Abstract
Indoor microbial communities play an important role in everyday human health, especially in the intensive care units (ICUs) of hospitals. We used amplicon pyrosequencing to study the ICU microbiome and were able to detect diverse sequences, in comparison to the currently used standard cultivation technique that only detected 2.5% of the total bacterial diversity. The phylogenetic spectrum combined species associated with the outside environment, taxa closely related to potential human pathogens, and beneficials as well as included 7 phyla and 76 genera. In addition, Propionibacterium spp., Pseudomonas spp., and Burkholderia spp. were identified as important sources of infections. Despite significantly different bacterial area profiles for floors, medical devices, and workplaces, similarities by network analyses and strains with identical molecular fingerprints were detected. This information will allow for new assessment of public health risks in ICUs, help create new sanitation protocols, and further our understanding of the development of hospital-acquired infections.
The majority of our life time is spent in indoor environments, but little is known about the bacterial communities with which we share indoors. Recently, the application of next generation sequencing techniques has allowed new insight into indoor microbial communities. In general, they are characterized by a high prokaryotic diversity and are comprised of diverse bacterial and archaeal phyla1,2,3,4. Indoor microbiomes originate mainly from the human skin or from outside air, and have even been known to include extremophiles. Furthermore, all of them contain potential human pathogens, but also beneficial bacteria that are characterized by a positive interaction with their host1,3. Kembel et al.3 were the first to analyze patient rooms and find a strong effect from both architecture and ventilation. In contrast to the majority of indoor environments, rooms in hospitals and especially intensive care units (ICUs) are routinely monitored5. Standard cultivation techniques, such as contact plates, are commonly used to monitor the microbial burden. However, culture collections contain a restricted spectrum and only a very small proportion of the total bacteria as already described in 1985 by Staley and Konopka6. A comparison of the bacterial diversity obtained by standard monitoring and next generation sequencing techniques in ICUs has unfortunately not yet been published. We hypothesize that the ICU microbiome is characterized by a much higher bacterial diversity and abundance than is currently thought.
Indoor microbial communities are an important component of everyday human health3,7, and are even partially composed of human-associated bacteria1 due to the high emission rate of up to 106 bacteria per person-hour8. In ICUs, sanitation protocols are stricter than in other areas of the hospital, yet many patients treated in ICUs are infected with hospital-acquired “nosocomial infections” often due to an underlying severe disease9,10. Moreover, these nosocomial infections remain among the leading causes of death in hospitals of developed countries. For example, they are a significant cause of morbidity and mortality in the United States; 1.7 million infections resulting in 99,000 deaths were reported in 200211. In Europe, the risk for nosocomial infections for patients in ICUs is reported as 45%9. Hospital surfaces are often overlooked reservoirs for this bacteria12,13,14, thus new sanitation standards are needed to drastically reduce this risk for hospital-acquired infections15. New sequencing techniques will allow for a greater understanding of whole ICU bacterial communities, including their beneficials, and contribute to a new perspective on hospital sterility.
The objective of this work was to analyse the structure of bacterial communities from the ICU of the Department of Internal Medicine at the University Hospital in Graz/Austria using a comparative approach between currently used standard cultivation of the ICU and 16S rRNA gene amplicon sequencing. Altogether, 34 surface samples obtained from three general areas (floors, medical devices, workplaces) inside the ICU were studied. 16S rRNA gene amplicons and isolates were identified and compared by a principal coordinate analysis. In addition, network analysis using Cytoscape 2.8 software16 was performed to identify the most abundant taxa and to compare their abundance across the three sampling areas.
Results
Structure and diversity of bacterial community profiles
From all surface samples of the three areas within the ICU - the floor environment (A: 5 samples), devices (B: 11 samples) and workplaces (C: 8 samples) - visualized in Fig. 1 - a high number of amplicons were obtained and sequenced. In total, the raw dataset of all 24 samples contained 356,571 sequences. After trimming, the final operational taxonomic unit (OTU) table consisted of 308,440 sequences. The docking station (MID53) contained the highest (16,137) and the floor of the patient room after cleaning (MID35) the fewest (5,321) amount of sequences respectively. Due to the different number of sequences among samples, the data was normalized to 5,321 sequences.
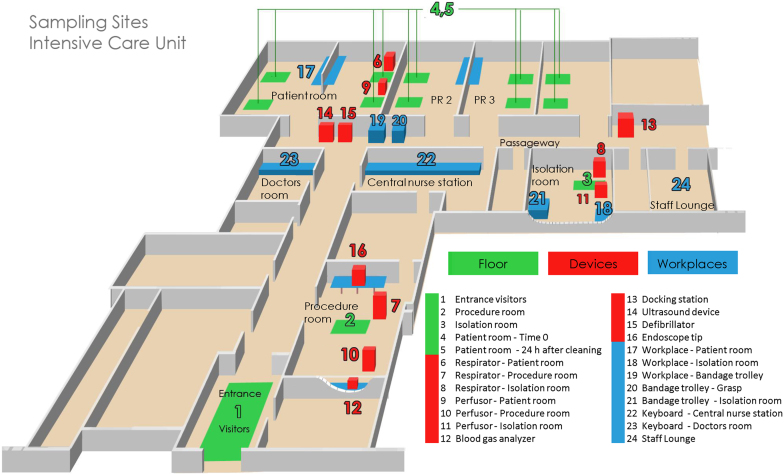
Figure 1. Cartoon illustration showing each sampling site of the intensive care unit (ICU).
Origin of the single samples is explained in Table 1.
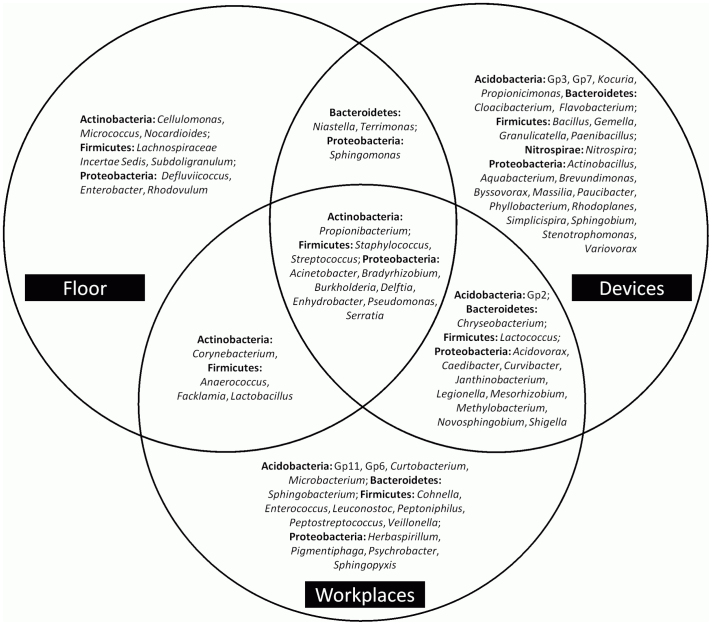
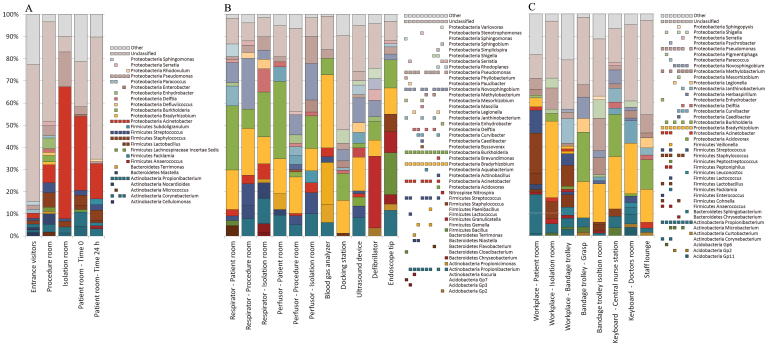
The composition of microbial communities included 7 different bacterial phyla: Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Nitrospira and Proteobacteria. Proteobacteria (64%) was the most abundant phylum across all samples. At genus level, differences between the communities of the various areas were detected (Fig. 2). The amplicon library of the ICU included sequences of 405 genera, 76 of them were reaching 1% of relative abundance. Only a minor part of sequences belonging to members of the genera Acinetobacter, Bradyrhizobium, Burkholderia, Delftia, Enhydrobacter, Propionibacterium, Pseudomonas, Serratia, Staphylococcus and Streptococcus were retrieved from surfaces of all three areas. Figure 2 also indicates that devices contained a greater variety of bacteria (23 genera) than the workplaces (15 genera) and the floor (8 genera). An overlap between the detected genera of the three areas was also observed; the highest was found between devices/workplaces (12 genera) and followed by floor/workplaces (4) and floor/devices (3). A comparison of the relative abundances confirmed differences between the three main areas (Fig. 3). Most notably, Pseudomonas and Propionibacterium were clearly most abundant on all sampling sites. On the floor (A), the most frequently present genus was Acinetobacter (24%) among the other commonly found genera Propionibacterium, Pseudomonas, Staphylococcus, and Streptococcus. The dominant genera on devices (B) and workplaces (C) were Pseudomonas (4% in B, 7% in C), Novosphingobium (10% in B, 5% in C), Burkholderia (14% in B, 15% in C), Bradyrhizobium (16% in B, 17% in C) and Propionibacterium (7% in B, 5% in C). The most abundant genus on floors, Acinetobacter, was less present in these two areas (3% in B, 2% in C). Chryseobacterium, Janthinobacterium, Legionella, Methylobacterium and Shigella were minimal on devices and workplaces. Corynebacterium was only present in some floor samples and workplaces, whereas Serratia was measured in low numbers in several samples of all areas. Gemella, Flavobacterium and Stenotrophomonas were only detected on several devices, while Bacillus, Granulicatella and Nitrospira were all observed in relatively high abundances. Taxonomic classification of each sampling site is shown in Fig. S1.
