 values of CA-12 compared to CA-7 may result from slower rotation (τR) of the Gd3+ chelates due to the attachment of the cRGD peptide to the icosahedral closo-B122− core.
Figure 2.
Relaxivity (r1) profile of CA-7, CA-12, and Omniscan in three different formulations at 25 °C and 7 T.
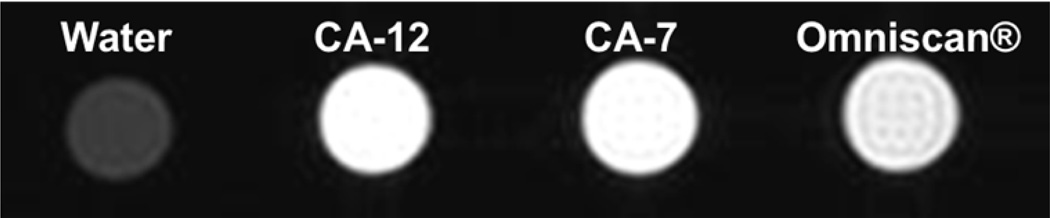
Molecular relaxivity values for CA-7 and CA-12 were calculated based on the number of Gd3+ ions per closomer. CA-7 and CA-12 have an average 11.4 and 10.6 Gd3+ ions per closomer, respectively, giving molecular relaxivity values of 54.7 and 62.5 mM−1s−1 per closomer. The T1-weighted MRI images of CA-12, CA-7 and Omniscan at 7T presented in figure 3 shows greater contrast enhancement for CA-12 and CA-7 compared to Omniscan due to their higher relaxivity values.
Figure 3.
T1-weighted MRI images of CA-12, CA-7, and Omniscan at 1mM Gd concentration in PBS (25 °C and 7 T).
Cell Binding Studies
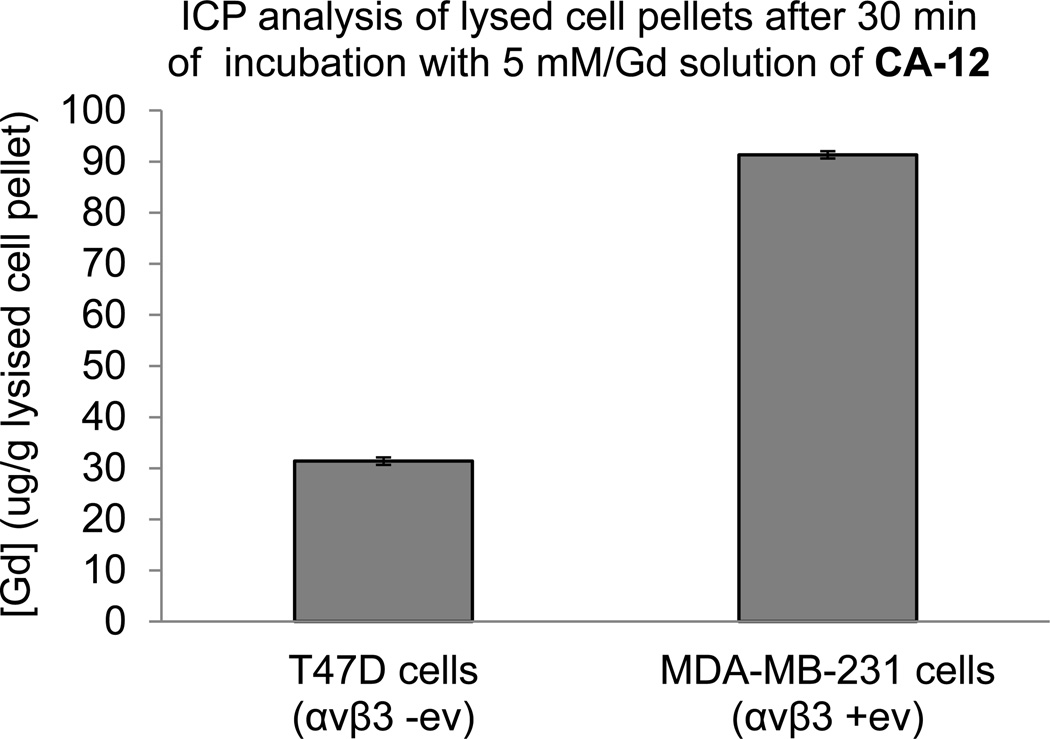
To establish the specificity of cRGD-closomer conjugate CA-12 for αvβ3 integrin receptors, in vitro binding experiments were performed using MDA-MB-231 and T47D cells. MDA-MB-231 cells express high levels of αvβ3 integrins, (αvβ3 positive) whereas T47D cells lack αvβ3 integrin receptors (αvβ3 negative).19 Cells were incubated with CA-12 for 30 min at 37 °C (Gd concentration 5 mM, PBS), and the gadolinium concentration in lysed cell pellets was measured using ICP-OES analysis.The Gd concentration was 300% higher in MDA-MB-231 cells relative to T47D cells (Figure 4), demonstrating the ability of cRGD-containing closomer conjugate CA-12 to target cells expressing αvβ3 integrin receptors.
Figure 4.
Gadolinium concentration in lysed pellets of cells incubated with CA-12 (5 mM Gd, PBS) determined by ICP-OES.
In vivo MRI studies
The ability of CA-12 to specifically target αvβ3 integrin-expressing tumors in vivo was also investigated by MRI of severely compromised immunodeficient (SCID) mice bearing human PC-3 prostate cancer xenografts. Eighteen mice were subjected to MRI scanning following intravenous injection of either cRDG-containing CA-12, non-targeted closomer CA-7, or Omniscan at a gadolinium dose of 0.04 mmol/kg body weight. Six mice were included in each of the three experimental groups.
