Abstract
Molecular dynamics are essential for life, and nuclear magnetic resonance (NMR) spectroscopy has been used extensively to characterize these phenomena since the 1950s. For the past 15 years, the Carr-Purcell Meiboom-Gill relaxation dispersion (CPMG RD) NMR experiment has afforded advanced NMR labs access to kinetic, thermodynamic, and structural details of protein and RNA dynamics in the crucial µs-ms time window. However, analysis of RD data is challenging because datasets are often large and require many non-linear fitting parameters, thereby confounding assessment of accuracy. Moreover, novice CPMG experimentalists face an additional barrier because current software options lack an intuitive user interface and extensive documentation. Hence, we present the open-source software package GUARDD (Graphical User-friendly Analysis of Relaxation Dispersion Data), which is designed to organize, automate, and enhance the analytical procedures which operate on CPMG RD data (http://code.google.com/p/guardd/). This MATLAB-based program includes a graphical user interface, permits global fitting to multi-field, multi-temperature, multi-coherence data, and implements χ2-mapping procedures, via grid-search and Monte Carlo methods, to enhance and assess fitting accuracy. The presentation features allow users to seamlessly traverse the large amount of results, and the RD Simulator feature can help design future experiments as well as serve as a teaching tool for those unfamiliar with RD phenomena. Based on these innovative features, we expect that GUARDD will fill a well-defined gap in service of the RD NMR community.
Keywords: Protein dynamics, RNA dynamics, Relaxation dispersion, Carr-Purcell Meiboom-Gill
Introduction
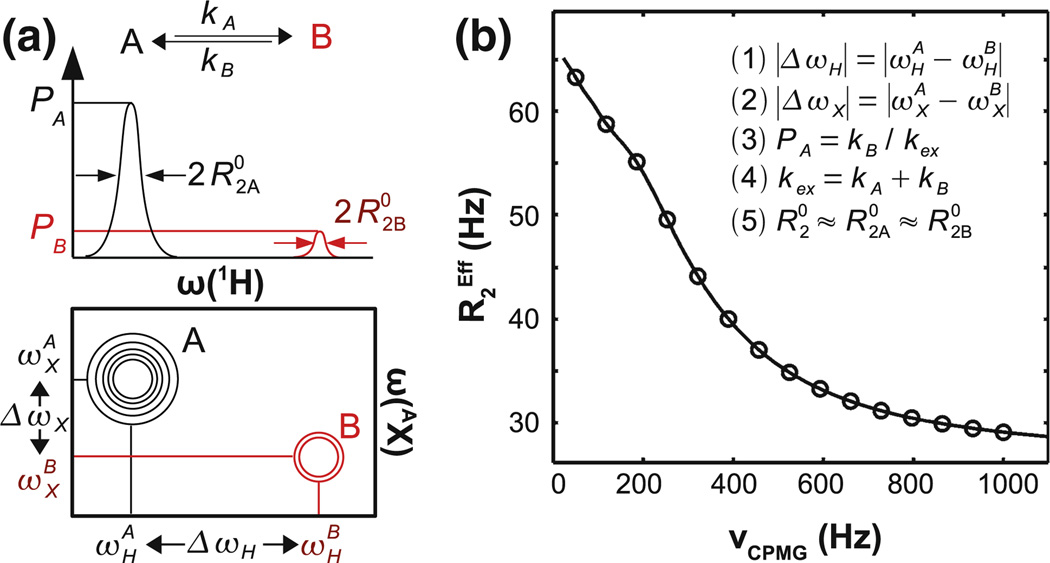
Macromolecular dynamics are essential for life, and nuclear magnetic resonance (NMR) spectroscopy has been used extensively to characterize these phenomena since the 1950s (Boehr et al. 2006; Kleckner and Foster 2011; McConnell 1958). For the past 15 years, the Carr-Purcell Meiboom-Gill relaxation dispersion (CPMG RD) NMR experiment has afforded advanced NMR labs access to kinetic, thermodynamic, and structural details of molecular dynamics in the crucial µs-ms time window for proteins (Kempf and Loria 2003; Kleckner and Foster 2011; Loria et al. 2008; Palmer et al. 2005), and more recently for RNA (Johnson and Hoogstraten 2008; Kloiber et al. 2011; Ren and Ghose 2011). CPMG RD enables researchers to extract information on certain dynamic processes by measuring the effective transverse relaxation rate of the NMR signal, , in response to the application of refocusing pulses applied at frequency νCPMG. Typically, the dynamic process is interpreted using the simplest description of exchange, the two-state A ↔ B model, which describes the RD curve using 4–5 parameters (Fig. 1).
Fig. 1.
The two-state exchange model requires 4–5 parameters to describe a single RD curve. a A single NMR probe (e.g., Ala 12NH) is considered to alternate between local structures designated A and B, which have distinct NMR chemical shifts in 1H and/or AX dimensions. Although two signals are shown here, the “minor” B state is not detected directly because its NMR signal is too weak and/or too broad. However, the A ↔ B exchange yields a quantitative effect on the measured RD curve of signal A. b A multiple quantum (MQ) RD curve is described by five parameters in the two-state model, which characterize the structural, thermodynamic, kinetic, and relaxation features of the exchanging states. (1) |ΔωH| is the magnitude of 1H chemical shift difference between states A and B (n.b., this is not detected in a single quantum (SQ) heteronuclear experiment). (2) |ΔωX| is the magnitude of AX chemical shift difference between states A and B (AX = 13C or 15N). (3) PA = kB/kex is the equilibrium population fraction of state A such that PA + PB = 1. (4) kex = − kA + kB is the total rate of exchange between states A and B. kA and kB are the rates of exchange from A → B and B → A respectively with kA = (1 − PA)kex and kB = PAkex. (5) is the transverse relaxation rate in the absence of exchange assuming (Ishima and Torchia 2006). The MQ curve shown here is simulated using |ΔωH| = 8 Hz, |ΔωX| = 201 Hz, kex = 1,000/s, PA = 90%, and
Analysis of CPMG RD data is challenged by (1) the non-linear nature of its mathematical formulation, (2) the multitude of parameters required for fitting compared to its relatively featureless appearance, (3) the reality that an RD curve is often under-sampled with noisy data (to spare expensive NMR time), and (4) the fact that fitting accuracy is not validated by mere visual inspection of the fit to the data, nor by merely minimizing the fitted residuals, because some curves are fit equally well with divergent combinations of parameters (Hansen et al. 2008b; Ishima and Torchia 2005, 2006; Kovrigin et al. 2006; Myint and Ishima 2009; Neudecker et al. 2006). To address these limitations, experimentalists often acquire large datasets encompassing multiple magnetic field strengths (Kovrigin et al. 2006) and/or temperatures (Boehr et al. 2010; Mulder et al. 2001; Vallurupalli and Kay 2006) and/or pressures (Korzhnev et al. 2006) and/or quantum coherences (QCs) (Korzhnev et al. 2005), and produce custom software to implement a global analysis assuming two-state exchange (Beach et al. 2005; Boehr et al. 2010; Grey et al. 2003; Hansen et al. 2008a, b; Henzler-Wildman et al. 2007; Jee et al. 2008; Kovrigin, June 30, 2011; Mulder et al. 2002; Namanja et al. 2010) or, less often, three-state exchange (Korzhnev et al. 2007, 2004b; Neudecker et al. 2006). Although these software tools are generally available from their respective authors, their complexity and specificity may overwhelm a novice spectroscopist, and more intuitive software has only begun to emerge (Bieri and Gooley 2011).
Hence, we present the software package GUARDD (Graphical User-friendly Analysis of Relaxation Dispersion Data), which is designed to facilitate scientific investigations by organizing, automating, and enhancing the analytical processes related to CPMG RD data, especially for novice spectroscopists. In addition to its user-friendly interface, GUARDD contains powerful features that engage the user in the fitting process and thus clarify assessment of fitting accuracy. Advanced labs can benefit from using GUARDD to easily manage data and display results, to plan experiments using the comprehensive RD Simulator, and to extend customized functionality, permitted via an open-source code license. Importantly, we have reduced the activation barrier for its use by establishing tutorials, extensive documentation, and an online community of support (http://code.google.com/p/guardd/).