Figure 2. Schematic drawing showing the detected bacterial genera found in the three different areas (floor, devices and workplaces) in the intensive care unit (ICU).
Overlaps between the facilities are indicated by the arrangement of the circles.
Figure 3. The bacterial communities of the floor environment (A), medical devices (B) and workplaces (C).
Relative clone composition of genera was determined by pyrosequencing of 16S rRNA from metagenomic DNA extracted from the intensive care unit. The identification of the closest strain based on 16S rRNA sequence similarity was achieved using the web server SnoWMAn 1.11 (http://snowman.genome.tugraz.at). Phylogenetic groups accounting for 1% of all quality sequences are summarized in the artificial group Other. Multi-colored charts at the legend are shown for each genus and sample correspondingly.
To determine richness and diversity, OTUs were identified at genetic distances of 3% (species level), 5% (genus level) and 20% (phylum level) by using quality sequences with a read length of ≥ 150 bp per sample. At 20% sequence divergence, most rarefaction curves showed saturation, indicating that the surveying effort covered almost the full extent of taxonomic diversity at this genetic distance (Fig. S2). Comparison of the rarefaction analyses with the number of OTUs determined by the Chao1 richness estimator revealed that 83% to 100% (20% genetic distance) of the estimated taxonomic richness was detected. At 3% and 5% genetic distance, the rarefaction curves were not saturated and the richness estimators indicated that 45% to 78% and 47% to 84% of the estimated richness were recovered. As a result, we did not survey the full extent of taxonomic diversity at these genetic distances, but a substantial fraction of the bacterial diversity within individual samples was assessed at species and genus level. The Shannon index of diversity (H′) was determined for all samples (Table 2). The highest bacterial diversity at a genetic distance of 3% was found on workplaces (3.46), followed by the floor (3.14) and devices (3.0). The Shannon index of each sampling site ranged from 2.29 to 4.64; the surfaces of the bandage trolley (MID38) and workplaces of the patient room (MID37) showed the highest diversity.
Table 1. Locations and characteristics of sampling points for cultivation-independent and -dependent methods of the intensive care unit (ICU).
| (A) Samples for cultivation-independent analysis | ||||
|---|---|---|---|---|
| Sampling location | Description | Sampling tool1 | Sample ID | MID |
| Entrance visitors | Floor | BiSKit | F1 | MID32 |
| Procedure room | Floor | BiSKit | F2 | MID33 |
| Isolation room | Floor | BiSKit | F3 | MID34 |
| Patient room - Time 0 | Floor | BiSKit | F4 | MID35 |
| Patient room - 24 h after cleaning | Floor | BiSKit | F5 | MID36 |
| Respirator - Patient room | Device | Nylon flocked swab | D6 | MID45 |
| Respirator - Procedure room | Device | Nylon flocked swab | D7 | MID46 |
| Respirator - Isolation room | Device | Nylon flocked swab | D8 | MID47 |
| Perfusor - Patient room | Device | Nylon flocked swab | D9 | MID48 |
| Perfusor - Procedure room | Device | Nylon flocked swab | D10 | MID49 |
| Perfusor - Isolation room | Device | Nylon flocked swab | D11 | MID50 |
| Blood gas analyzer | Device | Nylon flocked swab | D12 | MID51 |
| Docking station | Device | Nylon flocked swab | D13 | MID53 |
| Ultrasound device | Device | Nylon flocked swab | D14 | MID41 |
| Defibrillator | Device | Nylon flocked swab | D15 | MID40 |
| Endoscope tip | Device | Nylon flocked swab | D16 | MID52 |
| Workplace - Patient room | Working Surface | BiSKit | W17 | MID37 |
| Workplace - Isolation room | Working Surface | BiSKit | W18 | MID54 |
| Workplace - Bandage trolley | Working Surface | BiSKit | W19 | MID38 |
| Bandage trolley - Grasp | Working Surface | Nylon flocked swab | W20 | MID42 |
| Bandage trolley in isolation room | Working Surface | Nylon flocked swab | W21 | MID43 |
| Keyboard - Central nurse station | Working Surface | Nylon flocked swab | W22 | MID55 |
| Keyboard - Doctors room | Working Surface | Nylon flocked swab | W23 | MID56 |
| Staff Lounge | Working Surface | Nylon flocked swab | W24 | MID39 |
| (B) Samples for cultivation-dependent analysis | ||||
| Sampling location | Description | Sampling tool2 | Sample ID | CFU |
| Workplace - Patient room | Working Surface | Contact test | WS 42 | 2 |
| Workplace - Bandage trolley | Working Surface | Contact test | WB 43 | 21 |
| Respirator - Patient room | Device | Contact test | RS 44 | 15 |
| Perfusor - Patient room | Device | Contact test | PS 45 | 210 |
| Keyboard - Central nurse station | Device | Contact test | KO 46 | 512 |
| Keyboard - Doctors room | Device | Contact test | KD 47 | 48 |
| Blood gas analyzer | Device | Contact test | BG 48 | 72 |
| Docking station | Device | Contact test | DS 49 | 87 |
| Ultrasound device - Control panel | Device | Contact test | UU 50 | 6 |
| Ultrasound device - Probe | Device | Contact test | UU 51 | 25 |
Abbreviations: BiSKit, Biological Sampling Kit; MID, Multiplex Identifier Sequence; CFU, Colony Forming Units per sample.
1,2 Respective sample areas were 1 m2 for BiSKits, 25 cm2 for Nylon flocked swabs and for 10 cm2 contact tests.
Table 2. Species richness estimates obtained at 3%, 5% and 20% genetic dissimilarity from pyrosequencing of 16S rRNA from metagenomic DNA extracted from the floor, medical devices and workplaces.