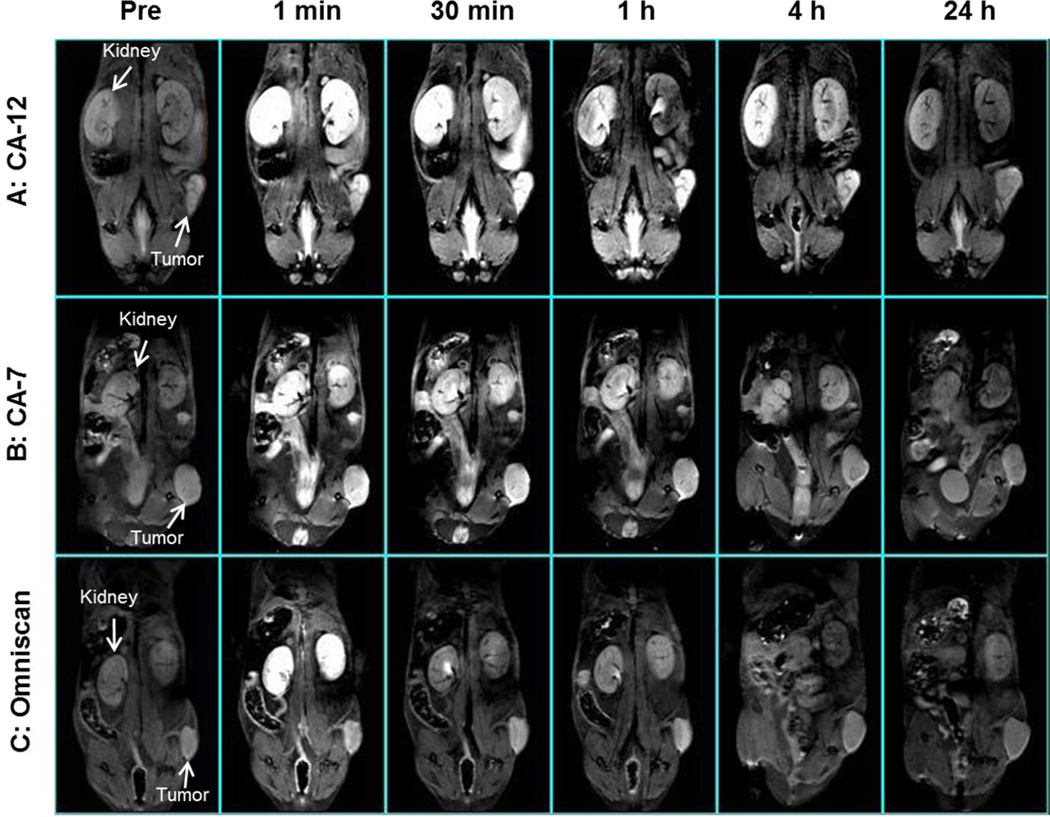
T1-weighted MRI scans showed that tumors of mice injected with targeted closomer CA-12 exhibited strong contrast enhancement for up to 4 h post-injection (Figure 5, panel A). Significant contrast enhancement by CA-7 persisted for only 1 hour post-injection (Figure 5, panel B). Contrast enhancement by Omniscan diminished within 30 min post-injection, and no enhancement was detectable by 4 h post-injection (Figure 5, panel C). MRI imaging also showed that contrast enhancement of the kidneys was higher and lasted longer in mice injected with CA-12 (Figure 5, panel A) compared to those injected with CA-7 or Omniscan (Figure 5, panels B and C), indicative of the relatively slower renal clearance of CA-12.
Figure 5.
Representative in vivo T1-weighted MRI scans of mice (n = 6 mice for each experimental group) pre- injection and at 1 min, 30 min, 1 h, 4 h and 24 h post-injection of (A) CA-12 (B) CA-7 and (C) Omniscan (gadolinium dose of 0.04 mmol/kg).
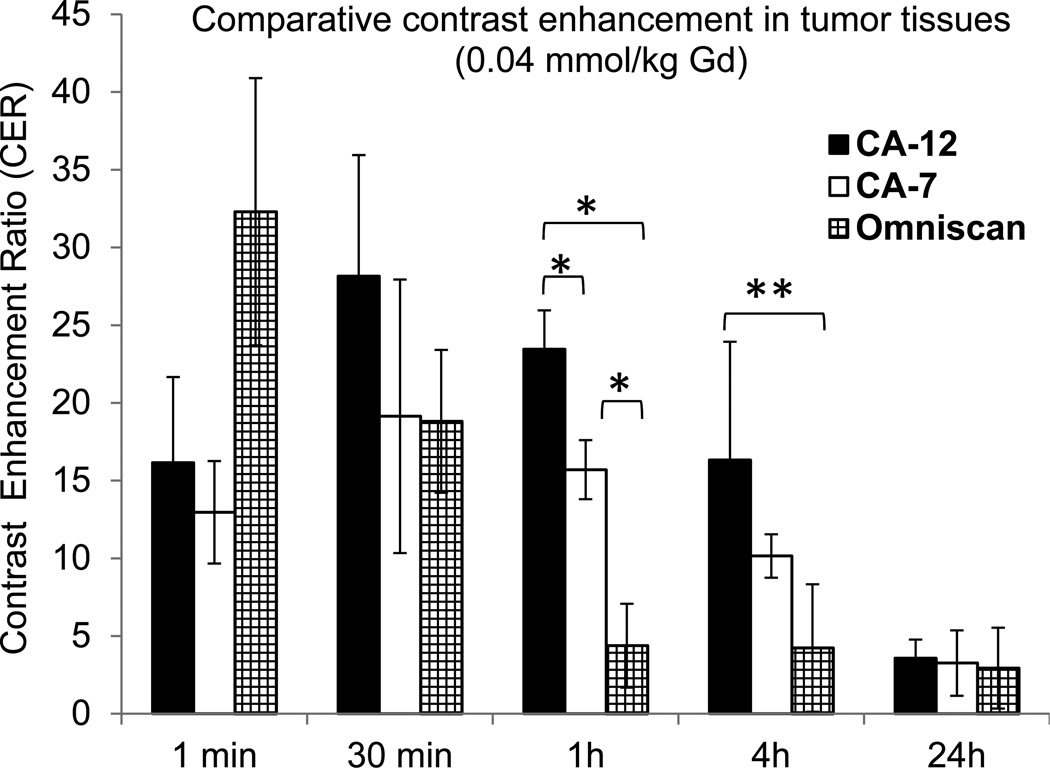
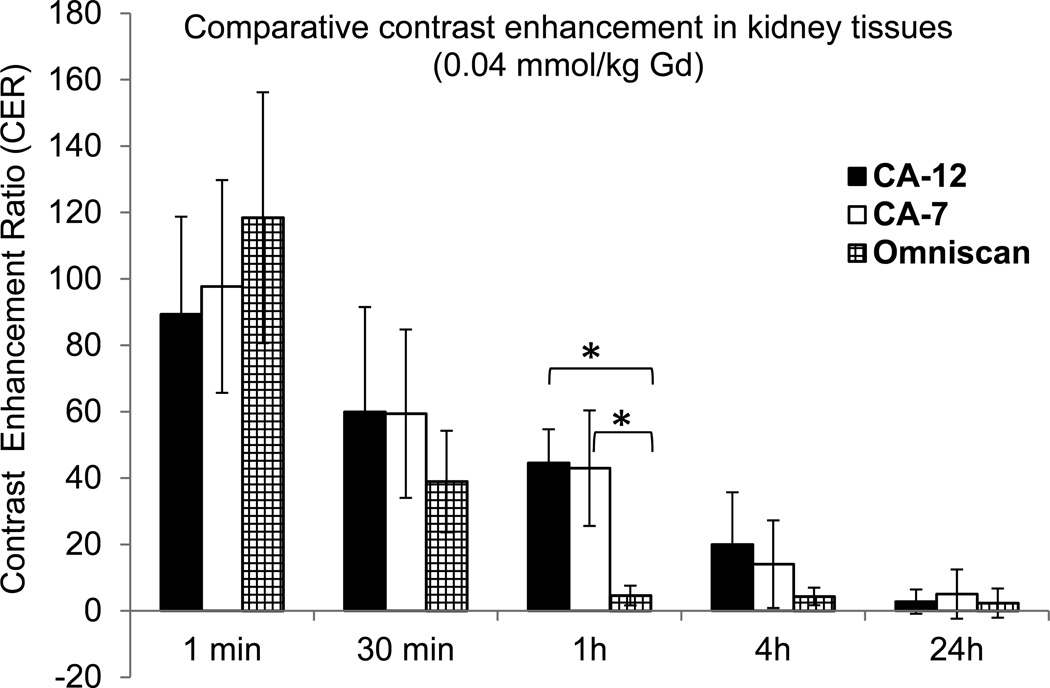
Calculation of the contrast enhancement ratios (CER) for tumor and kidney tissues permitted a quantitative assessment of the relative enhancement capacity of CA-12, the non-targeted closomer CA-7, and Omniscan. In tumor tissue, the CER did not decline significantly until 24 hours post-injection of CA-12, whereas it declined substantially by 4 h and 1 h post-injection of CA-7 and Omniscan, respectively (Figure 6). At 1 h and 4 h post injection of CA-12, tumor tissues had significantly higher CER compared to CA-7 (P < 0.01 at 1 h) and Omniscan (P < 0.01 at 1 h; P < 0.05 at 4 h). Similarly, the CER values for kidneys of mice injected with CA-12 and CA-7 initially increased then declined, though at a slower rate than for kidneys of mice injected with Omniscan (Figure 7). For example, the kidney signals are higher at 1 h post-injection of CA-12 compared to Omniscan (P < 0.01), and diminished to the baseline contrast at 24 h. The persistence of contrast in tumors and the slower renal clearance of CA-12 may be attributed to the specific binding to integrin receptors by the cRGD ligand.
Figure 6.