GUARDD software overview
GUARDD is designed for ease of use by implementing a graphical interface for key functions like managing data, performing and assessing fitting tasks, organizing and displaying results, and simulating RD data for experimental design and/or instruction. In the interest of space, only certain features are discussed, whereas comprehensive documentation and a user-friendly tutorial can be obtained via the GUARDD website (http://code.google.com/p/guardd/).
Data: hierarchical data objects are easily managed via graphical interface
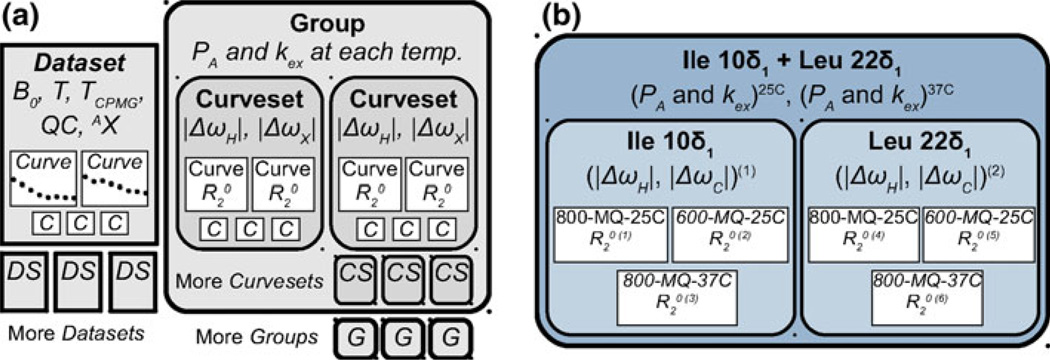
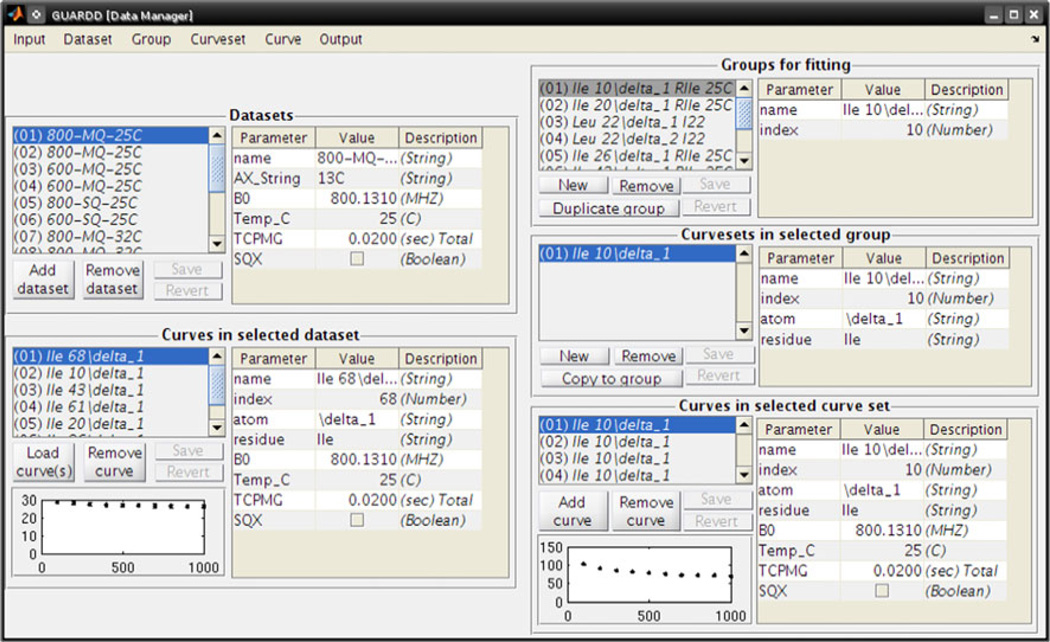
Because analyzing RD data in aggregate is necessary for an accurate description of molecular dynamics, GUARDD implements a hierarchical data management system which accommodates a variety of hypothetical two-state exchange models (Fig. 2). The lowest tier is an RD Curve, which describes a series of relaxation rates and uncertainties, each paired with a value of νCPMG, for a unique NMR signal/dynamic probe (i.e., , where σ designates uncertainty, for one or more values of νCPMG). Each Curve is typically loaded with Curves from other signals as a Dataset, which specifies the experimental conditions: magnetic field strength B0, temperature T, total time of the CPMG period TCPMG, and the nucleus AX and QC probed during that time (SQ = Single Quantum, MQ = Multiple Quantum). For convenience, Curves are loaded using a plain-text file that can easily be prepared using a spreadsheet program. For analysis, each Curve is designated its own relaxation rate, . At the middle tier, one or more Curves are aggregated by signal assignment into a Curveset, which designates the chemical shift differences, |ΔωH| and |ΔωX|, for that NMR probe. For each Curve, the QC determines the effect of the Curveset’s chemical shift differences (Korzhnev et al. 2005), such that MQ Curves are sensitive to |ΔωH| + |ΔωX| whereas SQ Curves are only sensitive to |ΔωX| and therefore fix ΔωH to zero. At the highest tier, one or more Curvesets are aggregated into a Group, which designates signals that share the kinetic parameters PA and kex at each temperature. Because each Curveset (and therefore Curve) within a Group is constrained to exhibit the same exchange kinetics, the highest tier is useful for segregating independent modes of exchange in the molecule (e.g., domains). Importantly, although GUARDD can auto-generate Curvesets and Groups, the user has complete control over how the data are organized, and is encouraged to create many combinations of Curvesets and Groups to test the validity of different descriptions of molecular dynamics. These essential tasks are easily accomplished using the GUARDD Data Manager (Fig. 3).
Fig. 2.
To accommodate many descriptions of molecular dynamics, GUARDD organizes the basic unit of RD data, the Curve (C), in a hierarchical manner, allowing for global fitting of multi-temperature, multi-field, multi-coherence data. a At the lowest tier, Curves are loaded in Datasets (DS), which specify experimental conditions during data acquisition of those Curves. For analysis, one or more Curves are aggregated into a Curveset (CS), and one or more Curvesets are aggregated into a Group (G). In this way, fitting parameters required for each Curve are shared, thus enhancing fitting accuracy to gain insight into molecular dynamics. More Curves (C), Datasets (DS), Curvesets (CS) and Groups (G) are shown as smaller icons to indicate that each of these data objects may contain an arbitrary number of lower-tier objects. b An example Group, containing dynamic probes Ile 10δ1 and Leu 22δ1, contains two Curvesets, which each contain three Curves. Fitting this Group using GUARDD tests the hypothesis that these two NMR probes report a common exchange process in the protein. The fitting procedure requires 14 parameters. (1–4) The Group requires PA and kex at each of two temperatures. (5–8) These Curves are MQ, and thus each Curveset requires specification of both |ΔωH| and |ΔωC| in ppm, which is converted to rad/s using each Curve’s field strength, B0. (9–14) Each Curve is designated its own transverse relaxation rate
Fig. 3.