| Shannon index (H′)a | Rarefactionb (No. of OTUs) | Chao1c (No. of OTUs) | Coverage (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dissimilarity cut-offd | 3% | 5% | 20% | 3% | 5% | 20% | 3% | 5% | 20% | 3% | 5% | 20% |
| Entrance visitors | 4.09 | 3.64 | 2.24 | 778.46 | 553.66 | 97.97 | 1713.009 | 1035.833 | 121.100 | 45.5 | 53.5 | 80.9 |
| Procedure room | 3.03 | 2.73 | 2.08 | 198.9 | 129.94 | 17 | 361.303 | 223.261 | 17.000 | 55.1 | 58.2 | 100.0 |
| Isolation room | 2.29 | 2.04 | 1.24 | 347.8 | 250.87 | 34 | 778.055 | 525.405 | 35.500 | 44.7 | 47.8 | 95.8 |
| Sick chamber - Time 0 | 3.54 | 3.02 | 2.23 | 571.68 | 412.8 | 56.99 | 1234.414 | 740.217 | 63.429 | 46.3 | 55.8 | 89.9 |
| Sick chamber - 24 h after cleaning | 2.74 | 2.40 | 1.60 | 200.9 | 131.94 | 24 | 354.270 | 222.048 | 26.000 | 56.7 | 59.4 | 92.3 |
| Respirator - Patient room | 2.86 | 2.51 | 1.80 | 163.94 | 98.97 | 13 | 210.923 | 117.913 | 13.000 | 77.8 | 84.0 | 100.0 |
| Respirator - Procedure room | 2.81 | 2.60 | 1.95 | 181.92 | 95.98 | 12 | 294.857 | 116.313 | 12.000 | 61.7 | 82.5 | 100.0 |
| Respirator - Isolation room | 3.07 | 2.70 | 2.05 | 167.93 | 94.97 | 14 | 233.022 | 122.750 | 17.000 | 72.1 | 77.4 | 82.4 |
| Perfusor - Patient room | 2.97 | 2.45 | 1.58 | 153.94 | 90.96 | 11 | 247.261 | 203.875 | 11.000 | 62.3 | 44.6 | 100.0 |
| Perfusor - Procedure room | 3.44 | 3.13 | 2.14 | 224.89 | 143.94 | 15 | 399.000 | 230.667 | 15.000 | 56.4 | 62.4 | 100.0 |
| Perfusor - Isolation room | 2.89 | 2.59 | 1.90 | 147.93 | 80.98 | 10 | 247.400 | 99.056 | 10.000 | 59.8 | 81.8 | 100.0 |
| Blood gas analyzer | 2.90 | 2.58 | 1.82 | 147.94 | 84.97 | 10 | 226.792 | 109.800 | 10.000 | 65.3 | 77.4 | 100.0 |
| Docking station | 3.10 | 2.81 | 2.06 | 242.9 | 160.95 | 28 | 397.895 | 220.111 | 29.500 | 61.1 | 73.1 | 94.9 |
| Ultrasound device | 3.11 | 2.90 | 1.90 | 181.92 | 119.96 | 16 | 297.161 | 154.440 | 16.000 | 61.2 | 77.7 | 100.0 |
| Defibrillator | 2.78 | 2.38 | 1.88 | 154.93 | 102.97 | 14 | 224.000 | 121.207 | 14.000 | 69.2 | 85.0 | 100.0 |
| Endoscope tip | 3.00 | 2.66 | 1.85 | 173.92 | 100.95 | 16 | 356.045 | 181.571 | 17.500 | 48.9 | 55.6 | 91.4 |
| Workplace - Patient room | 4.11 | 3.73 | 2.44 | 425.83 | 301.91 | 42 | 653.586 | 386.875 | 42.250 | 65.2 | 78.1 | 99.4 |
| Workplace - Isolation room | 3.05 | 2.77 | 1.75 | 169.93 | 101.97 | 14 | 266.871 | 146.400 | 14.000 | 63.7 | 69.7 | 100.0 |
| Workplace - Bandage trolley | 4.64 | 4.24 | 2.82 | 572.74 | 404.85 | 52.99 | 1073.039 | 579.690 | 63.500 | 53.4 | 69.9 | 83.5 |
| Bandage trolley - Grasp | 2.94 | 2.55 | 1.61 | 136.95 | 67.98 | 9 | 192.682 | 83.833 | 9.000 | 71.1 | 81.1 | 100.0 |
| Bandage vehicle isoltion room | 3.57 | 3.29 | 2.32 | 295.86 | 193.93 | 27 | 539.886 | 299.556 | 27.000 | 54.8 | 64.8 | 100.0 |
| Keyboard - Central nurse station | 3.34 | 3.04 | 1.76 | 181.94 | 111.97 | 15 | 235.318 | 137.143 | 15.000 | 77.3 | 81.7 | 100.0 |
| Keyboard - Doctors room | 3.45 | 3.14 | 2.03 | 197.92 | 118.96 | 16 | 306.243 | 153.167 | 16.000 | 64.7 | 77.7 | 100.0 |
| Staff Lounge | 2.60 | 2.26 | 1.50 | 141.95 | 74.98 | 7 | 224.650 | 117.857 | 7.000 | 63.2 | 63.6 | 100.0 |
aa higher number indicates more diversity.
bthe results from the rarefaction analyses are also depicted in Figure 4.
cnonparametric richness estimator based on the distribution of singletons and doubletons.
dClusters were obtained with 3%, 5% and 20% dissimilarity cut-offs which correspond to the taxonomic level of species, genera and phyla.
Differences between the bacterial community profiles
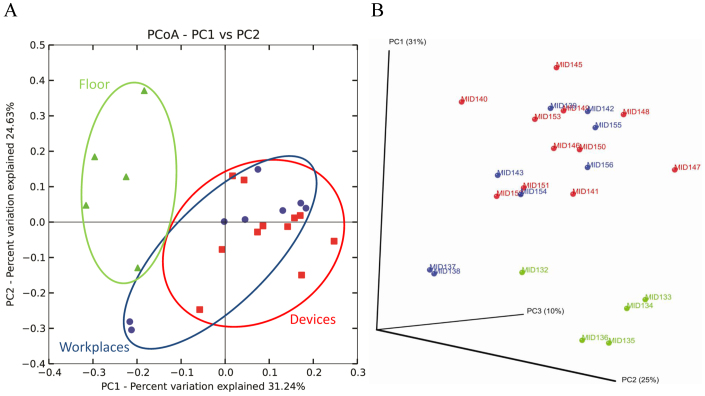
Using the software package QIIME17, the final OTU table for principal coordinate analysis (PCoA) comprised 3,925 OTUs and was distributed into 556 OTUs represented by more than 10 sequences. Beta diversity of the bacterial communities within the ICU revealed clear distinctions between bacterial populations among the three areas. Floor-associated bacterial communities formed clusters distinct from devices (Fig. 4), while the analyzed samples from workplaces and devices were similar. Samples from workplaces of the patient room (MID37) and bandage trolley (MID38) were significantly differentiated from those of the other workplaces and were closer to samples from the floor. Furthermore, the structure of the bacterial community found on the docking station (MID53) was completely distinct from other communities.
Figure 4. Bacterial communities associated with different areas of the intensive care unit by principal coordinate analysis (PCoA).
Two- (A) and three-dimensional (B) PCoA plot based on the weighted UniFrac distance matrix. Percentage of the diversity distribution explained by each axes is indicated on the figure. Samples associated with the floor (green triangle), medical devices (red rectangle) and workplaces (blue points) are shown as single points.
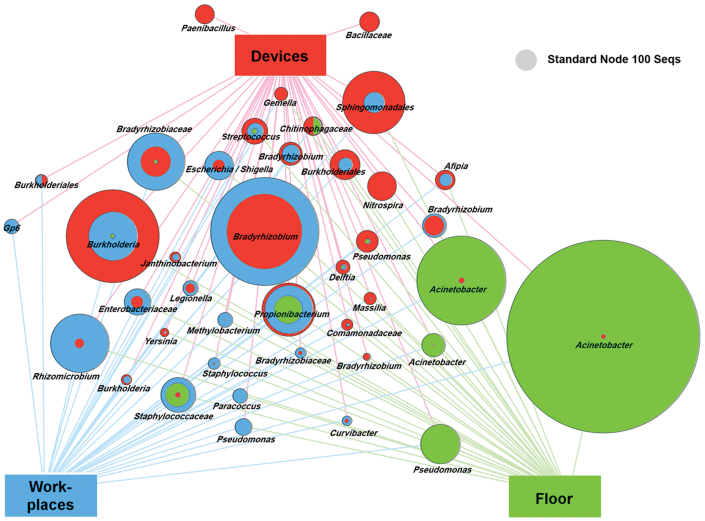
To gain better insight into the differences of the three areas, we applied a profile clustering network analysis (Fig. 5). This profile obtained by a Cytoscape network analysis showed the most abundant 40 OTUs and highlighted the relative distribution and abundances. Acinetobacter was the most abundant and ubiquitous bacterial genus with dominant occurrence on the floor. In addition, Bradyrhizobium and Burkholderia were among the dominant genera of all areas.
Figure 5. Profile clustering Cytoscape network visualize the 40 most abundant OTUs across the floor (green), medical devices (red) and workplaces (blue) of the ICU.
