Abstract
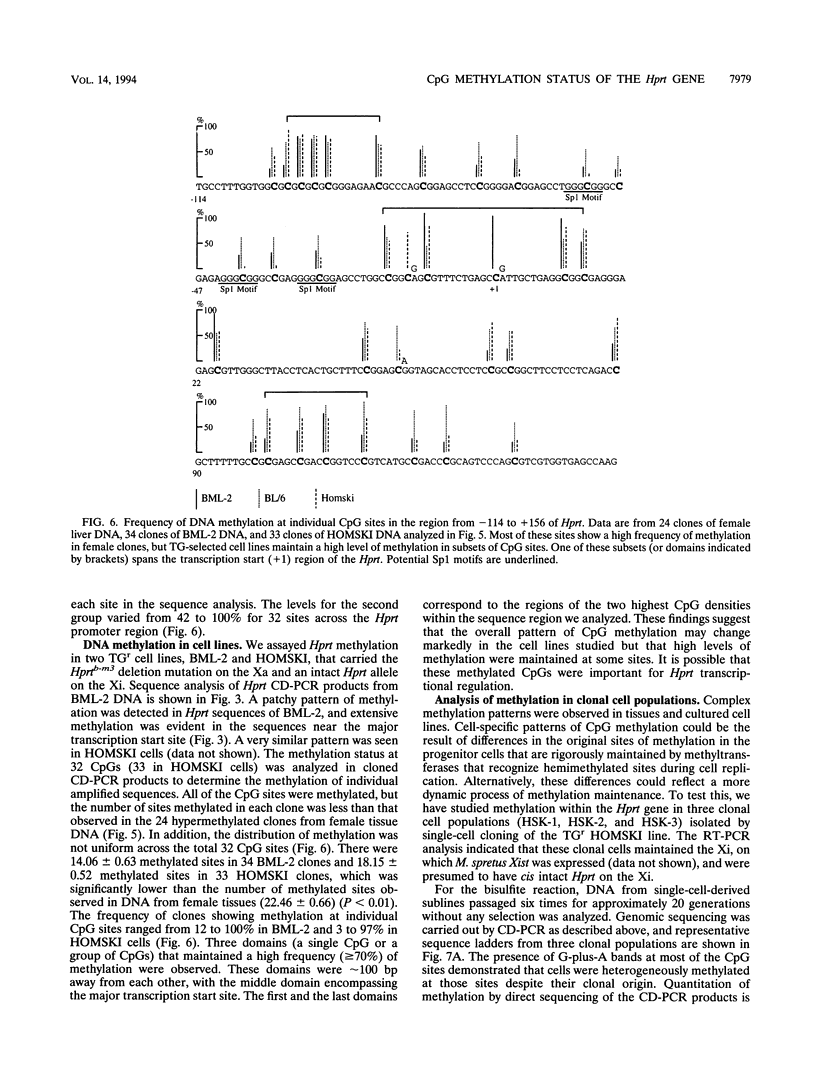
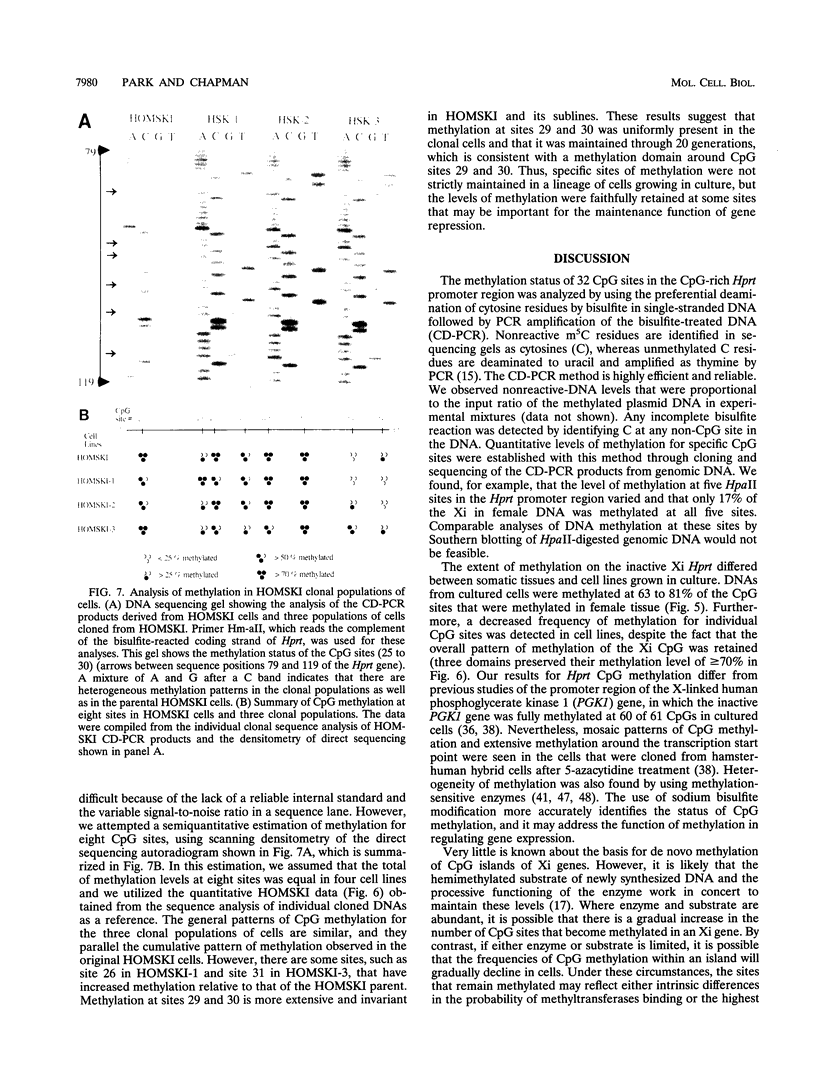
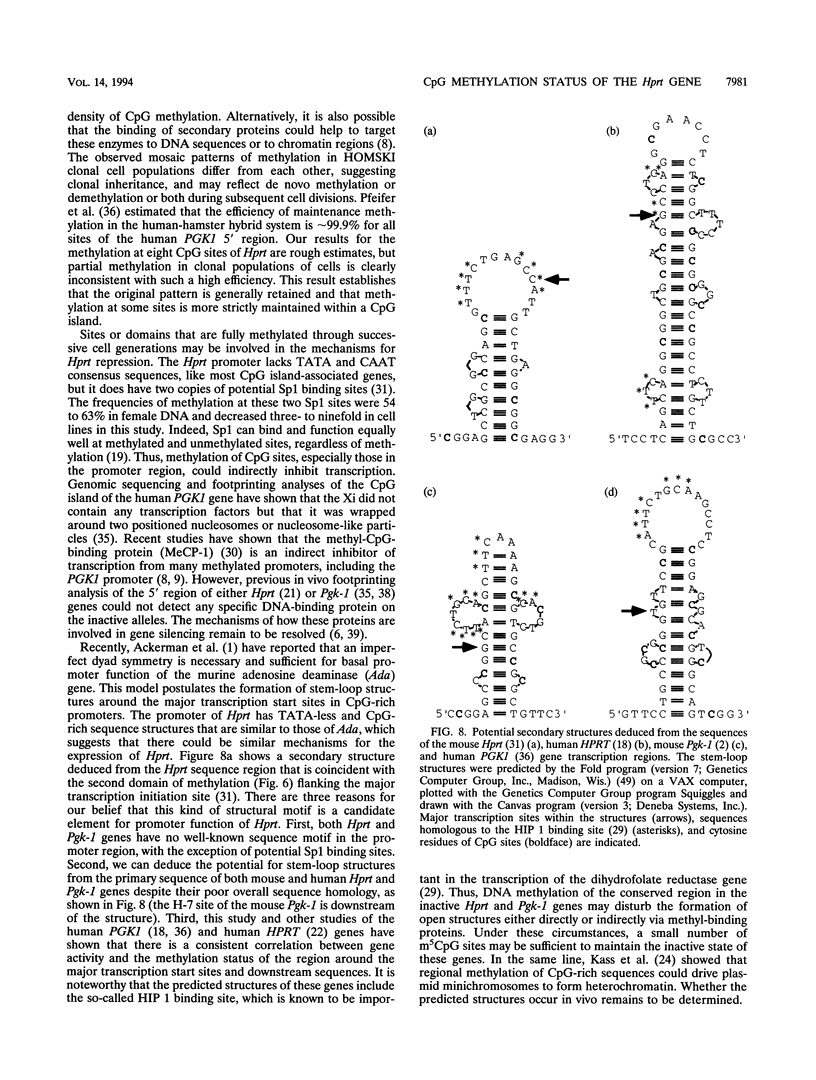
Inactive-X-chromosome genes in mammalian females have methylated CpG islands. We have questioned whether there are variable levels of cytosine methylation at different CpG sites within the island that might indicate the presence of primary sites of methylation which may be critical for the maintenance of gene repression and candidate sites for the initiation of inactivation. To address these questions, we have analyzed the methylation patterns of 32 CpG sites of the X-linked hypoxanthine phosphoribosyltransferase (Hprt) gene on the active and inactive X chromosomes of mouse tissues and cell lines, using genomic sequencing of bisulfite-treated genomic DNA. Cytosine is deaminated by bisulfite, but methylcytosine is not affected. Cell lines that were heterozygous for the Hprt deletion mutation (Hprtb-m3) and a functional Hprt allele were selected with 6-thioguanine. The resulting cell populations uniformly carry the intact Hprt allele on the inactive X chromosome. The methylation of these CpG sites was determined either by the direct sequence analysis of bisulfite-treated and amplified DNA or by the sequence analysis of clones derived from the amplified DNA. No CpG methylation was detected on the active Hprt genes from either males or the active X chromosome of females. On average, 22 CpGs were methylated in the other 50% of female DNA, and the level of methylation at individual sites varied from 42 to 100%. Analysis of the inactive Hprt gene in two cell lines showed that averages of 14 and 18 CpGs were methylated and that the frequency of methylation at 32 individual sites ranged from 