The GUARDD Data Manager makes it easy to browse and construct hierarchical fitting objects, which allow the user to test models of molecular dynamics probed by RD. Using this interface, Curves can be added or removed to or from Curvesets, which can be added, removed or copied to or from Groups. For convenience, GUARDD can auto-generate Groups and Curvesets from the all Datasets, or a specified subset, by segregating Curves according to NMR signal assignment. To read these data using an external program, a hierarchical tree of can be saved into a comprehensive plain-text file
Analysis: sharing fit parameters via two-state exchange at multiple temperatures
In order to fit a wide range of RD data, GUARDD uses the MQ Carver-Richards-Jones all-timescales dispersion equation (Korzhnev et al. 2004a). To accommodate multi-coherence data, MQ Curves allow both |ΔωH| and |ΔωX| to be optimized during fitting, whereas |ΔωH| is fixed to zero for SQ Curves. To accommodate multi-field data, the Curve-specific rad/s value of Δω is obtained from its parent Curveset using , where γX is the gyromagnetic ratio of nucleus X. To accommodate multi-temperature data, there are two fitting options. (A) Designate a Group-specific kex and PA for each of the NT temperatures, and optimize these 2NT parameters during fitting. After fitting, additional details of the dynamic process can be obtained from activation energies and exchange enthalpies (Boehr et al. 2010; Vallurupalli and Kay 2006) resulting from Arrhenius and van’t Hoff analyses, respectively (Kleckner and Foster 2011; Winzor and Jackson 2006). Briefly, Arrhenius analysis requires both PA and kex to quantify the temperature-dependence of exchange rate and van’t Hoff analysis requires only PA to quantify temperature-dependence of exchange populations. (B) The second fitting option uses the Arrhenius and van’t Hoff relations as part of the fitting process by optimizing four Group-specific parameters: kex and PA at one temperature as well as activation energy EAB = EB − EA and exchange enthalpy ΔH = HB − HA, which subsequently determine kex and PA at any temperature. These options are useful because when data are available at more than two temperatures, option (B) requires fewer parameters than option (A) (4 vs. 2NT), and therefore may be more appropriate for interpreting the RD dataset.
Analysis: grid search and Monte Carlo χ2 maps enhance and assess fitting accuracy
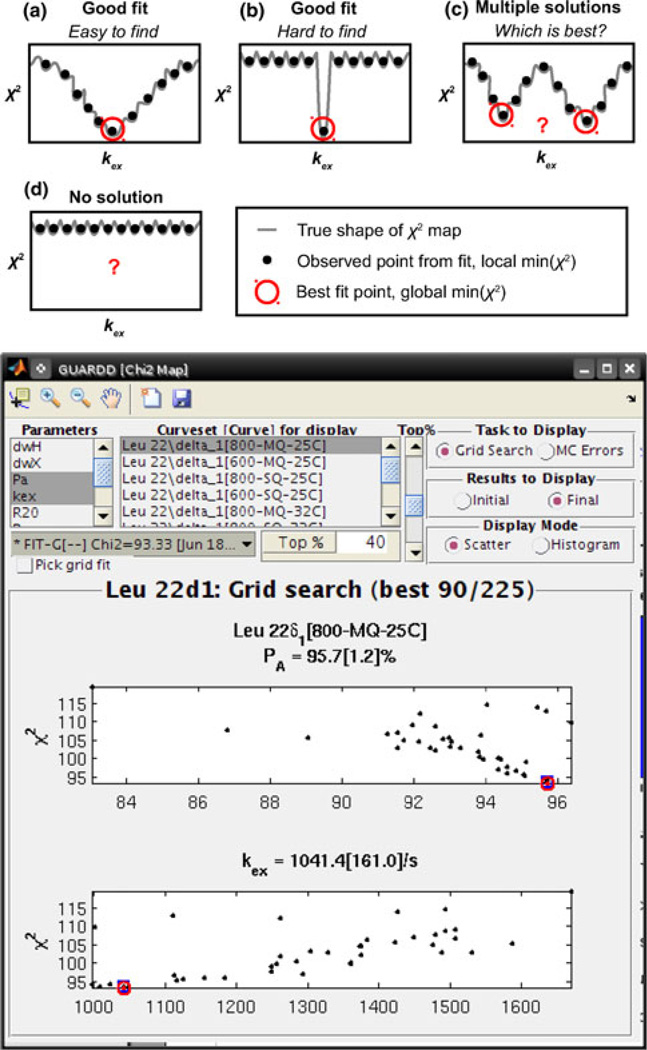
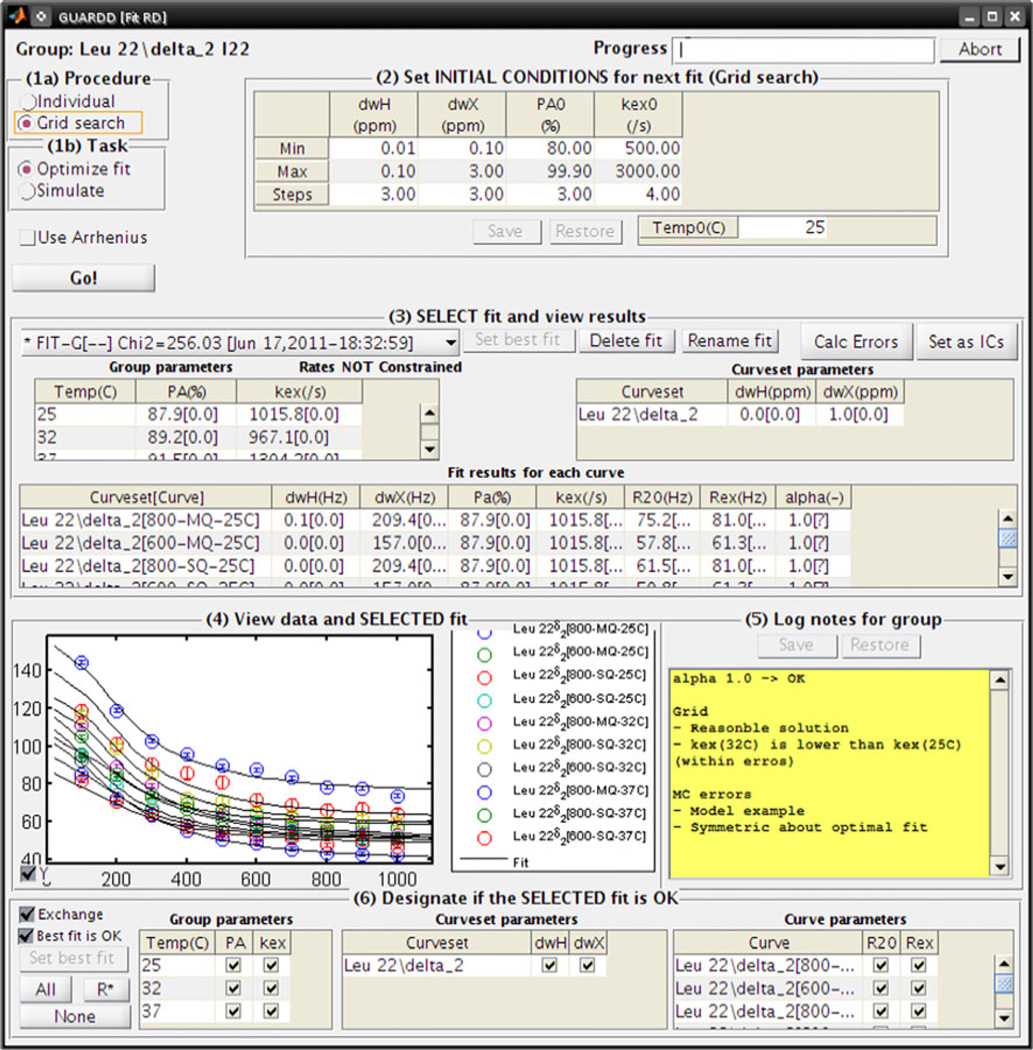
The goal of the fitting procedure is to obtain a set of parameters that accurately describes the trends in the data, which is typically pursued by minimizing the difference between the observed data and that calculated from the fit (Bevington and Robinson 2003; Motulsky and Christopoulos 2003). To accomplish this task, GUARDD invokes the MATLAB fmincon function with the interior-point algorithm (Byrd et al. 2000; though other options are available) to iteratively alter the fitting parameters p⃗ to minimize the target function , where designates an observed data point with error on a single Curve acquired with conditions B0, T, QC, AX, and designates the calculated point using the Curve conditions and the independent fitting parameters p⃗ for the Group: PA and kex for each temperature, |ΔωH| and |ΔωX| for each Curveset, and for each Curve. Unfortunately, the nonlinear nature of RD phenomena makes the relationship between χ2 and p⃗ difficult to predict, and therefore optimization algorithms often “fail” by finding a local minimum of χ2, which is sensitive to initial fitting conditions, instead of the intended global minimum (Fig. 4a). Hence, to assess the sensitivity of the final fit to initial conditions, GUARDD implements a grid search over a six-dimensional (6D) parameter space encompassing the most sensitive parameters: |ΔωH|, |ΔωX|, PA, kex, EAB, and ΔH. Each point on this 6D grid is used to set the initial conditions for subsequent optimization. Using GUARDD’s Fit RD window (Fig. 5), the user can specify the limits and number of steps for each dimension of the grid, the selection of which depends Group size, knowledge of the fitting solution, and use of the Arrhenius constraint which enables the EAB and ΔH dimensions (see section “Analysis: sharing fit parameters”, option (B)). The completed grid of Ngrid points contains Ngrid sets of optimized parameter values, with each set yielding one value of χ2. Because the grid for each Group is typically large (10–500 points in each of 5–50 dimensions), GUARDD’s Chi2 Map window is designed to make the otherwise-cumbersome navigation relatively easy (Fig. 4, Bottom).