Node sizes correspond to the mean relative abundance between the three sampling areas where two (three) sampling groups feature the same relative abundance over the respective samples and the full nodes are subdivided into two (three) areas. A comparative node (grey) indicates the size of a node that would represent 100 reads in a sample.
According to the statistical analysis, 330 out of 3,925 examined OTUs showed significant differences between the floor environment (A) and devices (B), and 336 between devices (B) and workplaces (C). A comparison of the floor environment (A) and the workplaces (C) resulted in a statistically significant difference between species for 155 OTUs. Statistical analysis of data is shown in Tab. S1. Most notably, in numerous samples of the floor environment (up to 59%) and in some samples from devices and workplaces, the relative abundance of Unclassified was particularly high. Comparison of the relative abundances indicated a correlation of Cyanobacteria at phylum level with the Unclassified at genus level. Additional BLAST analysis of the Unclassified detected that most sequences classified as Cyanobacteria-like are chloroplast sequences that originate from Pinaceae and from other plant components. Sequences were affiliated to species of conifer in the Pinaceae family containing Abies sp., Larix sp., Picea sp. or Pinus sp. with maximal identity of 99% and originated from Pinus pollen from outside air. Abundance of chloroplast sequences is shown in Tab. S2.
Comparison between standard cultivation and 16S pyrosequencing
Comparative samples were taken from 10 sampling sites of defined positions on devices and workplaces (Table 1). The colonies showed a low diversity in their morphology and colour (white approx. 80%; the others yellow and red). The highest number of colonies (512) was found on the keyboard in the central nurse station, but only two colonies were detected on the workplace of the patient room. The number of colony forming units (CFUs) of all sampling sites is listed in Table 1.
A total of 130 isolates obtained from contact plates were characterized by ARDRA (amplified ribosomal RNA gene restriction analysis using HhaI) and divided into 36 ARDRA groups at a cutoff level of 85%. Representative strains of each group were partially sequenced and identified by their 16S rRNA; the genera Aerococcus, Arthrobacter, Bacillus, Corynebacterium, Kocuria, Micrococcus, Paenibacillus, Planomicrobium, Roseomonas and Staphylococcus were identified. Twelve ARDRA groups include the majority of isolates and were identified as species belonging to the genus Staphylococcus. In contrast, 21 groups were represented by only one isolate. To analyze the genotypic diversity within the ARDRA groups at population level, BOX-PCR patterns of the whole bacterial genome were used. At 80% similarity, 24 unique populations were differentiated (Table 3). With the exception of the Roseomonas mucosa strain (ARDRA group 34), only Gram-positive species were found; 49 of them were identified as Staphylococcus epidermidis and Staphylococcus hominis. Other Staphylococcus sequences were classified as S. auricularis, S. caprae, S. cohnii, S. haemolyticus, S. lugdunensis and S. warneri. Less retrieved sequences belonged to Micrococcus antarcticus, M. luteus, Arthrobacter agilis, Bacillus circulans, B. idriensis and Kocuria palustris. Furthermore, a few sequences were identified as Aerococcus urinaeequi, Bacillus aerophilus, B. frigoritolerans, B. herbersteinensis, B. simplex, Corynebacterium propinquum, Kocuria rosea, Paenibacillus barcinonensis, Planomicrobium koreense and Roseomonas mucosa. Interestingly, the keyboard of the central nurse station showed the highest number of CFUs (512) containing six different species. In comparison, the respirator from the patient room contained only 15 colonies, but nine different species were identified. Isolates with identical and similar BOX patterns were detected on nearly all sampling sites (Fig. 6) indicating transmission between them or deposition of bacteria from identical or similar sources.
Table 3. Cultivation dependent ARDRA (amplified ribosomal RNA gene restriction analysis; cutoff level of 85%) and BOX analysis (cutoff level of 85%) of 130 isolates.
| ARDRA group | BOX group | Sampling site | Sample-ID | Closest NCBI database match | Accession No. | Max. identity |
|---|---|---|---|---|---|---|
| 1 | 1 | KD | 47/8 | Micrococcus luteus | NR_037113.1 | 99% |
| 2 | 1 | DS | 49/11 | Corynebacterium propinquum | NR_037038.1 | 99% |
| 3 | 1 | WB | 43/24 | Roseomonas mucosa | NR_028857.1 | 99% |
| 4 | 1 | DS | 49/16 | Staphylococcus hominis | NR_041323.1 | 99% |
| 4 | 2 | DS | 49/9 | Staphylococcus hominis | NR_041323.1 | 99% |
| 4 | 3 | DS | 49/1 | Staphylococcus hominis | NR_041323.1 | 99% |
| 4 | 4 | KD | 47/15 | Staphylococcus caprae | NR_024665.1 | 100% |
| 4 | 4 | DS | 49/13 | Staphylococcus caprae | ||
| 4 | 5 | UU | 51/12 | Staphylococcus epidermidis | NR_036904.1 | 99% |
| 4 | 5 | UU | 51/9 | Staphylococcus epidermidis | ||
| 4 | 5 | KD | 47/12 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 51/2 | Staphylococcus epidermidis | ||
| 4 | 5 | KD | 47/10 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 51/10 | Staphylococcus epidermidis | ||
| 4 | 5 | KD | 47/16 | Staphylococcus epidermidis | ||
| 4 | 5 | KD | 47/5 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 51/17 | Staphylococcus epidermidis | ||
| 4 | 5 | DS | 49/14 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 51/18 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 50/1 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 51/13 | Staphylococcus epidermidis | ||
| 4 | 5 | DS | 49/7 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 51/4 | Staphylococcus epidermidis | ||
| 4 | 5 | UU | 51/14 | Staphylococcus epidermidis | NR_036904.1 | 99% |
| 4 | 5 | KD | 47/14 | Staphylococcus epidermidis | ||
| 4 | 6 | KD | 47/17 | Staphylococcus hominis | NR_041323.1 | 99% |
| 4 | 7 | KD | 47/13 | Staphylococcus caprae | NR_024665.1 | 99% |
| 4 | 8 | DS | 49/10 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 4 | 9 | DS | 49/6 | Staphylococcus warneri | NR_025922.1 | 100% |
| 4 | 10 | DS | 49/4 | Staphylococcus hominis | ||
| 4 | 10 | UU | 50/3 | Staphylococcus hominis | ||
| 4 | 10 | UU | 50/2 | Staphylococcus hominis | ||