3 to 100%. The highest frequency of methylation in cell lines coincided with the sequences flanking transcription initiation sites. These results suggest that methylation patterns are heterogeneous within a tissue and even in clonal cell populations and that specific subsets of CpG sites sustain high methylation frequencies which may be critical for the maintenance of X-chromosome inactivation. The bisulfite method identified which CpG sites were methylated on the inactive X chromosome, and it provided a quantitative estimate of the frequency of methylation of these sites in genomic DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman S. L., Minden A. G., Yeung C. Y. The minimal self-sufficient element in a murine G+C-rich promoter is a large element with imperfect dyad symmetry. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11865–11869. doi: 10.1073/pnas.90.24.11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adra C. N., Boer P. H., McBurney M. W. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60(1):65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- Barlow D. P. Methylation and imprinting: from host defense to gene regulation? Science. 1993 Apr 16;260(5106):309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- Bartlett M. H., Adra C. N., Park J., Chapman V. M., McBurney M. W. DNA methylation of two X chromosome genes in female somatic and embryonal carcinoma cells. Somat Cell Mol Genet. 1991 Jan;17(1):35–47. doi: 10.1007/BF01233203. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Borsani G., Tonlorenzi R., Simmler M. C., Dandolo L., Arnaud D., Capra V., Grompe M., Pizzuti A., Muzny D., Lawrence C. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991 May 23;351(6324):325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992 Jan;11(1):327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M., Kafri T., Ariel M., Chaillet J. R., McCarrey J., Razin A., Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993 Sep;12(9):3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler S. L., Lee K. H., Adler D. A., Chapman V. M., Disteche C. M. Maintenance of X inactivation of the Rps4, Zfx, and Ube1 genes in a mouse in vitro system. Somat Cell Mol Genet. 1993 Jan;19(1):29–37. doi: 10.1007/BF01233952. [DOI] [PubMed] [Google Scholar]
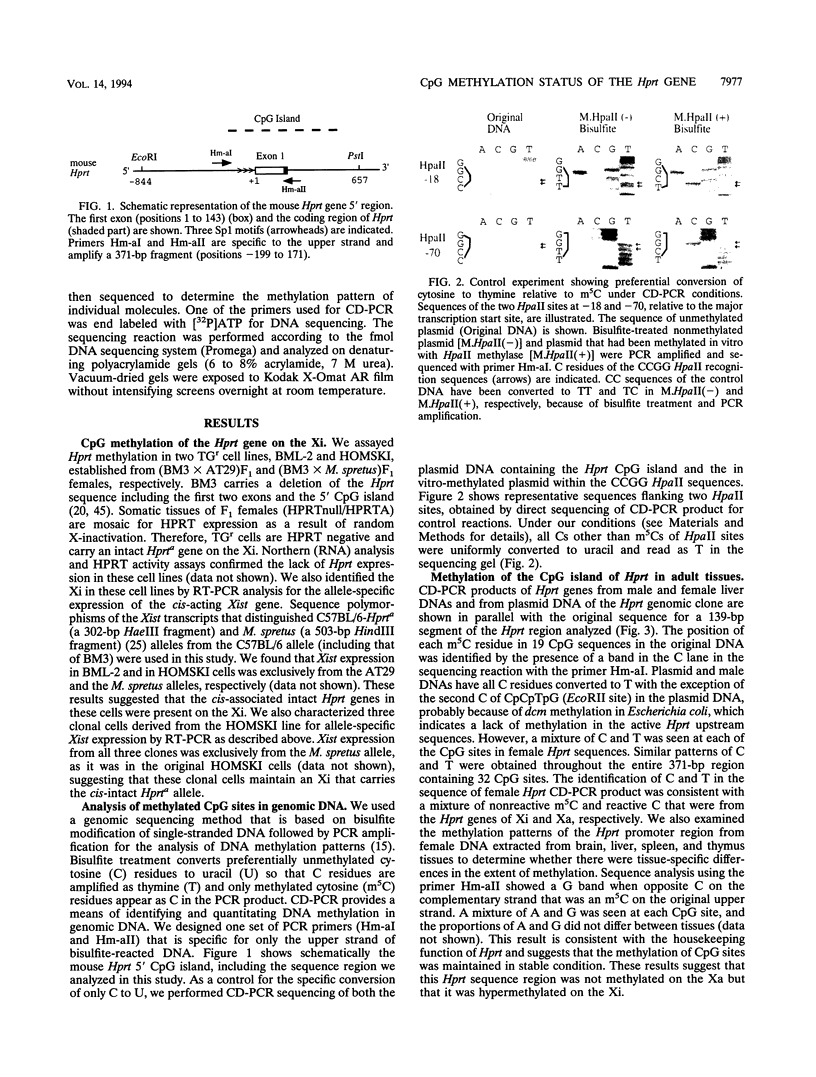
- Chapman V. M., Kratzer P. G., Siracusa L. D., Quarantillo B. A., Evans R., Liskay R. M. Evidence for DNA modification in the maintenance of X-chromosome inactivation of adult mouse tissues. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5357–5361. doi: 10.1073/pnas.79.17.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman V. M., Miller D. R., Armstrong D., Caskey C. T. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1292–1296. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., Keshet I., Shani M., Levine A., Razin A., Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991 May 16;351(6323):239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
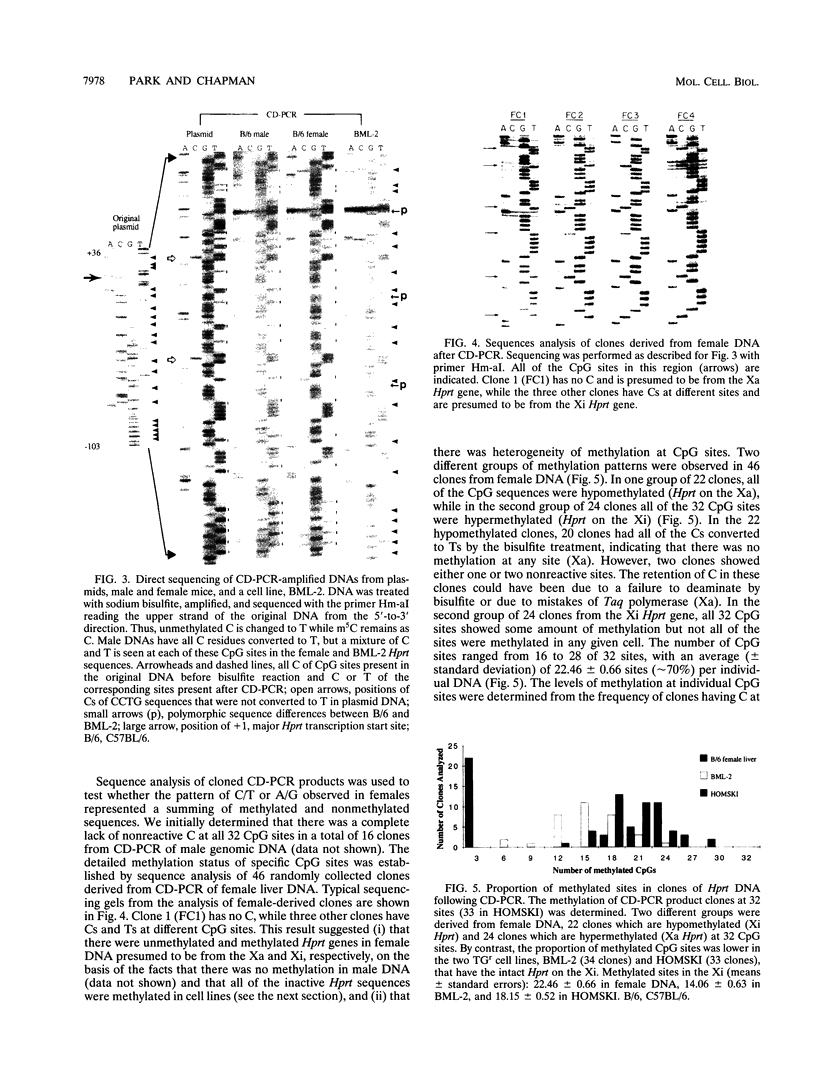
- Grant S. G., Chapman V. M. Mechanisms of X-chromosome regulation. Annu Rev Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Ellis N. A., Gartler S. M. Demethylation of specific sites in the 5' region of the inactive X-linked human phosphoglycerate kinase gene correlates with the appearance of nuclease sensitivity and gene expression. Mol Cell Biol. 1988 Nov;8(11):4692–4699. doi: 10.1128/mcb.8.11.