Contrast Enhancement Ratios (CER) for tumor tissues of mice as a function of time measured from the in vivo imaging experiments after intravenous injection of CA-12, CA-7, or Omniscan with a gadolinium dose of 0.04 mmol/kg. *P < 0.01, **P < 0.05.
Figure 7.
Contrast Enhancement Ratios or CER) for kidney tissues of mice as a function of time measured from the in vivo imaging experiments after intravenous injection of CA-12, CA-7, or Omniscan with a gadolinium dose of 0.04 mmol/kg. *P < 0.01.
After 24 h, all mice were euthanized, and tail, blood, heart, lungs, liver, spleen, stomach, large and small intestines, kidneys, brain, and muscle tissues were analyzed for the presence of gadolinium ions using ICP-OES. No gadolinium was detectable in any of the tissues, indicating complete elimination of the CAs.
CONCLUSIONS
We have shown for the first time the utility of a vertex-differentiated icosahedral closo-B122− core in the synthesis of a novel αvβ3 integrin-targeted polyfunctional MRI CA. The closomer CA-12 carries a single cRGD peptide attached to the closo-B122− core via a long PEG linker and holds 11 Gd3+-DOTA chelates attached through a HEG linker via carbamate linkages. This unique MRI CA exhibits a higher relaxivity per Gd than the small molecular CA Omniscan in PBS at 7T.
In vitro cell binding experiments and in vivo MRI analysis of tumor-bearing mice demonstrated the ability of CA-12 to selectively target integrin αvβ3-expressing cells. Binding studies showed that the Gd content was 3-fold higher in cells expressing αvβ3 receptors compared to cells in which receptor expression was absent. Serial T1-weighted MRI of mice possessing human PC-3 prostate cancer xenographs showed that higher and more persistent enhancement of tumors was achieved with CA-12 than with Omniscan or with CA-7, a closomer carrying 12 Gd3+-DOTA chelates but lacking the cRGD peptide. Mice exhibited no apparent toxic effects from any of the CA for 24 hours post-injection, and post-mortem ICP-OES analysis of numerous tissues found no detectable residual Gd.
The methodology presented here is versatile and should be highly useful in the development of a wide variety of high-payload, receptor-specific diagnostic and therapeutic agents. We are currently pursuing work in this direction.