| 4 | 10 | UU | 50/5 | Staphylococcus hominis | NR_041323.1 | 98% |
| 4 | 10 | DS | 49/15 | Staphylococcus hominis | ||
| 4 | 11 | DS | 49/3 | Staphylococcus caprae | NR_024665.1 | 100% |
| 4 | 12 | DS | 49/2 | Staphylococcus cohnii | NR_036902.1 | 100% |
| 4 | 13 | DS | 49/8 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 4 | 14 | UU | 51/6 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 4 | 15 | UU | 50/4 | Staphylococcus hominis | NR_041323.1 | 98% |
| 5 | 1 | KD | 47/11 | Staphylococcus epidermidis | NR_036904.1 | 99% |
| 6 | 1 | BG | 48/18 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 7 | 1 | KO | 46/17 | Staphylococcus warneri | NR_025922.1 | 100% |
| 7 | 2 | KO | 46/21 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 7 | 3 | KO | 46/18 | Staphylococcus hominis | ||
| 7 | 3 | KO | 46/19 | Staphylococcus hominis | NR_036956.1 | 99% |
| 8 | 1 | KO | 46/20 | Staphylococcus epidermidis | ||
| 8 | 1 | KO | 46/22 | Staphylococcus epidermidis | NR_036904.1 | 99% |
| 9 | 1 | KD | 47/1 | Staphylococcus epidermidis | NR_036904.1 | 98% |
| 9 | 1 | BG | 48/5 | Staphylococcus epidermidis | ||
| 9 | 1 | BG | 48/1 | Staphylococcus epidermidis | ||
| 9 | 2 | BG | 48/10 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 9 | 3 | BG | 48/7 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 9 | 4 | BG | 48/6 | Staphylococcus haemolyticus | NR_036955.1 | 99% |
| 9 | 5 | BG | 48/11 | Staphylococcus hominis | NR_041323.1 | 99% |
| 9 | 5 | BG | 48/12 | Staphylococcus hominis | ||
| 9 | 5 | BG | 48/9 | Staphylococcus hominis | ||
| 9 | 5 | BG | 48/13 | Staphylococcus hominis | ||
| 9 | 6 | BG | 48/3 | Staphylococcus lugdunensis | NR_024668.1 | 100% |
| 10 | 1 | BG | 48/14 | Staphylococcus hominis | NR_041323.1 | 99% |
| 10 | 2 | BG | 48/15 | Staphylococcus hominis | NR_041323.1 | 99% |
| 11 | 1 | BG | 48/2 | Planomicrobium koreense | ||
| 11 | 1 | BG | 48/8 | Planomicrobium koreense | NR_025011.1 | 100% |
| 12 | 1 | UU | 51/16 | Bacillus idriensis | NR_043268.1 | 99% |
| 12 | 1 | UU | 51/3 | Bacillus idriensis | NR_043268.1 | 99% |
| 13 | 1 | KO | 46/23 | Paenibacillus barcinonensis | NR_042272.1 | 99% |
| 14 | 1 | UU | 51/15 | Staphylococcus auricularis | NR_036897.1 | 99% |
| 14 | 2 | UU | 51/7 | Staphylococcus auricularis | NR_036897.1 | 99% |
| 15 | 1 | KD | 47/3 | Arthrobacter agilis | NR_026198.1 | 98% |
| 16 | 1 | UU | 51/8 | Bacillus herbersteinensis | NR_042286.1 | 96% |
| 17 | 1 | WS | 42/2 | Staphylococcus hominis | NR_036956.1 | 99% |
| 17 | 2 | RS | 44/11 | Staphylococcus haemolyticus | NR_036955.1 | 99% |
| 17 | 3 | KO | 46/7 | Staphylococcus warneri | NR_025922.1 | 100% |
| 17 | 4 | WB | 43/18 | Staphylococcus hominis | ||
| 17 | 4 | PS | 45/5 | Staphylococcus hominis | ||
| 17 | 4 | PS | 45/8 | Staphylococcus hominis | ||
| 17 | 4 | WB | 43/13 | Staphylococcus hominis | NR_041323.1 | 99% |
| 17 | 5 | WB | 43/17 | Staphylococcus epidermidis | ||
| 17 | 5 | KO | 46/16 | Staphylococcus epidermidis | ||
| 17 | 5 | WB | 43/2 | Staphylococcus epidermidis | ||
| 17 | 5 | KO | 46/3 | Staphylococcus epidermidis | ||
| 17 | 5 | KO | 46/15 | Staphylococcus epidermidis | ||
| 17 | 5 | RS | 44/3 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 17 | 6 | KO | 46/6 | Staphylococcus epidermidis | NR_036904.1 | 99% |
| 17 | 7 | PS | 45/6 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 17 | 8 | PS | 45/7 | Staphylococcus hominis | NR_041323.1 | 99% |
| 17 | 9 | PS | 45/3 | Staphylococcus hominis | NR_041323.1 | 98% |
| 17 | 10 | KO | 46/13 | Staphylococcus hominis | ||
| 17 | 10 | KO | 46/2 | Staphylococcus hominis | ||
| 17 | 10 | KO | 46/11 | Staphylococcus hominis | NR_041323.1 | 98% |
| 17 | 11 | KO | 46/14 | Staphylococcus lugdunensis | NR_024668.1 | 100% |
| 17 | 12 | RS | 44/18 | Staphylococcus hominis | ||
| 17 | 12 | KO | 46/10 | Staphylococcus hominis | ||
| 17 | 12 | RS | 44/12 | Staphylococcus hominis | ||
| 17 | 12 | RS | 44/2 | Staphylococcus hominis | ||
| 17 | 12 | PS | 45/11 | Staphylococcus hominis | ||
| 17 | 12 | PS | 45/4 | Staphylococcus hominis | ||
| 17 | 12 | WS | 42/1 | Staphylococcus hominis | ||
| 17 | 12 | RS | 44/1 | Staphylococcus hominis | NR_036956.1 | 99% |
| 17 | 12 | WB | 43/12 | Staphylococcus hominis | ||
| 17 | 12 | WB | 43/1 | Staphylococcus hominis | ||
| 17 | 13 | KO | 46/9 | Staphylococcus warneri | NR_025922.1 | 99% |
| 18 | 1 | WB | 43/10 | Staphylococcus epidermidis | NR_036904.1 | 100% |
| 18 | 2 | RS | 44/17 | Staphylococcus epidermidis | NR_036904.1 | 97% |
| 19 | 1 | RS | 44/10 | Staphylococcus caprae | NR_024665.1 | 99% |
| 20 | 1 | WB | 43/4 | Kocuria rosea | ||
| 20 | 1 | WB | 43/5 | Kocuria rosea | NR_044871.1 | 99% |
| 21 | 1 | WB | 43/23 | Bacillus circulans | NR_042726.1 | 99% |
| 21 | 2 | WB | 43/7 | Bacillus circulans | NR_042726.1 | 99% |
| 21 | 3 | WB | 43/16 | Bacillus aerophilus | GU339271.1 | 79% |
| 22 | 1 | RS | 44/14 | Bacillus frigoritolerans | ||
| 22 | 1 | RS | 44/4 | Bacillus frigoritolerans | NR_042639.1 | 100% |
| 22 | 2 | PS | 45/10 | Bacillus simplex | NR_042136.1 | 100% |
| 23 | 1 | BG | 48/17 | Staphylococcus hominis | NR_041323.1 | 99% |
| 24 | 1 | RS | 44/16 | Micrococcus luteus | ||
| 24 | 1 | RS | 44/5 | Micrococcus luteus | NR_037113.1 | 97% |
| 24 | 2 | WB | 43/6 | Micrococcus luteus | NR_037113.1 | 92% |
| 24 | 3 | WB | 43/3 | Micrococcus luteus | NR_037113.1 | 99% |
| 25 | 1 | RS | 44/8 | Arthrobacter agilis | NR_026198.1 | 100% |
| 26 | 1 | DS | 49/5 | Micrococcus luteus | NR_037113.1 | 98% |
| 26 | 2 | UU | 51/1 | Micrococcus luteus | NR_037113.1 | 97% |
| 27 | 1 | KD | 47/4 | Kocuria palustris | NR_026451.1 | 98% |
| 28 | 1 | BG | 48/16 | Micrococcus antarcticus | NR_025285.1 | 99% |
| 29 | 1 | BG | 48/4 | Micrococcus antarcticus | NR_025285.1 | 99% |
| 30 | 1 | WB | 43/9 | Micrococcus antarcticus | NR_025285.1 | 99% |
| 31 | 1 | KD | 47/7 | Micrococcus luteus | NR_037113.1 | 99% |
| 32 | 1 | RS | 44/9 | Micrococcus antarcticus | NR_025285.1 | 99% |
| 33 | 1 | KD | 47/9 | Micrococcus luteus | NR_037113.1 | 99% |
| 34 | 1 | RS | 44/13 | Kocuria palustris | NR_026451.1 | 99% |
| 35 | 1 | PS | 45/9 | Aerococcus urinaeequi | NR_043443.1 | 99% |
| 36 | 1 | KO | 46/4 | Micrococcus luteus | NR_037113.1 | 99% |
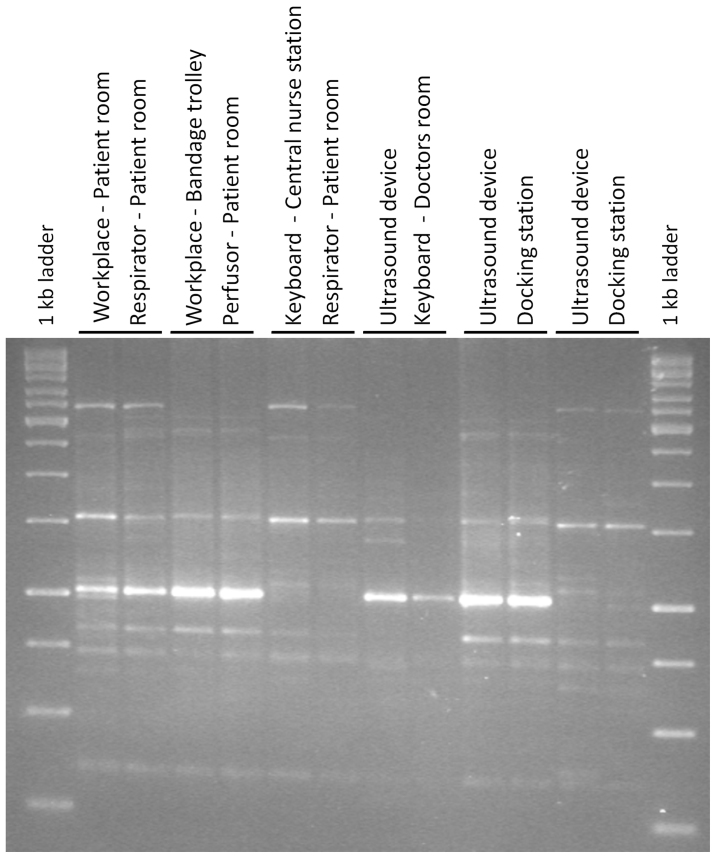
Figure 6. BOX PCR fingerprints of genetically very similar Staphylococcus species isolated from nearly all sampling sites.