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper M., Hardy K., Handyside A., Hunter S., Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987 Mar 19;326(6110):292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Hornstra I. K., Yang T. P. High-resolution methylation analysis of the human hypoxanthine phosphoribosyltransferase gene 5' region on the active and inactive X chromosomes: correlation with binding sites for transcription factors. Mol Cell Biol. 1994 Feb;14(2):1419–1430. doi: 10.1128/mcb.14.2.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstra I. K., Yang T. P. Multiple in vivo footprints are specific to the active allele of the X-linked human hypoxanthine phosphoribosyltransferase gene 5' region: implications for X chromosome inactivation. Mol Cell Biol. 1992 Dec;12(12):5345–5354. doi: 10.1128/mcb.12.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höller M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988 Sep;2(9):1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Johnson G. G., Larsen T. A., Blakely P., Chapman V. M. Elevated levels of erythrocyte hypoxanthine phosphoribosyltransferase associated with allelic variation of murine Hprt. Biochemistry. 1985 Sep 10;24(19):5083–5089. doi: 10.1021/bi00340a019. [DOI] [PubMed] [Google Scholar]
- Kass S. U., Goddard J. P., Adams R. L. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol. 1993 Dec;13(12):7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay G. F., Penny G. D., Patel D., Ashworth A., Brockdorff N., Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993 Jan 29;72(2):171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- Kratzer P. G., Chapman V. M., Lambert H., Evans R. E., Liskay R. M. Differences in the DNA of the inactive X