Fig. 4.
The nonlinear nature of the RD phenomena yields a “rough” χ2 map that can trap an optimization routine in any of many local minima, instead of the intended global minimum, and GUARDD is designed to address and illustrate these potential pitfalls. a–d The χ2 map is a hypersurface with amplitude χ2 and one dimension for each independent fitting parameter (e.g., 14 parameters yields a 14D hypersurface). Here, the response of χ2 to just one parameter, kex, produces a 2D slice through the hypersurface to illustrate four commonly encountered shapes that pose distinct challenges in obtaining an accurate fit. (Bottom) GUARDD addresses this challenge using a grid search to fit the data using many different initial conditions, thus sampling the χ2 map. The Chi2 Map window enables the user to navigate each dimension of the map, which helps assess fitting accuracy and select the best fit. Here, two parameters, PA(25°C) and kex(25°C), most closely correspond to the “Good fit” shape in a, though other parameters may require user-directed optimization, as in c, or may not be defined at all, as in d. To focus the display of this example χ2 map, only the best 40% of fits are shown, where each black point designates the result of a single fit optimization and the red circle shows the “best” fit currently selected by the user (n.b., each fit can be displayed individually for more careful examination)
Fig. 5.
For a given Group, the fitting window specifies grid search limits, enables refinement via user-specified initial fitting conditions, and organizes and displays the array of candidate fits. In the lower half of the window, the selected fit is displayed, the user designates which parameters are valid for subsequent analyses, and plain-text notes can be logged for reference and aggregate export
When seeking the best fit for the Group, one must often select one χ2 minimum out of several candidates (Fig. 4c). This issue is exacerbated by noisy RD data and/or small Rex resulting from exchange near a detection limit of the CPMG RD method. To this end, GUARDD is designed to accommodate three general strategies at the discretion of the user. (1) Select one of the fits, but mark the ill-defined parameters as “Not OK,” thus preventing their mis-interpretation in subsequent analyses. For example, in the fast-exchange regime (kex ≫ Δν), kex is often valid despite that PA and Δω are not independently defined, but are instead convoluted into the quantity Φex = PAPBΔω2 (Ishima and Torchia 1999; Luz and Meiboom 1963). (2) Alter the Group and re-fit. The user may remove noisy Curves and/or add additional Curvesets to help constrain the values of kex and PA. (3) Acquire more data and re-fit the new Group. The RD Simulator can help determine optimal conditions of temperature, magnetic field strength, and/or QC for efficient use of spectrometer time (see section “Optimization and education”). Importantly, NMR probes that sample more than two states may not fit well to the two-state exchange model used by GUARDD, and hence relatively poor fits (large χ2) may reflect this limitation.
As mentioned previously, RD data are often noisy, and this can further widen the variety of fitting solutions surrounding the global minimum of χ2. Hence, GUARDD implements a well-established Monte Carlo (MC) procedure to bootstrap from the residuals (Bevington and Robinson 2003; Motulsky and Christopoulos 2003). For a given Group, the MC algorithm generates many synthetic Groups, each randomly differing in values from the original Group, yet still within their noise, and fits them individually to assess the sensitivity of the final fit to noise in the data. Much like the results of the grid search, the MC results are often large (100 s of fits with 5–50 parameters each), but can be easily navigated using the Chi2 Map window.
The computational time required for these fitting tasks depends on the number of Curves in the Group, where 1, 3, or 10 Curves requires about 5, 15, or 60 s per step using a desktop PC with a 2.6 GHz CPU and 3 Gb of RAM running MATLAB R2011a. Because these tasks are repeated hundreds of times for tens of Groups, GUARDD includes a Batch Task function which allows lengthy computations to be queued for consecutive processing, while incremental progress is automatically saved to the hard drive.
Optimization and education: RD Simulator explores the nature of RD phenomena
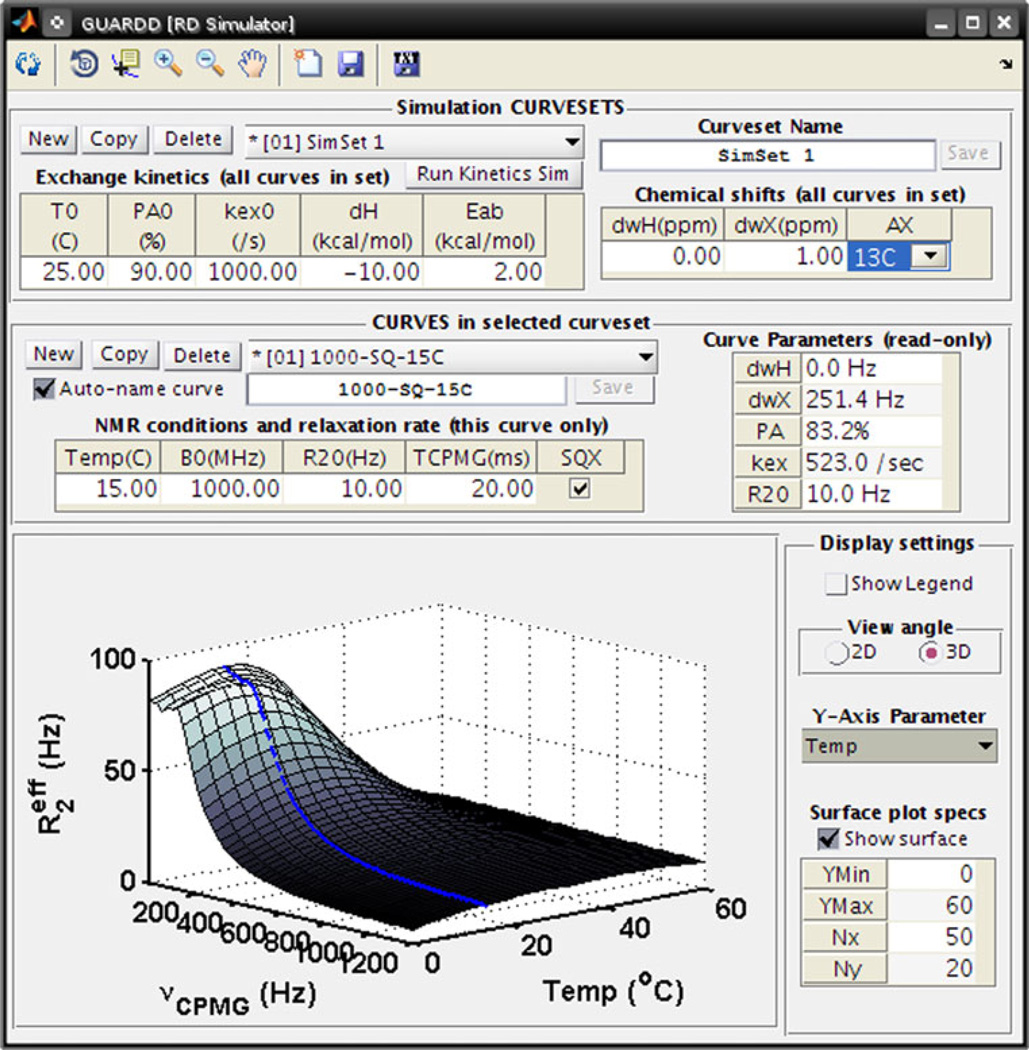
GUARDD includes an interactive RD Simulator that models multi-temperature, multi-coherence, multi-field RD data (Fig. 6). The related Kinetic Simulator models the temperature-dependence of population PA and rates kex, kA, and kB for two-state exchange, provided enthalpy change ΔH and activation energy EAB. These features are designed to serve three important functions.