EXPERIMENTAL SECTION
General information
Common reagents and chromatographic solvents were obtained from commercial suppliers (Sigma-Aldrich, Fisher Scientific, and Acros Organics) and used without any further purification. Lipophilic Sephadex LH-20 was obtained from GE Healthcare. DO3A-t- Bu-ester and cRGD peptide were purchased from Macrocyclics, Inc. and Peptides International, respectively. NMR spectra were recorded on Bruker Avance 400 and 500 MHz spectrometers. The high-resolution mass spectrometry analysis was performed using Applied Biosystems Mariner ESI-TOF. IR spectra were recorded on Thermo Nicolet FT-IR spectrometer. The dynamic light scattering analysis was performed on a Microtrac Zetatrac particle size analyzer. Gadolinium (Gd3+) concentrations of the samples were measured by inductively coupled plasma optical emission spectroscopy (ICP-OES) on a PerkinElmer OptimaTM 7000 DV instrument. All the Gd3+ and boron concentrations obtained via ICP analysis are the average of three measurements. The error bars provided in the figures are standard deviations.
Synthesis of 1
To a mixture of Et3N (2.70 g, 26.6 mmol) and hexaethylene glycol (5.00 g, 17.7 mmol) in DCM (50 mL) stirring at 0 °C, a solution of MsCl (2.03 g, 17.7 mmol) in DCM (20 mL) was added over 1 h period by syringe pump. After addition of MsCl, the reaction was allowed to proceed at 0 °C for 1 h, and then at RT for 2 h. The reaction mixture was concentrated to dryness. The residue was dissolved in DCM (100 mL) and washed with 3% HCl-water (100 mL) and brine (100 mL). The organic layer was separated, dried over Na2SO4, then concentrated and dried under high vacuum. The dried sample was dissolved in DMF (30 mL) to which potassium phthalimide (4.26 g, 23.0 mmol) was added, and the mixture was stirred at 110 °C for 15 h. Following concentration under high vacuum to remove the DMF, the resulting solid was dissolved in Et2O and filtered through a celite pad. The filtrate was concentrated, and the crude material was purified on a silica gel column to obtain pure product as a colorless oil. Yield: 3.2 g (44%). 1H NMR (400 MHz, CDCl3): δ 7.70 (dd, 2H, J = 2.8 & 5.2 Hz, −CH-Pth), 7.59 (dd, 2H, J = 2.8 & 5.2 Hz, −CH-Pth), 4.28 (brs, 1H, −OH), 3.77 (t, 2H, J = 5.6 Hz, −OCH2HEG), 3.60 (m, 4H, −OCH2HEG), 3.52-3.45 (m, 18H, −OCH2HEG). 13C NMR (100.6 MHz, CDCl3): δ 167.50, 161.98, 133.43, 133.03, 131.45, 122.53, 122.21, 72.03, 69.98-69.44 (multiple peaks), 67.21, 60.79, 36.68, 35.83, 30.73. HRMS (ESI): m/z calcd for C20H29NO8 [M+Na]+ 434.1785; found 434.1575; for C20H29NO8 [M+K]+ 450.1525; found 450.1038.
Synthesis of 2
To a mixture of Et3N (1.10 g, 10.9 mmol) and 1 (3.00 g, 7.29 mmol) in DCM (30 mL) stirring at 0 °C, a solution of MsCl (1.08 g, 9.48 mmol) in DCM (10 mL) was added over 1 h by syringe pump. After addition, the reaction was allowed to proceed at 0 °C for 1 h and then at room temperature for 2 h. The reaction mixture was concentrated to dryness, and the residue was then dissolved in DCM (100 mL). After successive washing with 3% HCl-water (100 mL) and brine (100 mL), the organic layer was separated, dried over Na2SO4, then concentrated and dried under high vacuum. The dried sample was then dissolved in acetone, sodium iodide (4.37 g, 29.1 mmol) was added to it, and the resulting mixture was stirred at 65 °C for 15 h. The reaction mixture was then concentrated to dryness, DCM (100 mL) was added, and the solution filtered through a celite pad. The filtrate was concentrated and purified by silica gel chromatography to obtain pure product as a colorless oil. Yield: 3.6 g (95%). 1H NMR (400 MHz, CDCl3): δ 7.76 (dd, 2H, J = 3.2 & 5.6 Hz, −CH-Pth), 7.65 (dd, 2H, J = 2.8 & 5.2 Hz, −CHPth), 3.81 (t, 2H, J = 6.0 Hz, −OCH2HEG), 3.67 (m, 4H, −OCH2HEG), 3.59-3.50 (m, 16H, −OCH2HEG), 3.18 (t, 2H, J = 7.2 Hz, −OCH2HEG). 13C NMR (100.6 MHz, CDCl3): δ 167.96, 133.77, 131.93, 123.02, 71.75, 70.45-69.92 (multiple peaks), 67.70, 37.10, 2.98. HRMS (ESI): m/z calcd for C20H28INO7 [M+Na]+ 544.0803; found 544.0283.
Synthesis of 3
To a mixture of DO3A-t-Bu-ester (2.00 g, 3.88 mmol) and 2 (3.04 g, 5.83 mmol) in DMF (25 mL), KHCO3 (0.78 g, 7.77 mmol) was added, and the mixture was stirred at 60 °C for 15 h. The reaction mixture was cooled and DCM (100 mL) added. The mixture was washed with brine (100 mL), and the organic layer was separated, dried over Na2SO4, and concentrated. After column chromatography over alumina (IV), the pure product was obtained as a viscous oil. Yield: 3.4 g (96%). 1H NMR (400 MHz, CDCl3): δ 7.63-7.60 (m, 4H, −CH-Pth), 3.68 (t, 2H, J = 5.6 Hz, −OCH2HEG), 3.55 (t, 2H, J = 5.6 Hz, −OCH2HEG), 3.45-3.36 (m, 18H, −OCH2HEG), 3.10-2.00 (m, 24H, −OCH2HEG & −CH2DOTA), 1.28 (m, 27H, −tBut-DOTA). 13C NMR (100.6 MHz, CDCl3): δ 171.95, 167.61, 133.69, 131.41, 122.60, 81.63, 69.69-69.12 (multiple peaks), 67.24, 66.57, 55.81, 55.06, 53.37, 51.60, 49.49, 49.17, 36.65, 27.48. HRMS (ESI): m/z calcd for C46H77N5O13 [M+Na]+ 930.5410; found 930.5362.