chromosomes of fetal and extraembryonic tissues of mice. Cell. 1983 May;33(1):37–42. doi: 10.1016/0092-8674(83)90332-x. [DOI] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993 Nov 25;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Lock L. F., Takagi N., Martin G. R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987 Jan 16;48(1):39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Means A. L., Farnham P. J. Transcription initiation from the dihydrofolate reductase promoter is positioned by HIP1 binding at the initiation site. Mol Cell Biol. 1990 Feb;10(2):653–661. doi: 10.1128/mcb.10.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Melton D. W., Konecki D. S., Brennand J., Caskey C. T. Structure, expression, and mutation of the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2147–2151. doi: 10.1073/pnas.81.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas T., Sparkes R. S., Shapiro L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981 Jan 23;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Monk M., Grant M. Preferential X-chromosome inactivation, DNA methylation and imprinting. Dev Suppl. 1990:55–62. [PubMed] [Google Scholar]
- Paterno G. D., Adra C. N., McBurney M. W. X chromosome reactivation in mouse embryonal carcinoma cells. Mol Cell Biol. 1985 Oct;5(10):2705–2712. doi: 10.1128/mcb.5.10.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Riggs A. D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991 Jun;5(6):1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Hansen R. S., Gartler S. M., Riggs A. D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Tanguay R. L., Steigerwald S. D., Riggs A. D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990 Aug;4(8):1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Saluz H. P., Jiricny J., Jost J. P. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J., Ward K., White R. Mosaic methylation in clonal tissue. Dev Biol. 1993 Apr;156(2):391–398. doi: 10.1006/dbio.1993.1086. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J., Grant M., LeBon J. M., Okuyama K., Chapman V., Monk M., Riggs A. D. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990 Sep;10(9):4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöger R., Kubicka P., Liu C. G., Kafri T., Razin A., Cedar H., Barlow D. P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993 Apr 9;73(1):61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., Warren S. T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992 Sep;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Thompson S., Clarke A. R., Pow A. M., Hooper M. L., Melton D. W. Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell. 1989 Jan 27;56(2):313–321. doi: 10.1016/0092-8674(89)90905-7. [DOI] [PubMed] [Google Scholar]
- Viegas-Pequignot E., Dutrillaux B., Thomas G. Inactive X chromosome has the highest concentration of unmethylated Hha I sites. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7657–7660. doi: 10.1073/pnas.85.20.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S. F., Migeon B. R. Studies of X chromosome DNA methylation in normal human cells. Nature. 1982 Feb 25;295(5851):667–671. doi: 10.1038/295667a0. [DOI] [PubMed] [Google Scholar]
- Yen P. H., Mohandas T., Shapiro L. J. Stability of DNA methylation of the human hypoxanthine phosphoribosyltransferase gene. Somat Cell Mol Genet. 1986 Mar;12(2):153–161. doi: 10.1007/BF01560662. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]