Fig. 6.
The RD Simulator can be used to explore the nature of RD phenomena to plan future experiments and to enhance understanding of RD. For example, by modeling the temperature-dependence of two-state exchange, one can identify the limits of detection for a particular NMR probe, thus avoiding uninformative experiments. The blue Curve corresponds to the “Curve Parameters” in the middle of the window, and the surface corresponds to how this Curve would appear at different temperatures, provided the “Exchange kinetics” in the upper panel. Data can be exported, either directly to GUARDD as a Group or to a plain-text file, with an arbitrary number of νCPMG points and signal to noise ratio
First, they are helpful for selecting appropriate conditions for future experiments, thus optimizing the use of expensive NMR time. For example, if RD data are already available at two temperatures, the simulator can determine what third temperature yields the largest dispersions, which often yield the most accurate and informative fits used to describe the molecular dynamics.
Second, provided conditions for a future experiment, the simulator can identify the quantity and quality of new data required for an informative fit. This is important because if noisy Curves are added to a Group, the fit quality can be compromised. This assessment is performed by creating Groups of original data plus simulated Curves, each with different νCPMG points and signal to noise ratios (e.g., 10 points at 10% noise, 15 points at 10% noise, 15 points at 5% noise). By comparing fits of these new Groups, the investigator can ensure that a sufficient number spectra and scans are acquired, thus avoiding the “noisy new data” pitfall.
Finally, these features are excellent teaching tools for learning basic principles of RD. Simply by editing fields in a table, the user can explore how the Curve reports on Δω, PA, kex, and how it is influenced by B0. Similarly, two-state exchange kinetics can be monitored in response to temperature, ΔH, and EAB. These tools can be used to create custom figures as teaching aids, or can engage a student directly with a “hands-on” approach.
Presentation: massive datasets are easy to visualize for analysis and dissemination
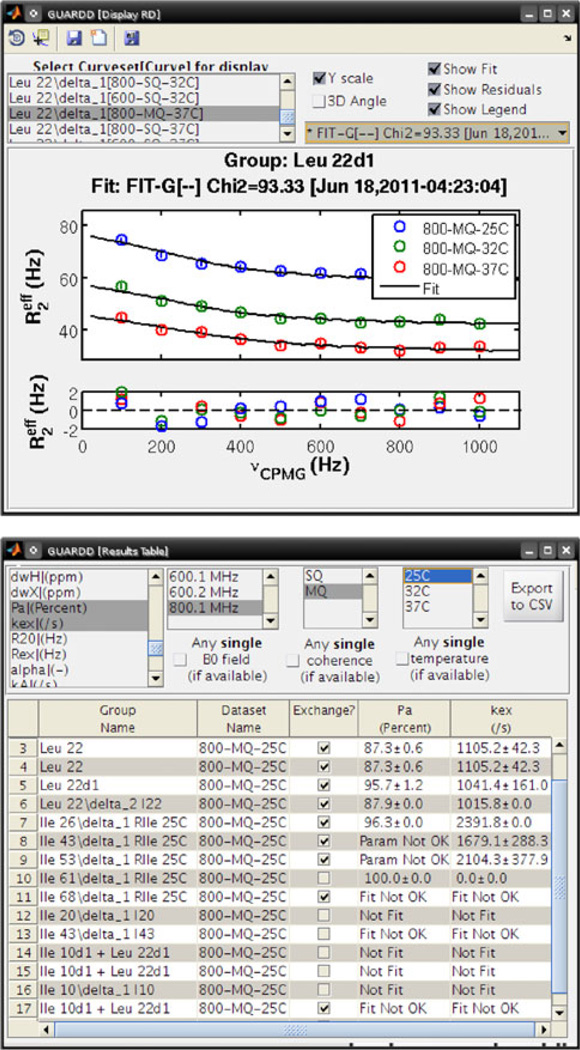
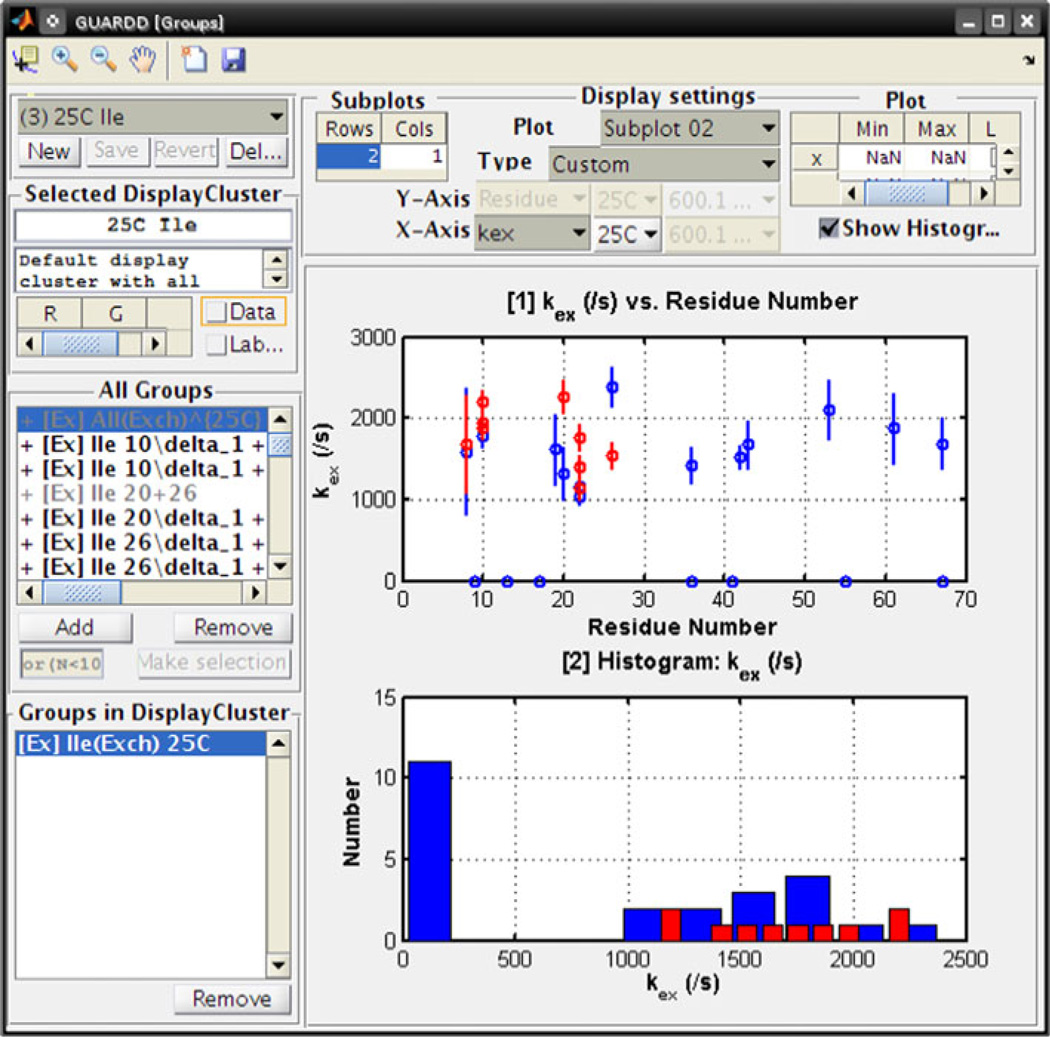
Because analysis of RD data typically yields many results, organization is challenging using a spreadsheet, for example. Hence, GUARDD is designed with versatile presentation features that allow the user to easily explore their results. In the Display RD window (Fig. 7, Top), the user can view the Curves from a Group with any of its candidate fits and residuals. Next, a table of results can be dynamically generated based on the user’s selection criteria from an arbitrary set of RD parameters and Curves (e.g., display only PA and kex at one temperature, instead of all RD parameters for all Curves for all Groups) (Fig. 7, Bottom). This customized table can be exported to plain-text for publication or external analysis. Finally, the Group Display window allows the user to seamlessly prepare scatter plots and histograms of arbitrary RD parameters (Fig. 8). This is helpful for identifying outlying results, comparing candidate fits, and producing publication-quality figures. Moreover, any figures prepared by GUARDD can easily be altered using MATLAB’s figure editor (e.g., adding and moving text, specifying symbols, colors, and font sizes).
Fig. 7.
GUARDD includes powerful features to easily view fitting results. (Top) The Display RD window plots the selected Curves from one Group with a selected fit and its residuals. (Bottom) The Results Table window dynamically generates a table encompassing selected RD parameters from selected subsets of the data (e.g., magnetic field strength, quantum coherence, temperature). This table can be exported to a plain-text file for publication and/or external analysis
Fig. 8.
After fitting, the Group Display window visually organizes results to seek the nature of molecular motions. Here, subsets of Groups can be displayed as scatter-plot correlations between arbitrary RD parameters (e.g., Residue Number, kex, PA, Rex, kA, α, ΔH). To this end, a set of Groups can be color-coded to designate a hypothetical concerted motion (e.g., protein domain A vs. B) or a particular fitting scenario (e.g., all Ile fit together vs. each Ile fit individually). Here, for two sets of Groups (red and blue), the upper subplot displays kex along the protein sequence and the bottom subplot displays kex as a histogram. This can help address the possible presence of multiple dynamic “modes” that each exchange at a distinct rate
Limitations and comparison to alternative software
Although GUARDD inherits desirable features from the MATLAB framework, this dependency imposes certain limitations. First, because MATLAB is an interpreted language, and because its graphical interface uses Java, GUARDD is slower than if it were coded using C or Python, for example. Although this lag is not prohibitive, it can be experienced while drawing the display, while reading or writing large session files, or while fitting data. Interested users may partially alleviate this drawback by using the MATLAB compiler, which generates a single executable that is not subject to runtime interpretation, and/or by programming consecutive fitting tasks to run in parallel. Second, malfunctions in MATLAB may hinder functionality of GUARDD (however, enhancements to MATLAB may imbue enhancements to GUARDD). Third, although MATLAB is a convenient cross-platform solution for dissemination of software, the user must have access to MATLAB and the MATLAB Optimization Toolbox (i.e., GUARDD is not a standalone program).
There are also limitations to the current features of GUARDD. First, the selection of exchange model is restricted to two-state using the all-timescales MQ Carver-Richards-Jones formulation (Korzhnev et al. 2004a). Although these equations are very general, certain cases may benefit from simplifications assuming skewed populations (PB < PA; Ishima and Torchia 1999) or assuming fast-exchange (kex > Δν; Kempf and Loria 2003; Kleckner and Foster 2011; Luz and Meiboom 1963). Also, more advanced labs may feel limited by the lack of analytical options like three-state exchange models (Neudecker et al. 2006), ZQ or DQ coherences (Korzhnev et al. 2005), pressure-dependence (Korzhnev et al. 2006), Anti-TROSY/TROSY analysis (Hansen et al. 2008a), or temperature-dependence using transition state theory (Boehr et al. 2010; Korzhnev et al. 2004b).
Regarding fitting accuracy, we expect GUARDD will match softwares that implement a grid search (Grey et al. 2003; Ishima and Torchia 2005; Kovrigin et al. 2006; Namanja et al. 2010), but will surpass ones that do not (Bieri and Gooley 2011), due to the ruggedness of RD χ2 maps (Ishima and Torchia 2005; Kovrigin et al. 2006) and the effectiveness of the grid search procedure (Bevington and Robinson 2003; Motulsky and Christopoulos 2003). Regarding error estimation, the MC bootstrap procedure used by GUARDD is very effective, but not always the most computationally efficient option. For example, depending on the size of the dataset, the jackknife approach (Choy et al. 2005; Mosteller and Tukey 1968, 1977) or the covariance matrix method (Korzhnev et al. 2004b) may be superior, but GUARDD does not currently implement those options.
As detailed in prior sections, GUARDD is distinguished from other RD software for its user-friendly graphical interface, versatile data management and presentation features, and comprehensive simulator. These qualities are important because they fill a well-defined void in the domain of RD analysis software. Furthermore, because our goal is to enhance the analysis of CPMG RD data, limitations to GUARDD can be addressed in future updates.
Discussion and conclusions
Because CPMG RD is widely used, and because current analytical software options are limited, we expect GUARDD will be of broad interest to the NMR community. Especially for novice spectroscopists, the user-friendly interface, tutorials, organizational features, and RD Simulator will enhance and accelerate both knowledge of RD phenomena and analysis of their data. For experienced spectroscopists, the innovative display features will allow them to navigate results with ease, and the RD Simulator will help them plan future experiments with quantitative precision. Although GUARDD can accomplish important analytical procedures from start to finish, we encourage users to provide feedback, to request additional functionality, and to edit and share the source code, as we continue to serve the needs of RD experimentalists.
Materials and methods
Alternative software for fitting CPMG RD NMR data
Although many labs will supply CPMG RD analysis software upon request, some is already publicly accessible: CPMG Fit (Palmer Lab), http://biochemistry.hs.columbia.edu/labs/palmer/software/cpmgfit.html; CATIA from F. Hansen (Kay Lab), http://pound.med.utoronto.ca/~flemming/catia/; NESSY from M. Bieri (Gooley Lab), http://home.gna.org/nessy/.
Extracting RD data from signal intensity
GUARDD can receive RD Curve data as NMR signal intensities, which is used to calculate relaxation rates , where I(νCPMG) is the signal intensity in the 2D spectrum acquired with refocusing frequency νCPMG, I0 is the reference signal intensity obtained in the spectrum with no refocusing block, and TCPMG is the duration of the refocusing block. Errors in intensities are estimated via standard deviation from repeat measures of I(νCPMG).
Modeling MQ relaxation dispersion
The fitting equations for MQ dispersions, which simplify to SQ dispersions provided ΔωH = 0, are as follows using δ = 1/(4νCPMG) and n = TCPMGνCPMG (Korzhnev et al. 2004a):
| (1a) |
| (1b) |
| (1c) |
| (1d) |
| (1e) |
| (1f) |
| (1g) |
| (1h) |
| (1i) |
| (1j) |
| (1k) |
Exchange broadening
The exchange broadening Rex quantifies the height of the dispersion curve and is estimated from the fit as .
Scaling factor for chemical exchange timescale
The scaling factor α quantifies the timescale of chemical exchange, and is calculated α = d(ln(Rex))/d(ln(Δω)) (Millet et al. 2000).