Synthesis of 4
A mixture of 3 (4.4 g, 4.84 mmol) and hydrazine (1.55 g, 48.4 mmol) in EtOH (60 mL) was stirred at 65 °C for 3 h. Upon reaction completion, the reaction mixture was concentrated to dryness. The residue was dissolved in DCM (100 mL) and filtered using a sintered glass funnel. The filtrate was concentrated and dried under high vacuum. Following column chromatography of the crude product over alumina (IV), pure product was obtained as a viscous oil. Yield: 3.6 g (96%). 1H NMR (400 MHz, CDCl3): δ 3.24-3.16 (m, 20H), 2.88 (m, 2H), 2.78-2.59 (m, 4H), 2.56-1.83 (m, 20H), 1.07 (m, 27H, −tBut-DOTA). 13C NMR (100.6 MHz, CDCl3): δ 171.68, 171.40, 81.01, 80.98, 71.35, 69.35-69.04 (multiple peaks), 68.74, 68.55, 66.10, 55.28, 54.53, 53.11, 51.10, 49.47, 48.67, 40.35, 27.09, 26.83. HRMS (ESI): m/z calcd for C38H75N5O11 [M+Na]+ 800.5355; found 800.5723.
Synthesis of closomer 6
A mixture of 12-fold 4-nitrophenyl carbonate closomer 5 (0.20 g, 0.07 mmol) and DOTA ligand 4 (3.33 g, 4.28 mmol) in ACN (30 mL) was stirred for 7 days at RT under an argon atmosphere. The reaction mixture was then concentrated and purified via size-exclusion chromatography over Lipophilic Sephadex LH-20 using MeOH as the eluent. The product was obtained as a sticky, light-brown solid. M.P.: 65–68 °C. Yield: 0.60 g (80%). 1H NMR (400 MHz, CD3CN): δ 6.57 (brs, 10H), 3.79-2.13 (m, 554H), 1.47 (m, 324H). 13C NMR (100.6 MHz, CD3CN): δ 173.8, 173.66, 171.50, 171.25, 82.74, 81.75, 71.02-70.35 (multiple peaks), 68.1, 57.19, 56.97, 56.31, 55.92, 55.28, 53.62, 53.17, 51.15, 50.66, 41.80, 28.34, 28.26, 28.14. 11B NMR (160.4 MHz, CD3CN): δ −18.19. HRMS (m/z): Average mass calcd. for C468H888B12N60O156 [M+6H]4++2K+Na+TBA 2838.7986. Found: 2838.5745, calcd. for C468H888B12N60O156 [M+6H]4++2K+2Na+TBA 2861.7986. Found 2861.6614.
Synthesis of 12-fold DOTA-Gd closomer CA-7
Closomer 6 (0.33 g, 0.03 mmol) was treated with 80% TFA/DCM (10 mL) for 6 h at RT under an argon atmosphere. The reaction mixture was then concentrated under reduced pressure. The resulting residue was dissolved in a minimum amount of methanol, and the product was precipitated by adding ether. The solid product was filtered, washed with ether, and dried under high vacuum overnight. The now deprotected closomer 6 was dissolved in 1 M citrate buffer (15 mL, pH 7) and added to a solution of GdCl3.6H2O (1.40 g, 3.78 mmol, in 15 mL of 1 M citrate buffer, pH 7) over 5 h at RT with vigorous stirring. The reaction mixture was maintained at approximately pH 7 using 0.3 N NaOH. When addition of 6 was complete, the reaction mixture was stirred for an additional 24 h at RT. To avoid possible aggregation and intermolecular chelation of Gd3+, the reaction mixture was sonicated a few times over the entire course of the reaction. To remove excess GdCl3, the reaction mixture was dialyzed in deionized water for 48 h using 1000 MWCO dialysis tubing (Spectra/Por). Following lyophilization, the product CA-7 was obtained as an off-white solid. M.P.: 255–260 °C (decomposed). Yield: 0.22 g (71%). ICP-OES analysis: calcd. average Gd per closomer 12. Found: 11.4. The mass spectrometry analysis attempts were unsuccessful.
Synthesis of alkyne-terminated cRGD conjugate 8
Alkyne-terminated cRGD conjugate 8 was synthesized according to our recently published procedure.19
Synthesis of 11-fold 4-nitrophenyl carbonate closomer 9
Closomer 9 was synthesized according to our recently described method.16
Synthesis of closomer 10
Eleven-fold 4-nitrophenyl carbonate closomer 9 (0.03 g, 9.90 × 10−3 mmol), alkyne-terminated cRGD conjugate 8 (10.3 mg, 9.90 × 10−3 mmol), and copper (I) iodide (2.80 mg, 14.7 × 10−3 mmol) were dissolved in a 1:1 mixture of THF and ACN (6 mL). After addition of DIPEA (20 mg, excess), the reaction mixture was vigorously stirred at RT for 15 h under an argon atmosphere. Upon reaction completion, the reaction mixture was concentrated to remove THF and ACN. The residue was dissolved in ethyl acetate and filtered through a celite pad. The filtrate was concentrated, and the crude product was purified via size-exclusion column chromatography on Lipophilic Sephadex LH-20 using ACN as the eluent. The purified product was obtained as a sticky, light-yellow solid. Yield: 35.0 mg (87%). 1H NMR (500 MHz, CD3CN): δ 8.72 (m, 22H), 7.94 (s, 1H), 7.39 (m, 24H), 7.13 (m, 2H), 6.90 (m, 1H), 4.65-4.58 (m, 4H), 4.21 (s, 2H), 3.99-3.91 (m, 6H), 3.62 (m, 76H), 3.16 (m, 13H), 2.30 (m, 6H), 2.06 (m, 8H), 1.94 (m, 2H), 1.68 (m, 10H), 1.44-1.31 (m, 15H), 1.16 (m, 2H), 1.05 (m, 14H) (Fewer proton counts for TBA+ is due to exchange with H3O+ and Na+). 13C NMR (125.7 MHz, CD3CN): δ 156.63, 150.96, 150.69, 145.12, 127.63, 125.09, 122.31, 70.13-68.93 (multiple peaks), 63.78, 58.33, 46.93, 23.30, 19.33, 12.80, 8.23. 11B NMR (160.4 MHz, CD3CN): δ −14.34, −18.12. HRMS (m/z): Average mass calcd. for C140H150B12N22O81 [M]2− 1783.4705. Found: 1783.4463.