The similarity of BOX patterns between them was more than 80%.
Culture-dependent identification was compared with 16S rRNA gene 454-pyrosequencing analysis. While OTUs taken from the amplicon libraries were affiliated with 405 different genera (76 genera ≥ 1% of relative abundance), standard cultivation obtained only 10 bacterial genera corresponding to 2.5% of the total bacterial diversity. Most of the reference sequences of isolates presented an exact match with the pyrosequencing data, but some MIDs did not reach 1% of the relative abundance. Complete linkage clustering indicated that sequences of Micrococcus luteus (Sequence ID: 43/6; max. identity: 92%), Corynebacterium propinquum (49/11; 99%) and Bacillus aerophilus (43/16; 79%) were not present in the cultivation-independent data because their similarity values were lower (Table 3) in comparison to the other sequences.
Discussion
In this study we found a much higher diversity of bacterial communities in the ICU by using the 16S pyrosequencing approach than compared to the standard cultivation technique. Distinct profiles between the floor environment, medical devices and workplaces were found using both strategies. However, various ubiquitous taxa as well as genotypically identical strains were frequently observed.
Although the surfaces in ICUs were characterized by highly diverse bacterial communities, they were actually reduced in comparison with other indoor environments such as living, patient, class or rest rooms3. Similar to other indoor microbial communities, these communities were partially colonized by human-associated bacteria. Although it is impossible to predict the pathogenicity of a strain based on 16S rRNA sequences, the proportion of bacteria identified as those genera/species closely related to human pathogens was very high. They are known for their facultative pathogenic and nosocomial character, e.g. Acinetobacter, Stenotrophomonas, Burkholderia, Flavobacterium, Propionibacterium, Pseudomonas, Staphylococcus and Escherichia/Shigella18,19. Humans are not only the most important dispersal vectors for bacteria inside rooms11; their bacterial fingerprint represents a unique mix of bacteria including pathogens20. Therefore, patients in the ICU may have contributed to this high proportion of potential pathogens. To evaluate this hypothesis, we compared our data with the infections acquired by patients during this time (February-May 2011) in the ICU and found several overlaps which could potentially confirm the potential pathogenic character of several surface-associated bacteria. Altogether, from 101 bacterial infections, the majority was caused by Staphylococcus (40; S. aureus, S. epidermidis and spec. div.). However, Gram-negative pathogens were also identified, e.g. according to their abundance: E. coli, Klebsiella, Pseudomonas, Serratia, Enterobacter, Edwardsiella, Proteus and Chryseobacterium. Conversely, we found bacterial genera, e.g. Burkholderia, Pseudomonas, Lactobacillus, or Methylobacterium which contain plant-associated taxa that can also undergo bivalent interactions with humans. Although they can cause facultative infections in those with certain predispositions, they can also live in symbiosis with plants or can be used as pro- and prebiotics for both plants and humans21,22,23,24. Their origin and function in hospital environments is still unclear. However, one method of transmission could be the transport via pollen into the hospital environment due to the detection of pollen as a vector for specific plant-associated bacteria25. We found a high proportion of chloroplast sequences from Pinus pollen - Pinus trees were among the most common plants outside - in the floor environment and on several devices. These sequences were also detected and discussed in other pyrosequencing-based studies1,3. In addition to air conditioning, the investigated ICU was also window-ventilated, which has been known to result in an increased abundance of chloroplast DNA than in exclusively mechanically ventilated rooms3.
The bacterial communities from three general area floors, medical devices, and workplaces were characterized by a specific and distinct composition. Skin-associated genera (Propionibacterium, Corynebacterium, Staphylococcus, Streptococcus, Bradyrhizobium)26 were highly abundant on medical devices and working surfaces, which was expected considering that they are frequently touched by hands of hospital staff: a typical hand surface harbored on average more than 150 unique species-level bacterial phylotypes20. Interestingly, genera of Burkholderia and Bradyrhizobium were most abundant on devices and workplaces and are both originally plant-associated genera with the potential to fix nitrogen, but are also able to colonize hospital water supplies and surfaces27. In contrast, the floor environment contained genera that are typically distributed in environmental sources, such as soil or water. Acinetobacter was the dominant genus of the floor sample but was also present on almost all devices. During recent years, antibiotic-resistant Acinetobacter infections have become an increasingly common nosocomial problem28,29. Another emerging nosocomial pathogen Clostridium difficile was fortunately not detected in our amplicon libraries. Due to its long-living spores, this bacterium is often found and can cause large-scale outbreaks of nosocomial diarrhea5.
Although bacterial communities in the ICU could be effectively differentiated, connections and transmissions were also detected by principal coordinate- and network analyses. For example, samples of the bandage trolley (MID38) and the workplace in the patient room (MID37) are located between clusters of devices and floor. This bandage trolley is used the whole day in several rooms by different persons of the staff. Additionally, the workplace of the patient room is frequently in contact with the hospital staff, which can explain the transfer. In addition, these two sampling sites were characterized by the highest bacterial diversity (Shannon diversity indices: MID38 4.64; MID37 4.11). Transmission of bacteria from one site to the other was also found comparing molecular fingerprints of the isolated strains. For example, similar BOX fingerprints of staphylococci were identified on nearly all sampling sites. However, deposition of bacterial strains from identical or similar sources, e.g. by personal staff, can also explain this finding. Surface sanitation is an often overlooked, yet crucial component of transmission15, which should be considered more in sanitation protocols. No differences were observed between the bacterial communities of the isolation room for patients who were temporarily isolated with the risk of spreading an infectious disease or were severely immunocompromised and the other patient rooms. This observation is most likely because the isolation room was not in use at the time of sampling.
Although it is well-known that cultivation-dependent techniques capture only a small part of the microbiome6,30, we found an unexpected high difference between the bacterial diversity using both methods – standard cultivation and amplicon sequencing. While in the 16S rRNA gene amplicon library the amount of Gram-positive and Gram-negative bacteria was nearly the same, we detected almost exclusively Gram-positive bacteria by cultivation. With the exception of Roseomonas mucosa known to be associated with bacteremia and other human infections31, cultivation failed to capture the Gram-negative spectrum. This similar proportion of Gram-negative and Gram-positive bacteria as in our libraries was also found in the bacterial infections acquired during this time in the ICU. While the Gram-positive spectrum comprised mainly Staphylococcus and Enterococcus species, the Gram-negative pathogens were characterized by a higher taxonomic diversity. The Gram-positive diversity was well represented in the isolate collection, where the majority of the obtained cultivation-based sequences belonged to Staphylococcus (S. epidermidis, S. hominis, S. auricularis, S. caprae, S. cohnii, S. haemolyticus, S. lugdunensis, S. warneri), which are already described as the most common bacteria in hospitals. Several of them such as S. epidermidis, S. haemolyticus or S. warneri emerged as causal agents of nosocomial infections with diverse resistances against antibiotics32. Interestingly, identical 16S rRNA gene sequences were found for isolates as well as in the amplicon library. Moreover, all sequences from isolates could be found in the amplicon libraries. However, due to the fact that the cultivation of Gram-negatives ultimately failed, new standard protocols should be developed to assess the overall diversity. For this cultivation, we used CASO agar plates that are synonymous to Tryptic Soy Agar (TSA) and Soybean Casein Digest Agar (CSA) suggested by both the European (EP) and United States Pharmacopoeia (USP). While it is difficult to explain why colonies of the well-cultivable bacteria genera such as Burkholderia and Pseudomonas were not isolated from plates, our cultivation results were highly similar to those obtained from the weekly routine monitoring and our 16S amplicon library was comparable with other pyrosequencing based studies1,2,3,4. However, we must consider that pyrosequencing based on DNA may also detect DNA from non-living and living bacteria. Light-activated ethidium monoazide or propidium monoazide can help to remove free extracellular DNA from environmental samples in the future33.