Temperature-dependence of exchange kinetics
Provided reasonable fitted exchange rates at multiple temperatures, Arrhenius and van’t Hoff analyses are performed (Winzor and Jackson 2006). Arrhenius analysis requires both PA and kex to quantify temperature dependence of exchange rate via where k = kA = (1 − PA)kex (or kB = PAkex) = kinetic rate of exchange from A → B (or B → A), P = PAB (or P = PBA) pre-exponential rate, the exchange rate from A → B (or B → A) at infinite temperature, E = EAB (or E = EBA) = activation energy (≈ enthalpy) required to exchange from A → B (or B → A), R = gas constant, and T = absolute temperature. van’t Hoff analysis only requires PA to quantify temperature dependence of exchange populations via where K = (1 − PA)/PA = kA/kB = equilibrium constant for exchange, ΔS = system entropy change from A → B, ΔH = system enthalpy change from A → B. Errors in Arrhenius and van’t Hoff analyses are estimated from errors from MATLAB’s fit routine (provided data at more than two temperatures), or from propagation of relative error from the fitting variables (when limited to data at only two temperatures).
Kinetic simulator
The two-state kinetic simulator itemizes all kinetic parameters of interest: ΔH, ΔS, EAB, PAB, EBA, PBA, kex(T), PA(T), kA(T), and kB(T) where T is an arbitrary temperature, provided ΔH, EAB, , and T0. By extension of the Arrhenius and van’t Hoff relations above, this is accomplished as follows:
Using ΔH and PA(T0), the van’t Hoff relation yields ΔS
which, with ΔH, determines PA at any temperature via van’t Hoff
Next, using PA and kex at T0 determines kA and kB at T0
and using EAB and kA at T0, the Arrhenius relation yields PAB
which, with EAB, yields kA at any temperature via Arrhenius
Next, knowledge of PA and kA at any temperature yields kex at any temperature
and therefore kB at any temperature kB(T) = kex(T) − kA(T). Knowledge of kB at any temperature yields EBA via the Arrhenius relation and selection of any two temperatures T1 and T2 (e.g., 280 and 320 K)
Finally, using kB(T0) and EBA, the Arrhenius relation yields PBA
Monte Carlo bootstrapping from residuals (Motulsky and Christopoulos 2003)
Residuals are calculated for each observed νCPMG value in a given dispersion Curve as . The set of residuals is used to create a normal distribution N(mean(ε⃗), var(ε⃗)) for the Curve. Next, a synthetic dispersion Curve is created using the best fit at each observed νCPMG value plus a random sample from this normal distribution: . This is repeated for each Curve in the Group such that a synthetic Group is produced. The synthetic Group is fit using initial conditions from the best fit of the actual data. This is repeated 100 times to create 100 synthetic Groups and 100 sets of optimized fit parameters. The error in a given parameter is estimated as the standard deviation of the optimized fit parameter from its 100 element distribution. Errors in subsequent quantities (e.g., kA, kB, ln(kA), etc.) are estimated using propagation of error assuming all parameters are uncorrelated (zero covariance).
Acknowledgments
This work is funded by National Institutes of Health grant R01GM077234. The authors are thankful to investigators who shared their RD data, including Z. Wu, P. Vallurupali, F. Mulder, and D. Korzhnev. The authors are thankful for developmental feedback from all Foster Lab members, as well as E. Kovrigin, and D. Kern. Special thanks to A. Simmons for careful review of this manuscript.
Abbreviations
- NMR
Nuclear magnetic resonance
- RD
Relaxation dispersion
- QC
Quantum coherence
- SQ
Single quantum
- MQ
Multiple quantum
- ZQ
Zero quantum
- DQ
Double quantum
- s
Second
- ms
Millisecond (10−3 s)
- µs
Microsecond (10−6 s)
- Rex
NMR exchange broadening
Effective transverse relaxation rate, as measured by RD NMR
- Δω
Chemical shift difference upon exchange from A state to B state (rad/s)
- Δν
Chemical shift difference upon exchange from A state to B state (Hz)
- PA
Population of A state
- kex
Total exchange rate between A and B states
- kA
Exchange rate from A to B
- kB
Exchange rate from B to A
- ΔH
Enthalpy change from A to B
- EAB
Activation energy upon exchange from A to B
- 6D
Six-dimensional
- MC
Monte Carlo
Contributor Information
Ian R. Kleckner, Biophysics Program, The Ohio State University, 484 West 12th Ave Room 776, Columbus, OH 43210, USA, ian.kleckner@gmail.com
Mark P. Foster, Biochemistry Department, The Ohio State University, 484 West 12th Ave Room 776, Columbus, OH 43210, USA, Foster.281@osu.edu
References
- Beach H, Cole R, Gill ML, Loria JP. Conservation of mus-ms enzyme motions in the apo- and substrate-mimicked state. J Am Chem Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. http://view.ncbi.nlm.nih.gov/pubmed/15969595. [DOI] [PubMed] [Google Scholar]
- Bevington P, Robinson D. Data reduction and error analysis for the physical sciences. Boston, MA: McGraw-Hill; 2003. [Google Scholar]
- Bieri M, Gooley P. Automated NMR relaxation dispersion data analysis using NESSY. BMC Bioinform. 2011;12:421. doi: 10.1186/1471-2105-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. http://view.ncbi.nlm.nih.gov/pubmed/16895318. [DOI] [PubMed] [Google Scholar]
- Boehr DD, McElheny D, Dyson HJ, Wright PE. Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc Natl Acad Sci USA. 2010;107:1373–1378. doi: 10.1073/pnas.0914163107. http://view.ncbi.nlm.nih.gov/pubmed/20080605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd RH, Gilbert JC, Nocedal J. A trust region method based on interior point techniques for nonlinear programming. Math Program. 2000;89:149–185. [Google Scholar]
- Choy W, Zhou Z, Bai Y, Kay LE. An 15N NMR spin relaxation dispersion study of the folding of a pair of engineered mutants of apocytochrome b562. J Am Chem Soc. 2005;127:5066–5072. doi: 10.1021/ja042560u. http://view.ncbi.nlm.nih.gov/pubmed/15810841. [DOI] [PubMed] [Google Scholar]
- Grey MJ, Wang C, Palmer AG., 3rd Disulfide bond isomerization in basic pancreatic trypsin inhibitor: multisite chemical exchange quantified by CPMG relaxation dispersion and chemical shift modeling. J Am Chem Soc. 2003;125:14324–14335. doi: 10.1021/ja0367389. http://view.ncbi.nlm.nih.gov/pubmed/14624581. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B. 2008a;112:5898–5904. doi: 10.1021/jp074793o. http://view.ncbi.nlm.nih.gov/pubmed/18001083. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Lundström P, Neudecker P, Kay LE. Probing chemical shifts of invisible states of proteins with relaxation dispersion NMR spectroscopy: how well can we do? J Am Chem Soc. 2008b;130:2667–2675. doi: 10.1021/ja078337p. http://view.ncbi.nlm.nih.gov/pubmed/18237174. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hübner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. http://view.ncbi.nlm.nih.gov/pubmed/18026086. [DOI] [PubMed] [Google Scholar]
- Ishima R, Torchia DA. Estimating the time scale of chemical exchange of proteins from measurements of transverse relaxation rates in solution. J Biomol NMR. 1999;14:369–372. doi: 10.1023/a:1008324025406. http://view.ncbi.nlm.nih.gov/pubmed/10526408. [DOI] [PubMed] [Google Scholar]
- Ishima R, Torchia DA. Error estimation and global fitting in transverse-relaxation dispersion experiments to determine chemical-exchange parameters. J Biomol NMR. 2005;32:41–54. doi: 10.1007/s10858-005-3593-z. http://view.ncbi.nlm.nih.gov/pubmed/16041482. [DOI] [PubMed] [Google Scholar]