Synthesis of closomer 11
A mixture of closomer 10 (0.03 g, 7.40 × 10−3 mmol) and DOTA ligand 4 (0.32 g, 0.40 mmol) in ACN (10 mL) was stirred for 7 days at RT under an argon atmosphere. The reaction mixture was concentrated and purified via size-exclusion chromatography over Lipophilic Sephadex LH-20 using MeOH as the eluent. The pure product was obtained as a sticky, light-brown solid. M.P.: 72–75 °C. Yield: 65.0 mg (83%). 1H NMR (400 MHz, CD3OD): δ 8.09 (s, 1H), 7.30-7.16 (m, 5H), 7.05 (m, 2H), 6.80 (m, 2H), 6.70 (m, 1H), 4.66 (m, 7H), 4.52-2.35 (m, 590H), 1.54-1.51 (m, 300H), 1.16 (m, 6H), 0.95 (m, 6H). 13C NMR (100.6 MHz, CD3CN): δ 173.78, 173.66, 171.25, 156.05, 130.28, 128.82, 124.9, 82.74, 82.72, 81.85, 71.02-69.87 (multiple peaks), 68.20, 57.19 (multiple peaks), 56.31, 55.71, 55.15, 53.55-53.23 (multiple peaks), 50.66 (multiple peaks), 48.43, 41.80, 28.26 (multiple peaks). 11B NMR (160.4 MHz, CD3CN): δ −14.54, −18.32. HRMS (m/z): Average mass calcd. for C492H920B12N66O169 [M+6H]4+ + Na 2671.647030. Found: 2670.9688.
Synthesis of cRGD conjugated 11-fold DOTA-Gd closomer CA-12
Closomer 11 (0.09 g, 8.40 × 10−3 mmol) was treated with 80% TFA/DCM (10 mL) for 6 h at RT under an argon atmosphere. The reaction mixture was concentrated under reduced pressure, and the residue was dissolved in a minimum amount of methanol. Ether was added to precipitate the solid product. The product was filtered, washed with additional ether, and dried under high vacuum overnight. The now-deprotected closomer 11 was dissolved in 1 M citrate buffer (15 mL, pH 7) and added to a solution of GdCl3.6H2O (0.34 g, 0.92 mmol, in 15 mL of 1 M citrate buffer, pH 7) over 5 h at RT with vigorous stirring. The reaction mixture was maintained at approximately pH 7 using 0.3 N NaOH. When addition of 11 was complete, the reaction mixture was stirred at RT for an additional 24 h. To avoid possible aggregation and intermolecular chelation, the reaction mixture was sonicated a few times over the entire course of the reaction. The reaction mixture was then dialyzed in deionized water for 48 h using 1000 MWCO dialysis tubing (Spectra/Por). Following lyophilization, the product CA-12 was obtained as an off-white solid. M.P.: 207–210 °C (decomposed). Yield: 63.0 mg (69%). ICP-OES analysis: calcd. average Gd per closomer 11. Found: 10.6. Attempts at mass spectrometry analysis were unsuccessful.
Relaxivity Determinations
Solutions of CA-7 and CA-12, and Omniscan were prepared in PBS, 2% Tween-80/PBS and bovine calf serum. T1 relaxation rates were determined for each of the three formulations of CA-12 and CA-7 at Gd3+ ion concentrations of 0.25, 0.5 and 1 mM. The Gd3+ ion concentration of Omniscan was 1 mM. The measurements were repeated on two or more independently prepared samples to ensure consistency. A buffer matched blank sample (0 mM) was also used in the relaxivity measurements of each sample.
Measurements were performed using a 7 Tesla Varian Unity Inova MRI system (Varian Inc./Agilent Technologies) at RT. A T1-weighted MRI pulse sequence was applied with TE = 15 ms, and TR = 500 ms, slice thickness = 1 mm, matrix = 256 × 256, and FOV = 30 × 30 mm. A series of inversion recovery (IR) spin-echo images were acquired using TR = 3 s, TE = 15 ms, and the following inversion delays: 0.082, 0.1, 0.12, 0.16, 0.24, 0.32, 0.64, 1.28, 2.56, 3.6 s. Water signal intensities were measured using VnmrJ2.1D software (Varian Inc. /Agilent Technologies, 2005). Relaxation rates were calculated using a three-parameter experiential recovery fitting in Origin 8.5.0 (OriginLab Corporation, 2012).
Cell Binding Assays
MDA-MB-231 and T47D cells were obtained from the American Type Culture Collection (Manassas, VA). Cell culture reagents were purchased from Fisher Scientific (Chicago, IL) or Sigma-Aldrich (St. Louis, MO). Cells were cultured in complete growth medium [DMEM/F12 containing 10% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT, USA), 1% antibiotics] in a humidified incubator at 37 °C and 5% CO2. When cells had reached approximately 70% confluency (3 days), they were treated with trypsin-EDTA solution and washed twice with cold PBS. The cells were counted using a hemacytometer and diluted with PBS to a concentration of 1 × 107 cell/mL. Cells were incubated with CA-12 (Gd concentration 5 mM) in a 37 °C water bath for 30 min. After incubation, cells were centrifuged at 800×g at 4 °C for 4 min, and the supernatant removed. The pellet was washed twice with PBS, and cells were resuspended in 200 µL PBS. ICP-OES was used to measure the gadolinium concentration of the cells.
In vivo MRI Analysis
In vivo MRI studies were performed in severely compromised immunodeficient (SCID) mice bearing human PC-3 prostate cancer xenografts. Animal studies were conducted in accordance with the highest standards of care as outlined in the NIH’s guide for care and use of laboratory animals and in accordance with policy and procedures for animal research at the Harry S. Truman Memorial Veterans Hospital. Four-to-five-week-old SCID mice were obtained from Taconic (Germantown, NY). Mice were housed four animals per cage in sterile micro-isolator cages in a temperature- and humidity-controlled room with a 12-hour light/dark schedule. The animals were fed autoclaved rodent chow (Ralston Purina Company, St. Louis, MO) and provided water ad libitum. Mice were inoculated with human prostate cancer PC-3 tumor cells on the right flank. Eighteen mice, six mice in each experimental group, were used in MRI experiments between 3–4 weeks post inoculation of tumor cells. Average body weight of mice was 25–30 grams at the time of MRI.
Imaging was performed using a 7T Varian Unity Inova MRI equipped with a Millipede quadrature RF coil (40 mm ID). A T1-weighted multi-slice spin echo sequence was performed to record images pre- and post-injection of CAs. The imaging parameters were TE = 15 ms, TR = 500 ms, slice thickness = 1 mm, 17 slices, average = 4, matrix = 128 × 256, and FOV = 35 × 70 mm (coronal). Mice were anesthetized using 2.5% isoflurane in oxygen via a nose cone. Once pre-injection imaging had been completed, CA-12, CA-7, or Omniscan dissolved in 2% Tween-80/PBS was injected through a tail vein (gadolinium dose 0.04 mmol/kg body weight). Mice were imaged immediately after introduction of the CAand again at 30 min, 1, 4, and 24 h post-injection. A water tube placed under the animal was treated as a reference signal. Vitals were continually monitored, and mice were released to their cage after each imaging session in order to fully recover.
Image analysis and processing were performed with VnmrJ2.1D software (Varian Inc. /Agilent Technologies, 2005). Regions of interest (ROIs) were manually drawn on the tumor tissue, muscle near the tumor, kidney cortex, and liver for each time point. Signal intensity (SI) was measured as the mean of the intensity over the segmented ROIs. SIs were normalized to the SI of muscle with the assumption that signal enhancement of muscle tissue is zero at 4 h and 24 h post-injection of CA. The contrast enhancement ratio (CER) for a tissue was calculated according to the equation
CER = (SIpost – SIpre) / SIpre × 100%.
To assess persistence of the CAs in tissues, mice were euthanized 24 hours post-injection. Tumor, blood, heart, lungs, liver, spleen, stomach, large intestine, small intestine, kidney, brain, and muscle samples were collected, weighed and immediately frozen at −20 °C. Prior to the ICP-OES analysis, tissue samples were allowed to warm up to room temperature, acid digested and the gadolinium and boron content of the tissues was determined.
Statistical analysis
Quantitative data were expressed as the mean ± standard deviation (SD). Means were compared by analysis of a student’s t-test using t-test function in Microsoft Excel (2007). P values of less than 0.05 were considered to be statistically significant.
Supplementary Material
ACKNOWLEDGMENT
This research was funded by National Cancer Institute (Grant R21-CA114090). Authors acknowledge the support provided by the Veterans’ Affairs Biomolecular Imaging Center at the Harry S. Truman VA Hospital and the University of Missouri-Columbia. We thank Brett Meers, Sarah Higdon, Lisa Watkinson and Terry Carmack for technical assistance and Pamela Cooper for manuscript editing.
Footnotes
ASSOCIATED CONTENTS
Supporting Information.
Supporting information contains copies of IR spectra, dynamic light scattering, and SE-HPLC analysis data for CA-7 and CA-12. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Mansfield P. Angew. Chem. Int. Ed. 2004;43:5456–5464. doi: 10.1002/anie.200460078. [DOI] [PubMed] [Google Scholar]; (b) Lauffer RB. Chem. Rev. 1987;87:901–927. [Google Scholar]; (c) Hermann P, Kotek J, Kubíček V, Lukeš I. Dalton Trans. 2008:3027–3047. doi: 10.1039/b719704g. [DOI] [PubMed] [Google Scholar]; (d) Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Chem. Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]; (e) Werner EJ, Datta A, Jocher CJ, Raymond KN. Angew. Chem. Int. Ed. 2008;47:8568–8580. doi: 10.1002/anie.200800212. [DOI] [PubMed] [Google Scholar]; (f) Caravan P. Acc. Chem. Res. 2009;42:851–862. doi: 10.1021/ar800220p. [DOI] [PubMed] [Google Scholar]
- 2.Major JL, Meade TJ. Acc. Chem. Res. 2009;42:893–899. doi: 10.1021/ar800245h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Caravan P. Chem. Soc. Rev. 2006;35:512–523. doi: 10.1039/b510982p. [DOI] [PubMed] [Google Scholar]
- 3.(a) Park J-A, Lee J-J, Jung J-C, Yu D-Y, Oh C, Ha S, Kim T-J, Chang Y. ChemBioChem. 2008;9:2811–2813. doi: 10.1002/cbic.200800529. [DOI] [PubMed] [Google Scholar]; (b) Verwilst P, Eliseeva SV, Carron S, Vander Elst L, Burtea C, Dehaen G, Laurent S, Binnemans K, Muller RN, Parac-Vogt TN, De Borggraeve WM. Eur. J. Inorg. Chem. 2011:3577–3585. doi: 10.1021/ic200726t. [DOI] [PubMed] [Google Scholar]; (c) Verwilst P, Eliseeva SV, Elst LV, Burtea C, Laurent S, Petoud S, Muller RN, Parac-Vogt TN, De Borggraeve WM. Inorg. Chem. 2012;51:6405–6411. doi: 10.1021/ic300717m. [DOI] [PubMed] [Google Scholar]; (d) Dijkgraaf I, Rijnders AY, Soede A, Dechesne AC, van-Esse GW, Brouwer AJ, Corstens FHM, Boerman OC, Rijkers DTS, Liskamp RMJ. Org. Biomol. Chem. 2007;5:935–944. doi: 10.1039/b615940k. [DOI] [PubMed] [Google Scholar]
- 4.(a) Weinmann HJ, Ebert W, Misselwitz B, Schmitt-Willich H. Eur. J. Radiol. 2003;46:33–44. doi: 10.1016/s0720-048x(02)00332-7. [DOI] [PubMed] [Google Scholar]; (b) Nair SA, Kolodziej AF, Bhole G, Greenfield MT, McMurry TJ, Caravan P. Angew. Chem. Int. Ed. 2008;47:4918–4921. doi: 10.1002/anie.200800563. [DOI] [PubMed] [Google Scholar]; (c) Marom H, Miller K, Bechor-Bar Y, Tsarfaty G, Satchi-Fainaro R, Gozin M. J. Med. Chem. 2010;53:6316–6325. doi: 10.1021/jm100289b. [DOI] [PubMed] [Google Scholar]
- 5.(a) Chen Y. Theranostics. 2011;1:28–29. doi: 10.7150/thno/v01p0028. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tan M, Lu Z-R. Theranostics. 2011;1:83–101. doi: 10.7150/thno/v01p0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sipkins DA, Cheresh DA, Kazemi MR, Nevin LM, Bednarski MD, Li KC. Nat. Med. 1998;4:623–626. doi: 10.1038/nm0598-623. [DOI] [PubMed] [Google Scholar]
- 7.(a) Mousa SA. Curr. Opin. Chem. Biol. 2002;6:534–541. doi: 10.1016/s1367-5931(02)00350-2. [DOI] [PubMed] [Google Scholar]; (b) Jin H, Varner J. Brit. J. Cancer. 2004;90:561–565. doi: 10.1038/sj.bjc.6601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K, Chen Y. Theranostics. 2011;1:189–200. doi: 10.7150/thno/v01p0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benedetto S, Pulito R, Crich SG, Tarone G, Aime S, Silengo L, Hamm J. Magn. Reson. Med. 2006;56:711–716. doi: 10.1002/mrm.21023. [DOI] [PubMed] [Google Scholar]
- 10.(a) Boswell CA, Eck PK, Regino CAS, Bernardo M, Wong KJ, Milenic DE, Choyke PL, Brechbiel MW. Mol. Pharmaceutics. 2008;5:527–539. doi: 10.1021/mp800022a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) De León-Rodríguez LM, Lubag A, Udugamasooriya DG, Proneth B, Brekken RA, Sun X, Kodadek T, Sherry AD. J. Am. Chem. Soc. 2010;132:12829–12831. doi: 10.1021/ja105563a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zarabi B, Borgman MP, Zhuo J, Gullapalli R, Ghandehari H. Pharm Res. 2009;26:1121–1129. doi: 10.1007/s11095-009-9830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Li W, Su B, Meng S, Ju L, Yan L, Ding Y, Song Y, Zhou W, Li H, Tang L, Zhao Y, Zhou C. Eur. J. Radiol. 2011;80:598–606. doi: 10.1016/j.ejrad.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 11.(a) Bryson JM, Chu W, Lee J, Reineke TM. Bioconjugate Chem. 2008;19:1505–1509. doi: 10.1021/bc800200q. [DOI] [PubMed] [Google Scholar]; (b) Song Y, Kohlmeir EK, Meade TJ. J. Am. Chem. Soc. 2008;130:6662–6663. doi: 10.1021/ja0777990. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Livramento JB, Helm L, Sour A, O’Neil C, Merbach AE, Tóth É. Dalton Trans. 2008:1195–1202. doi: 10.1039/b717390c. [DOI] [PubMed] [Google Scholar]; (d) Langereis S, de Lussanet QG, van Genderen MHP, Backes WH, Meijer EW. Macromolecules. 2004;37:3084–3091. [Google Scholar]; (e) Torchilin V, Babich J, Weissig V. J. Liposome Res. 2000;10:483–499. [Google Scholar]; (f) Kielar F, Tei L, Terreno E, Botta M. J. Am. Chem. Soc. 2010;132:7836–7837. doi: 10.1021/ja101518v. [DOI] [PubMed] [Google Scholar]; (g) Floyd WC, Klemm PJ, Smiles DE, Kohlgruber AC, Pierre VC, Mynar JL, Fréchet JMJ, Raymond KN. J. Am. Chem. Soc. 2011;133:2390–2393. doi: 10.1021/ja110582e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Hawthorne MF, Maderna A. Chem. Rev. 1999;99:3421–3434. doi: 10.1021/cr000001+. [DOI] [PubMed] [Google Scholar]; (b) Hawthorne MF, Farha OK, Julius R, Ma L, Jalisatgi SS, Li T, Bayer MJ. ACS Symp. Ser. 2006;917:312–324. [Google Scholar]; (c) Issa F, Kassiou M, Rendina LM. Chem. Rev. 2011;111:5701–5722. doi: 10.1021/cr2000866. [DOI] [PubMed] [Google Scholar]
- 13.Bayer MJ, Hawthorne MF. Inorg. Chem. 2004;43:2018–2020. doi: 10.1021/ic030289w. [DOI] [PubMed] [Google Scholar]
- 14.(a) Jalisatgi SS, Kulkarni VS, Tang B, Houston ZH, Lee MW, Hawthorne MF. J. Am. Chem. Soc. 2011;133:12382–12385. doi: 10.1021/ja204488p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Goswami LN, Chakravarty S, Lee MW, Jalisatgi SS, Hawthorne MF. Angew. Chem. Int. Ed. 2011;50:4689–4691. doi: 10.1002/anie.201101066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goswami LN, Ma L, Chakravarty S, Cai Q, Jalisatgi SS, Hawthorne MF. Inorg. Chem. 2012 doi: 10.1021/ic3017613. (Article ASAP) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goswami LN, Houston ZH, Sarma SJ, Li H, Jalisatgi SS, Hawthorne MF. J. Org. Chem. 2012;77:11333–11338. doi: 10.1021/jo3021314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barge A, Cravotto G, Gianolio E, Fedeli F. Contrast Med. Mol. Imaging. 2006;1:184–188. doi: 10.1002/cmmi.110. [DOI] [PubMed] [Google Scholar]
- 18.Goswami LN, Houston ZH, Sarma SJ, Jalisatgi SS, Hawthorne MF. Org. Biomol. Chem. 2013;11:1116–1126. doi: 10.1039/c2ob26968f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Takayama S, Ishii S, Ikeda T, Masamura S, Doi M, Kitajima M. Anticancer Res. 2005;25:79–84. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.