In support of our hypothesis, we found an unexpected high diversity from the bacterial communities with bacteria closely related to human pathogens, but also taxa known for their beneficial interaction with eukaryotes. By using 16S pyrosequencing and corresponding network analysis, we have the tools to evaluate existing sanitation concepts. We suggest that the whole bacteria community should be considered in the assessment of these concepts because diversity within these communities often correlates with the ecosystem function of disease suppression34. If this can be proved for indoor microbiomes, the demand to “Stop killing beneficial bacteria” by Blaser35 should be expanded.
Methods
Experimental design and sampling procedure
Samples were taken from selected surface areas and devices of the intensive care unit (ICU, Department of Internal Medicine, University hospital) in Graz/Austria. The intensive care unit contains 15 beds, including one isolation unit for severe immunocompromised patients. In this ICU the critically ill patients from all internal medicine subspecialities as well as neurologic patients are treated. All sampling locations and their characteristics are given in Table 1. During sampling, all employees and devices of the ICU were in full operation and eight large surface samples (1 m2) were performed by using biological sampling kits (BiSKits; QuickSilver Analytics, Abingdon, MD, USA). For wet sampling of 1 m2, BiSKits were premoistened with the manufacturer-provided sterile buffer36 and the selected area was wiped in three different directions while rotating the sampling device2. Afterwards, samples were stored and chilled (4 to 8°C) during transportation, and frozen immediately at −70°C upon arrival at the laboratory.
Nylon flocked swabs (MicroRheologics, Copan, Brescia, Italy) were used to take samples of 16 devices. For sampling, swabs were moistened briefly in a 15 ml Falcon test tube containing 2.5 ml of sterile water37. After sampling of provided surfaces (5 × 5 cm), the swabs were broken into the Falcon test tubes and were kept cool (4 to 8°C). In the laboratory, microorganisms were extracted by vortexing (5 seconds) and sonication (84 W; 35 kHz; Sonorex super DK) for 120 seconds. Lastly, all samples were stored at −70°C. In addition, samples for cultivation were taken using CASO agar plates (Heipha Diagnostika, Eppelheim, Germany). In our study, ten contact tests of these CASO plates were performed according to the guidelines of the quarterly controls of the ICU. Samples were taken on defined positions and incubated at room temperature (RT) for 4 days. Colony forming units (CFU) were counted and 130 isolates were selected and subcultured on CASO agar. The isolates were purified and stored at −70°C in a nutrient broth (NB) containing 50% glycerol.
Total community DNA isolation
UV sterilized Amicon Ultra-15 filters (cutoff 50 kDa; Millipore GmbH, Schwalbach, Germany) were used for concentration of the sampling liquid from BiSKits. The resulting BiSKit suspension of each sampling liquid from swabs were subjected to DNA extraction using the modified XS buffer method2: XS buffer (2 x) was freshly prepared as follows: (20 ml stock solution): 1 M Tris/HCl (pH 7.4) (4 ml); 7 M ammonium acetate (4.56 ml); 250 mM ethylene diamine tetraacetic acid (3.2 ml); 10% sodium dodecyl sulfate (w/v) (4 ml); potassium ethyl xanthogenate (0.4 g); PCR-grade water (4.99 ml). For completely dissolving the xanthogenate, the buffer was incubated at 65°C for 15 min. Starting with 1 ml sample, in total, 1 ml of 2 × XS buffer was added, and the mixture was stirred gently (short vortex). After an incubation of 2 h at 65°C, and mixing by hand every 30 min, the suspension was vortexed for 10 seconds. The tube was placed on ice for 10 min and centrifuged afterwards (100 g, 5 min, 4°C). The supernatant was transferred into a PhaseLock Gel tube (Eppendorf, Hamburg, Germany), and an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added. The suspension was mixed gently and centrifuged (2000 g, 5 min, 15°C). The aqueous layer was transferred into a new tube. To precipitate DNA, the same volume of cold 100%isopropanol and 1/10 volume of 4 M ammonium acetate was added and gently mixed. After incubation at −20°C overnight, the suspension was centrifuged at 13500 g at 4°C for 30 min. The (invisible) pellet was washed with 1 ml 70% ethanol (ice cold) and centrifuged (13500 g, 30 min, 4°C). The pellet was then dried completely and dissolved in 20 μl PCR-grade water.
16S rRNA gene amplicon pyrosequencing
For pyrosequencing, the 16S rRNA gene was amplified in a nested PCR approach with the universal bacterial primer set 27f (5′-GAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-AAGGAGGTGATCCANCCRCA-3′). The PCR reaction mixture (10 μl) contained 1 × Taq&Go, 0.25 μM of each primer and 1 μl of template DNA (95°C, 5 min; 30 cycles of 95°C, 30 s; 57°C, 30 s; 72°C, 90 s; and elongation at 72°C, 5 min). In a second PCR, 1 μl of the amplicon was used. 16S rRNA gene sequences were amplified by using the forward primer Unibac-II-515f (5′-GTGCCAGCAGCCGC-3) containing the 454-pyrosequencing adaptors (MIDs) and the reverse primer UnibacII927r_454 (5′-CCCGTCAATTYMTTTGAGTT-3′). Sequences of MIDs are listed in Tab. S3. The reaction mixture for the second PCR (60 μl) contained 1 × Taq&Go, 0.25 μM of each primer and 6 μl of the PCR product solution (95°C, 5 min; 32 cycles of 95°C, 20 s; 54°C, 15 s; 72°C, 30 s; and elongation at 72°C, 10 min). PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, USA). The partial 16S rRNA genes were sequenced using a Roche GS FLX+ 454 pyrosequencer (GATC Biotech, Konstanz, Germany).
Sequences that were shorter than 150 bp in length or of low quality were removed from the pyrosequencing-derived data sets using the pipeline initial process of the RDP pyrosequencing pipeline (http://pyro.cme.msu.edu)38. Due to the different number of sequences among samples, the data was normalized. The webserver SnoWMAn 1.11 (http://snowman.genome.tugraz.at)39 for taxonomic-based analysis was used with the following settings: analysis type: BLAT pipeline; reference database: greengenes_24-Mar-2010; rarefaction method: RDP; taxonomy: RDP; confidence threshold: 80%; include taxa covering more than: 1%. For rarefaction analysis, operational taxonomic units (OTUs) were clustered with 3% (species level), 5% (genus level) and 20% (phylum level) dissimilarity cut-offs40,41. Rarefaction curves were constructed by using the tools aligner, complete linkage clustering, and rarefaction of the RDP pyrosequencing pipeline. Shannon42 and Chao1 indices43 were calculated with the complete linkage clustering data. Principal coordinate analysis (PCoA) plots were generated using the open source software package QIIME (http://qiime.sourceforge.net), which allows analysis of high-throughput community sequencing data17. Network analysis was performed to visualize the most abundant taxa and to compare their abundance across the three sampling areas. The relative abundance of single OTUs was calculated for each sample and were used to generate an average value for each of the three areas. The open source software Cytoscape 2.816 was employed to visualize the 40 most abundant OTUs based on total read counts. To differentially detect abundant microbial clusters between the three areas clusters with ≥10 sequences were explored using Metastats web interface44. Bacterial populations of the ICU dataset were examined using a combination of the nonparametric t-test, exact Fisher's test, and the false discovery rate with 1000 permutations. P-values were determined for each cluster correspondingly.
Phylogenetic analysis of the isolates
Based on isolated colony DNA, Amplified ribosomal RNA gene restriction analysis (ARDRA) using the restriction enzyme HhaI (MP Biomedicals, Eschwege, Germany) was performed to cluster isolates with similar band pattern into genotypic groups according to Berg et al.45. Isolates with similar ARDRA patterns were clustered in one group and analyzed performing BOX-PCR fingerprinting. BOX-PCR was done using the BOX_A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′) as described by Rademaker and de Bruijn46. Computer-assisted evaluation of ARDRA- and BOX-PCR generated fingerprints were made using the GelCompar II software (version 5.1; Applied Math, Kortrijk, Belgium). Chosen isolates were identified by partial 16S rRNA gene sequencing at the sequencing core facility in Microsynth AG, Balgach, Switzerland. Obtained sequences were aligned using the NCBI sequence database and the BLAST algorithm. The 16S rRNA gene sequences of the isolates were deposited in the NCBI nucleotide sequence database under accession numbers HE962211-HE962235.