- Ishima R, Torchia DA. Accuracy of optimized chemical-exchange parameters derived by fitting CPMG R2 dispersion profiles when R2(0a) not = R2(0b) J Biomol NMR. 2006;34:209–219. doi: 10.1007/s10858-005-6226-7. http://view.ncbi.nlm.nih.gov/pubmed/16645811. [DOI] [PubMed] [Google Scholar]
- Jee J, Ishima R, Gronenborn AM. Characterization of specific protein association by 15N CPMG relaxation dispersion NMR: the GB1A34F monomer-dimer equilibrium. J Phys Chem B. 2008;112:6008–6012. doi: 10.1021/jp076094h. http://pubs.acs.org/doi/abs/10.1021/jp076094h. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Jr, Hoogstraten CG. Extensive backbone dynamics in the GCAA RNA tetraloop analyzed using 13C NMR spin relaxation and specific isotope labeling. J Am Chem Soc. 2008;130:16757–16769. doi: 10.1021/ja805759z. http://view.ncbi.nlm.nih.gov/pubmed/19049467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf JG, Loria JP. Protein dynamics from solution NMR: theory and applications. Cell Biochem Biophys. 2003;37:187–211. doi: 10.1385/cbb:37:3:187. http://view.ncbi.nlm.nih.gov/pubmed/12625627. [DOI] [PubMed] [Google Scholar]
- Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. http://view.ncbi.nlm.nih.gov/pubmed/21059410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloiber K, Spitzer R, Tollinger M, Konrat R, Kreutz C. Probing RNA dynamics via longitudinal exchange and CPMG relaxation dispersion NMR spectroscopy using a sensitive 13C-methyl label. Nucleic Acids Res. 2011;39:4340–4351. doi: 10.1093/nar/gkq1361. http://view.ncbi.nlm.nih.gov/pubmed/21252295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev DM, Kloiber K, Kay LE. Multiple-quantum relaxation dispersion NMR spectroscopy probing millisecond time-scale dynamics in proteins: theory and application. J Am Chem Soc. 2004a;126:7320–7329. doi: 10.1021/ja049968b. http://view.ncbi.nlm.nih.gov/pubmed/15186169. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004b;430:586–590. doi: 10.1038/nature02655. http://view.ncbi.nlm.nih.gov/pubmed/15282609. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Neudecker P, Mittermaier A, Orekhov VY, Kay LE. Multiple-site exchange in proteins studied with a suite of six NMR relaxation dispersion experiments: an application to the folding of a Fyn SH3 domain mutant. J Am Chem Soc. 2005;127:15602–15611. doi: 10.1021/ja054550e. http://view.ncbi.nlm.nih.gov/pubmed/16262426. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Bezsonova I, Evanics F, Taulier N, Zhou Z, Bai Y, Chalikian TV, Prosser RS, Kay LE. Probing the transition state ensemble of a protein folding reaction by pressure-dependent NMR relaxation dispersion. J Am Chem Soc. 2006;128:5262–5269. doi: 10.1021/ja0601540. http://view.ncbi.nlm.nih.gov/pubmed/16608362. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Religa TL, Lundström P, Fersht AR, Kay LE. The folding pathway of an FF domain: characterization of an on-pathway intermediate state under folding conditions by (15)N, (13)C(alpha) and (13)C-methyl relaxation dispersion and (1)H/(2)H-exchange NMR spectroscopy. J Mol Biol. 2007;372:497–512. doi: 10.1016/j.jmb.2007.06.012. http://view.ncbi.nlm.nih.gov/pubmed/17689561. [DOI] [PubMed] [Google Scholar]
- Kovrigin E. [Accessed 30 June 2011];BiophysicsLab. 2011 http://biophysicslab.net.
- Kovrigin EL, Kempf JG, Grey MJ, Loria JP. Faithful estimation of dynamics parameters from CPMG relaxation dispersion measurements. J Magn Reson (San Diego, Calif.: 1997) 2006;180:93–104. doi: 10.1016/j.jmr.2006.01.010. http://view.ncbi.nlm.nih.gov/pubmed/16458551. [DOI] [PubMed] [Google Scholar]
- Loria JP, Berlow RB, Watt ED. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc Chem Res. 2008;41:214–221. doi: 10.1021/ar700132n. http://view.ncbi.nlm.nih.gov/pubmed/18281945. [DOI] [PubMed] [Google Scholar]
- Luz Z, Meiboom S. Nuclear magnetic resonance study of protolysis of trimethylammonium ion in aqueous solution—order of reaction with respect to solvent. J Chem Phys. 1963;39:366–370. [Google Scholar]
- McConnell HM. Reaction rates by nuclear magnetic resonance. J Chem Phys. 1958;28:430–431. [Google Scholar]
- Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG. The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J Am Chem Soc. 2000;122:2867–2877. [Google Scholar]
- Mosteller F, Tukey J. Data analysis, including statistics. In: Lindzey G, Aronson E, editors. Handbook of social psychology. 2nd edn. vol 2. Reading: Addison-Wesley; 1968. pp. 80–203. [Google Scholar]
- Mosteller F, Tukey JW. Data analysis and regression: a second course in statistics. Reading: Addison Wesley; 1977. [Google Scholar]
- Motulsky HJ, Christopoulos A. Fitting models to biological data using linear and nonlinear regression. A practical guide to curve fitting. San Diego: Graphpad Software Inc; 2003. [Google Scholar]
- Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. http://view.ncbi.nlm.nih.gov/pubmed/11685237. [DOI] [PubMed] [Google Scholar]
- Mulder FAA, Hon B, Mittermaier A, Dahlquist FW, Kay LE. Slow internal dynamics in proteins: application of NMR relaxation dispersion spectroscopy to methyl groups in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2002;124:1443–1451. doi: 10.1021/ja0119806. http://view.ncbi.nlm.nih.gov/pubmed/11841314. [DOI] [PubMed] [Google Scholar]
- Myint W, Ishima R. Chemical exchange effects during refocusing pulses in constant-time CPMG relaxation dispersion experiments. J Biomol NMR. 2009;45:207–216. doi: 10.1007/s10858-009-9344-9. http://view.ncbi.nlm.nih.gov/pubmed/19618276. [DOI] [PubMed] [Google Scholar]
- Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson BD, Wilson KA, Etzkorn FA, Peng JW. Toward flexibility-activity relationships by NMR spectroscopy: dynamics of Pin1 ligands. J Am Chem Soc. 2010;132:5607–5609. doi: 10.1021/ja9096779. http://view.ncbi.nlm.nih.gov/pubmed/20356313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker P, Korzhnev DM, Kay LE. Assessment of the effects of increased relaxation dispersion data on the extraction of 3-site exchange parameters characterizing the unfolding of an SH3 domain. J Biomol NMR. 2006;34:129–135. doi: 10.1007/s10858-006-0001-2. http://view.ncbi.nlm.nih.gov/pubmed/16604422. [DOI] [PubMed] [Google Scholar]
- Palmer AG, 3rd, Grey MJ, Wang C. Solution NMR spin relaxation methods for characterizing chemical exchange in high-molecular-weight systems. Methods Enzymol. 2005;394:430–465. doi: 10.1016/S0076-6879(05)94018-4. http://view.ncbi.nlm.nih.gov/pubmed/15808232. [DOI] [PubMed] [Google Scholar]
- Ren Z, Ghose R. Slow conformational dynamics in the cystoviral RNA-directed RNA polymerase P2: influence of substrate nucleotides and template RNA. Biochemistry. 2011;50:1875–1884. doi: 10.1021/bi101863g. http://view.ncbi.nlm.nih.gov/pubmed/21244027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallurupalli P, Kay LE. Complementarity of ensemble and single-molecule measures of protein motion: a relaxation dispersion NMR study of an enzyme complex. Proc Natl Acad Sci USA. 2006;103:11910–11915. doi: 10.1073/pnas.0602310103. http://view.ncbi.nlm.nih.gov/pubmed/16880391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzor DJ, Jackson CM. Interpretation of the temperature dependence of equilibrium and rate constants. J Mol Recognit JMR. 2006;19:389–407. doi: 10.1002/jmr.799. http://view.ncbi.nlm.nih.gov/pubmed/16897812. [DOI] [PubMed] [Google Scholar]