Author Contributions
Conceived and designed the experiments: G.B., C.H., K.H.S. Performed the experiments: L.O., C.Z. Analyzed the data: L.O., S.L., G.B. Wrote the first draft of the manuscript: L.O., G.B. Contributed to the writing of the manuscript: L.O., C.H., G.B., I.C.M.J.E. criteria for authorship read and met: L.O., C.Z., S.L., C.H., K.H.S., G.B. Agree with manuscript results and conclusions: L.O., C.Z., S.L., C.H., K.H.S., G.B.
Supplementary Material
The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing
Acknowledgments
We thank Anastasia Bragina (Graz) and Christine Moissl-Eichinger (Regensburg) for fruitful discussions and valuable technical support. Furthermore, we would also like to thank Meg Starcher (Graz/Washington) for English revision of the manuscript. This work has been supported by a grant to G.B. from the Federal Ministry of Economy, Family and Youth (BMWFJ), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and ZIT - Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG.
References
- Flores G. E. et al.. Microbial biogeography of public restroom surfaces. PLoS ONE 6, e28132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissl-Eichinger C. Archaea in artificial environments: Their presence in global spacecraft clean rooms and impact on planetary protection. The ISME J 5, 209–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel S. W. et al.. Architectural design influences the diversity and structure of the built environment microbiome. The ISME Journal 6, 1469–1479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt K. M., Gerba C. P., Maxwell S. L. & Kelley S. T. Office space bacterial abundance and diversity in three metropolitan areas. PLoS ONE 7, e37849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman N., Stukel T. A., Ma X., Vermeulen M. & Guttmann A. Reduction in Clostridium difficile infection rates after mandatory hospital public reporting: findings from a longitudinal cohort study in Canada. PLoS Medicine 9, e1001268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J. T. & Konopka A. Measurements of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39, 321–346 (1985). [DOI] [PubMed] [Google Scholar]
- Aundel A. V., Sterling E. M., Biggin J. H. & Sterling T. D. Indirect health effects of relative humidity in indoor environments. Environ Health Perspect 65, 351–361 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Hospodsky D., Yamamoto N., Nazaroff W. W. & Peccia J. Size-resolved emission rates of airborne bacteria and fungi in an occupied class room. Indoor Air 22, 339–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. L. et al.. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee. JAMA 274, 639–644 (1995). [PubMed] [Google Scholar]
- Plowman R. The socioeconomic burden of hospital acquired infection. Euro Surveill 5, 49–50 (2000). [DOI] [PubMed] [Google Scholar]
- Klevens R. M. et al.. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122, 160–166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota B. Contamination, disinfection, and cross-colonization: Are hospital surfaces reservoirs for nosocomial infection? Clin Infectious Diseases 39, 1182–1189 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastmeier P., Schwab F., Bärwolff S., Rüden H. & Grundmann H. Correlation between the genetic diversity of nosocomial pathogens and their survival time in intensive care units. Journal of Hospital Infection 62, 181–186 (2005). [DOI] [PubMed] [Google Scholar]
- Kramer A., Schwebke I. & Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Diss 6, 130 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter J. A. & French G. L. Survival of nosocomial bacteria and spores on surfaces and inactivation by hydrogen peroxide vapor. J Clin. Microbiol 47, 205–207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M. E., Ono K., Ruscheinski J., Wang P. L. & Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 27, 431–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al.. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J. J., Baig W., Reynolds G. W. & Sandoe J. A. T. Endocarditis caused by Propionibacterium species: a report of three cases and a review of clinical features and diagnostic difficulties. J Med Microbiol 55, 981–987 (2006). [DOI] [PubMed] [Google Scholar]
- Das I. et al.. Carbapenem-resistant Acinetobacter and role of curtains in an outbreak in intensive care units. J Hosp Infection 50, 110–114 (2002). [DOI] [PubMed] [Google Scholar]
- Fierer N., Hamady M., Lauber C. L. & Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A 105, 17994–17999 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G., Eberl L. & Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol 7, 1673–1685 (2005). [DOI] [PubMed] [Google Scholar]
- Berg G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84, 11–18 (2009). [DOI] [PubMed] [Google Scholar]
- Ryan R. P. et al.. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol 7, 514–25 (2009). [DOI] [PubMed] [Google Scholar]
- Lai C. C. et al.. Infections caused by unusual Methylobacterium species. J. Clin Microbiol 49, 3329–3331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürnkranz M. et al.. Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb Ecol 63, 418–428 (2012). [DOI] [PubMed] [Google Scholar]
- Grice E. A. & Segre J. A. The skin microbiome. Nature Rev Microbiol 9, 244–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutala W. A. & Weber D. J. Water as a reservoir of nosocomial pathogens. Infect Control Hosp Epidemiol 18, 609–616 (1997). [PubMed] [Google Scholar]
- Carey R. B., Banerjee S. N. & Srinivasan A. Multidrug-resistant Acinetobacter infections, 1995–2004. Presented at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco (2006). [Google Scholar]
- Munoz-Price L. S. & Weinstein R. A. Acinetobacter infection. New England Journal of Medicine 358, 1271–81 (2008). [DOI] [PubMed] [Google Scholar]
- Schleifer K. Microbial diversity: facts, problems and prospects. Systematic Applied Microbiology 27, 3–9 (2004). [DOI] [PubMed] [Google Scholar]
- Christakis B. B., Perlorentzou S., Alexaki P., Megalakaki A. & Zarkadis I. K. Central line-related bacteraemia due to Roseomonas mucosa in a neutropenic patient with acute myeloid leukaemia in Piraeus, Greece. J Med Microbiol 55, 1153–1156 (2006). [DOI] [PubMed] [Google Scholar]
- Uçkay I. et al.. Foreign body infections due to Staphylococcus epidermidis. Ann Med 41, 109–119 (2009). [DOI] [PubMed] [Google Scholar]
- Wagner A. O., Malin C., Knapp B. A. & Illmer P. Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide. Appl Environ Microbiol. 74, 2537–2539 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elsas J. D. et al.. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci U S A 109, 1159–1164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature 476, 393–394 (2011). [DOI] [PubMed] [Google Scholar]
- Buttner M. P. et al.. Evaluation of the Biological Sampling Kit (BiSkit) for large-area surface sampling. Appl Environ Microbiol 70, 7040–7045 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A., Facius R., Wirth R. & Moissl-Eichinger C. Validation of a nylon-flocked-swab protocol for efficient recovery of bacterial spores from smooth and rough surfaces. Appl Environ Microbiol 76, 5148–5158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R. et al.. The ribosomal database project: improved alignments and new tools for rRNA analysis Nucleic Acids Res. 37, D141–D145 (2009). [DOI] [PMC free article] [PubMed]
- Stocker S. et al.. SnoWMAn: High-throughput phylotyping, analysis and comparison of microbial communities. Under revision (2011). [Google Scholar]
- Hur I. & Chun J. A method for comparing multiple bacterial community structures from 16S rDNA clone library sequences. J Microbiol 42, 9–13 (2004). [PubMed] [Google Scholar]
- Schloss P. D. & Handelsman J. Toward a census of bacteria in soil. PLoS Comput Biol. 2, 786–793 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C. E. The mathematical theory of communication. 1963. MD Comput 14, 306–317 (1997). [PubMed] [Google Scholar]
- Chao A. & Bunge J. Estimating the number of species in a sochastic abundance model. Biometrics 58, 531–539 (2002). [DOI] [PubMed] [Google Scholar]
- White J. R., Nagarajan N. & Pop M. Statistical methods for detecting deferentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5, e1000352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G. et al.. Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl Environ Microbiol 68, 3328–3338 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker J. L. W. & De Bruijn F. J. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer-assisted pattern analysis. In: Caetano-Anolle′s G, Gresshoff PM, eds. DNA markers: protocols, applications and overviews. New York: John Wiley & Sons. pp. 151–171 (1997).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing