Abstract
Background:
High-throughput evaluation of tissue biomarkers in oncology has been greatly accelerated by the widespread use of tissue microarrays (TMAs) and immunohistochemistry. Although TMAs have the potential to facilitate protein expression profiling on a scale to rival experiments of tumour transcriptomes, the bottleneck and imprecision of manually scoring TMAs has impeded progress.
Methods:
We report image analysis algorithms adapted from astronomy for the precise automated analysis of IHC in all subcellular compartments. The power of this technique is demonstrated using over 2000 breast tumours and comparing quantitative automated scores against manual assessment by pathologists.
Results:
All continuous automated scores showed good correlation with their corresponding ordinal manual scores. For oestrogen receptor (ER), the correlation was 0.82, P<0.0001, for BCL2 0.72, P<0.0001 and for HER2 0.62, P<0.0001. Automated scores showed excellent concordance with manual scores for the unsupervised assignment of cases to ‘positive' or ‘negative' categories with agreement rates of up to 96%.
Conclusion:
The adaptation of astronomical algorithms coupled with their application to large annotated study cohorts, constitutes a powerful tool for the realisation of the enormous potential of digital pathology.
Keywords: image analysis, immunohistochemistry, breast cancer, systems pathology, digital pathology
Immunohistochemistry (IHC) is the most widely used method for the assessment of protein expression in tissues in both the clinical and research setting. The advantages of IHC, which include preserved tissue morphology, quick turnaround time and ability to assay small amounts of tissue such as core biopsies, have established it as the principal ancillary study in diagnostic pathology. The coupling of IHC and tissue microarray (TMA) technology has enabled researchers to screen for candidate biomarkers in large study cohorts including clinical trials. However, this process continues to rely heavily on manual assessment of staining resulting in laboriously acquired semi-quantitative readouts of protein expression. In addition, TMAs and IHC have enabled the investigation of high-dimensional relationships between proteins expressed in cancers in a manner analogous to expression profiling using cDNA microarrays (Callagy et al, 2003; Makretsov et al, 2004; Abd El-Rehim et al, 2005; Jacquemier et al, 2005; Ali et al, 2011). However, these efforts are seriously limited by the bottleneck of manually assessing immunostains for tens of proteins across thousands of cases and the pathologist's ability to discriminate between small staining differences on this scale.
Astronomers have long been faced with the problem of automatically deriving objective, reproducible and continuous information from complex telescopic images of the sky. Driven by the large volume of data, image analysis in the field of astronomy has matured into a sophisticated, robust discipline. We therefore investigated the adaptation of algorithms used in astronomy to immunostained microscopic images of breast cancer in order to produce comparable measures of protein expression (Walton et al, 2010). We describe three algorithms developed for oestrogen receptor (ER), B-cell lymphoma protein 2 (BCL2) and human epidermal growth factor receptor 2 (HER2) representing examples of nuclear, cytoplasmic and membranous staining patterns, respectively. Our method includes a technique for dividing the study population into ‘positive' and ‘negative' subgroups in an unsupervised manner. The algorithms were tested in a cohort of over 2000 breast tumours represented in TMAs and compared with manual scores produced by pathologists.
This utilisation of digital pathology results in the production of continuous readouts of protein expression more typical of genomic experiments while retaining tissue morphology. Genomic research has been enormously advanced by the existence of public repositories of gene expression data. In the interest of transparency and in order to encourage continuing development, we have made all TMA images (over 6000 images) and algorithms used in this study available in a public repository. We hope that this resource will act as a hub for the collaborative development of image analysis algorithms by innovative researchers from diverse disciplines.
Materials and methods
Study population
The large population-based breast study SEARCH (studies of epidemiology and risk factors in cancer heredity) was used for this work. This study includes women diagnosed with breast cancer from the East Anglia region. Details of this study have been published previously (Lesueur et al, 2005). IHC data from 2258 patients were included in this study. Characteristics of the study cohort are detailed in Table 1. The SEARCH study is approved by the Cambridgeshire 4 Research Ethics Committee (02/5/42); all study participants provided written informed consent.
Table 1. Characteristics of study cohort.
| Variable | ||
|---|---|---|
| Median age (range) | 51 (24–73) | |
| Median follow-up in years (range) | 9.5 (0.4–18.6) | |
| Number of breast cancer deaths (%) | 384 (17) | |
| 5-year survival (%) |
89 |
|
|
Categories |
Number |
Percent |
|
Grade | ||
| 1 | 460 | 20 |
| 2 | 928 | 41 |
| 3 | 575 | 25 |
| Missing |
295 |
13 |
|
Node status | ||
| Negative | 1230 | 54 |
| Positive | 785 | 35 |
| Missing |
243 |
11 |
|
Tumour size | ||
| <2 cm | 1203 | 53 |
| 2–4.9 cm | 844 | 37 |
| ⩾5 cm | 72 | 3 |
| Missing |
139 |
6 |
|
ER status | ||
| Negative | 438 | 19 |
| Positive | 1331 | 59 |
| Missing |
489 |
22 |
|
BCL2 status | ||
| Negative | 327 | 14 |
| Positive | 1393 | 62 |
| Missing |
538 |
24 |
|
HER2 status | ||
| Negative | 1468 | 65 |
| Positive | 185 | 8 |
| Missing |
605 |
27 |
|
Chemotherapy | ||
| No | 1489 | 66 |
| Yes | 768 | 34 |
| Missing |
1 |
0 |
|
Endocrine therapy | ||
| No | 374 | 17 |
| Yes | 1884 | 83 |
| Missing | 0 | 0 |
Abbreviation: ER=oestrogen receptor.
TMAs, IHC and scoring
TMAs were constructed as previously described (Kononen et al, 1998). One 0.6 mm tissue core was used to represent each tumour. Following dewaxing in xylene and rehydration through graded alcohols, TMA sections were immunostained using a BondMax Autoimmunostainer (Leica, Bucks, UK). Details of antibodies and staining protocols are presented in Table 2. Bound primary antibody was detected using a polymer-conjugated secondary antibody as part of the Bond Polymer detection kit (Leica, Bucks, UK) and signal was developed using 3′-3′-diaminobenzidine (DAB) producing a brown stain. TMA slides were digitised using the Ariol platform (Genetix Ltd, Hampshire, UK) and images were subsequently extracted uncompressed (lossless) as .jpegs for downstream analysis. Scanned TMA images were manually scored by a pathologist using the Ariol user interface and blinded to patient or tumour characteristics; details of scoring systems are shown in Table 2.
Table 2. IHC reagents, protocols and scoring systems.
| Protein | Clone | Clonality | Source | Dilution | Antigen retrieval | Scoring system | Cutoff |
|---|---|---|---|---|---|---|---|
| ER | 6F11/2 | Mouse monoclonal | Novocastra | 1 in 70 | Citrate buffer pH6, 30 min | Allred | >2 |
| HER2 | c-erbB-2 | Humanised monoclonal | Dako | 1 in 250 | Citrate buffer pH6, 40 min | Herceptest | ⩾2* |
| BCL2 | 124 | Mouse monoclonal | Dako | 1 in 200 | Tris-EDTA buffer pH9, 20 min | Modified H-score | >10% |
| MCM2 | 1B10 | Mouse monoclonal | Novocastra | 1 in 25 | Citrate buffer pH6, 20 min | NA | NA |
Abbreviation: ER=oestrogen receptor.
Allred Scoring System: Staining intensity score: 1=weak, 2=moderate, 3=strong; Proportion score: 1=<1%, 2=1–10%, 3=11–33%, 4=34–66%, 5=>66% Total score=Intensity score+proportion score=0–8.
Modified H-score (0–300)=intensity (0–3) × percentage of stained cells.
HercepTest: 0=No staining or weak staining in ⩽10% of cells, 1=weak incomplete membranous staining in >10% of cells, 2=moderate circumferential membranous staining in >10% of cells, 3=strong circumferential membranous staining in >10% of cells.
Adaptation of astronomical algorithms
We first converted stained TMA images into a format compatible with astronomy processing techniques since they are based on positive going fluxes relative to some positive sky background. The flexible image transport system (FITS) (Wells et al, 1981) was used since the uncompressed JPEG colour images are equivalent to three channels (Red Green Blue (RGB)); the conversion extracts the three image planes and inverts the intensities.
Immunostains localising to the membrane (HER2)
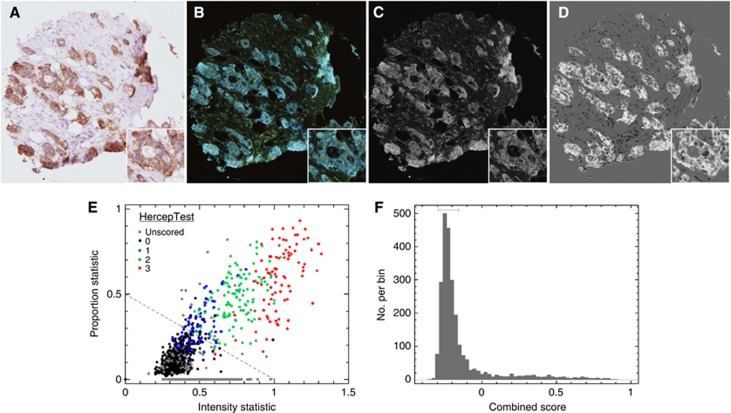
We used a top-level image processing approach consisting of forming a reference image by averaging the R+G channels and using this to form a difference image with respect to the B channels, that is, B−(R+G)/2 (Figure 1C and D). Estimates of the overall background level and random pixel noise in both reference and difference images were made. We used an iteratively clipped median for the level, and the median of the absolute deviations from the median (MAD) as the basis of the noise estimator (Hoaglin et al, 1983). A threshold k-sigma above the overall background was applied, in order to identify all significantly visible regions in the reference image and only those that were significantly stained in the difference image. The automated score was defined by two components: the proportion of pixels picked out in the difference image relative to the reference and the overall intensity (median) of these pixels in the difference image. Figure 1F illustrates how analysis of a histogram of the automated scores can be used to set a ‘blind' threshold for positivity, where the threshold was set at the 95% confidence point that staining was present based on the scatter of the unstained ensemble.
Figure 1.
Astronomical image analysis of membranous (HER2) immunostaining. (A) HER2 stained core scored 2+. (B) Converted to an astro-format with RGB channel intensities inverted such that the brown stained regions become blue regions in emission. (C) Reference image constructed from the average of the inverted red and green channels. (D) The difference image formed by subtracting the reference image in C from the inverted blue channel image. (E) Scatter plot of automated scores for HER2 images using measures of the overall intensity of staining (x-axis) and proportion of image (y-axis) that is stained. Most images unscored by the automated method lie along the proportion=0 boundary. (F) Histogram of the projection of the two-dimensional automated scores onto a one-dimensional continuous grid based on the perpendicular distance of each point from the fixed fiducial dashed line shown in (E).
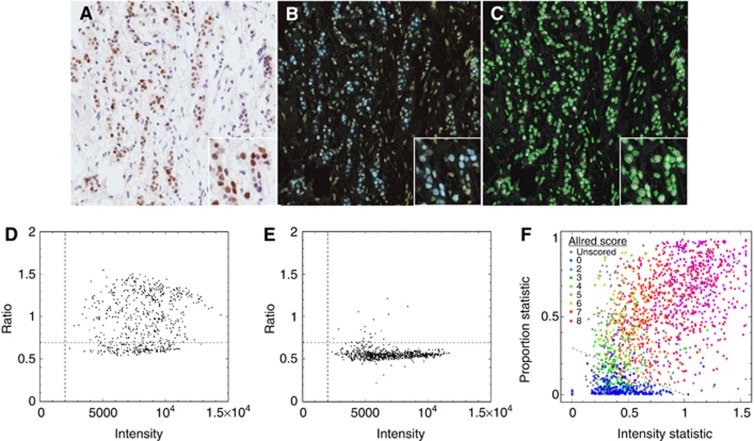
Immunostains localising to the nucleus (ER)
Immunostained tumour nuclei within complex tissue sections showed many similarities to astronomical images where small discrete objects, stars and distant galaxies, are superposed against a varying sky background. In astronomy this image segmentation problem has been well-studied (Irwin, 1985) and is composed of three stages: background estimation and tracking; detailed segmentation, that is, object detection using thresholded pixel connectivity to define objects; and finally object parameterisation, that is, generating measures such as position, shape, and intensity for each object.
A reference image was produced using the average of all three channels, with the intention of maximising overall signal-to-noise. Overall background variation in the reference image was tracked and removed to simplify image segmentation. Regions of contiguous connected pixels above some noise threshold were identified. These included isolated nuclei and clusters of closely packed nuclei, so a further step equivalent to ‘watershedding' (Tuominen et al, 2010) was required to segment individual nuclei (Figure 2C).
Figure 2.
Astronomical image analysis of nuclear (ER) immunostaining. (A) Example image from nuclear ER staining with Allred manual score of intensity 3 and proportion 5. (B) Converted to an astro-format. (C) Automatic segmentation at the nuclear level with each green ellipse denoting a potential nucleus for further scoring. (D) Ratio of blue channel flux (y-axis) to average reference red green flux (x-axis) for each detected nucleus. The horizontal dashed line is automatically determined from the complete set of objects for all cores in a TMA slide by defining a boundary between unstained and stained nuclei. The vertical dashed line defines a signal-to-noise requirement for nuclei to be considered for scoring. (E) The equivalent summary scatter plot for an example image with Allred manual score of intensity=1 and proportion=2; note the well-defined cluster of unstained nuclei. (F) Scatter plot of the results for manually scored ER images colour coded using the Allred score. The final automatic score is defined using the perpendicular distance of each point from the fixed fiducial dashed line.
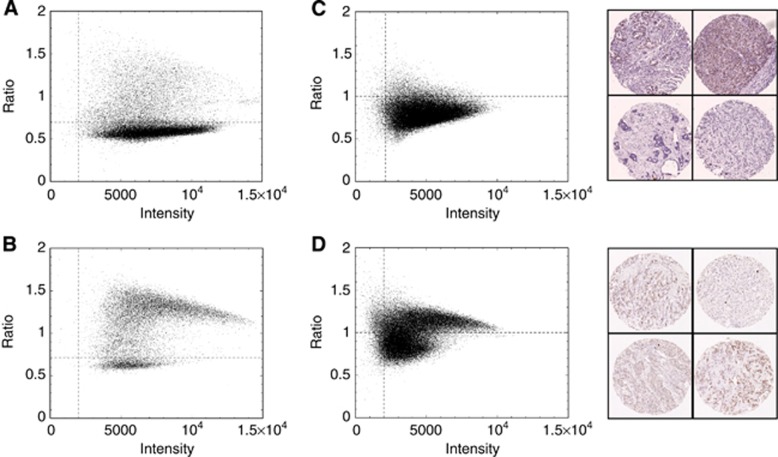
Object descriptors were computed for each nucleus. Figure 2D and E show diagnostic plots where the detected objects satisfied size limits and ellipticity/circularity constraints. The degree of staining for each nucleus (y-axis) was recorded as the ratio of the B channel intensity to the average of the R+G channels. This latter measure is shown on the x-axis revealing subtleties of the variation of the ratio as a function of the overall degree of staining. Figure 2D shows an ER+ example, while Figure 2E illustrates an ER− example. The vertical dashed boundaries are a minimum signal-to-noise limit requirement for inclusion in the final score, while the horizontal dashed boundary denotes the border between stained (above) and unstained (below) nuclei. The histogram of the distribution of automated scores was used to set a ‘blind' threshold for positivitiy. Nuclei that did not satisfy the selection requirements were flagged as ‘unscored'. We tried locating the locus (ratio) of unstained nuclei, and measuring the spread about this locus, to set a boundary independently for every image (tissue core). However, in some cases insufficient nuclei or complete lack of unstained nuclei led to dramatic differences in boundary location between individual tissue cores. Instead, we considered 172 cores of a single TMA slide as an ensemble, defining a single boundary for the set hence evading systematic variation due to small-number statistics. This also yields an overall quality check on the fidelity of the staining of a particular slide. This is illustrated in Figure 3A–D and Supplementary Figure S1. Figure 2F illustrates the distribution of manual Allred scores by the intensity and proportion components of the automated score. The final proportion statistic for each core is defined as the proportion of nuclei lying above the boundary compared with the total number of points on the plot, and the intensity as the ratio of the difference between the median ordinate values of the points above the boundary (stained) compared with below (unstained). This difference is then normalised by the median ordinate value of the unstained points to minimise dependency on image contrast.
Figure 3.
Summary plots of all objects (tumour nuclei) in a TMA slide with example tissue cores. Scatter plots illustrating the distribution of all objects (tumour nuclei) according to staining intensity for whole TMA slides containing 172 tissue cores for ER (A, B). Summary scatter plots for slides stained for MCM2 together with four example tissue cores alongside each plot, from the corresponding slides (C, D).
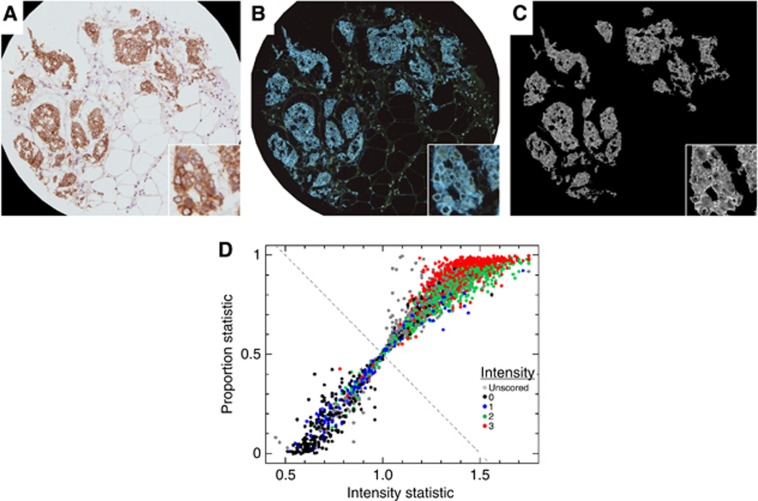
Immunostains localising to the cytoplasm (BCL2)
A hybrid approach based on the top-level fragmentation of an image from the nuclear analysis was chosen, where the segmentation was halted at the level of groups of contiguous connected pixels.
The top-level segmentation was based on a background-corrected reference image composed of the average of the R+G channels, to avoid introducing a bias against unstained regions. Due to the complexity of the shapes involved, segments were retained for analysis based on a size criterion (number of connected contiguous pixels) (Figure 4C). Each remaining pixel was coded with the ratio of the background-corrected B channel flux to the average of the background-corrected R+G channels. This method reduced the impact of varying degrees of contrast, while the background correction reduced the sensitivity to overall background pollution. The final score was based on the proportion of segmented pixels with a flux ratio >1 compared with the total number of segmented pixels and the median value for the ratio of fluxes, labelled as the intensity statistic in Figure 4D. The funnel-like appearance of the scatter plot of automated scores (Figure 4D) arises as a consequence of the method. The neck at coordinates (1.0, 0.5) is a result of using the median ratio as the intensity score, by definition for a proportion of 0.5 the median ratio must be unity. The split between ‘positive' and ‘negative' scores was defined by the ‘neck' point at (1.0, 0.5).
Figure 4.
Astronomical image analysis of cytoplasmic (BCL2) immunostaining. (A) Example of a BCL2 stained image manually scored with intensity 3 and proportion 100%. (B) Image in (A) converted to an astro-format. (C) Automatic segmentation of the reference image, formed from the average of the inverted red and green channels, to pick out large contiguous regions of complex structure. These regions are then coded with the ratio of (inverted) blue channel intensity to the reference intensity level. A summary score for each image, akin to the manual score, is then made based on the proportion of the segmented structures that are stained, and the median intensity ratio of the staining. (D) Scatter plot of automated scores for BCL2. Manually scored BCL2 images are colour coded using the manual intensity scoring. The final automatic score is defined using the perpendicular distance of each point from the fixed fiducial dashed line.
Statistical analyses
Spearman's correlation coefficient was used to assess correlation between continuous automated scores and ordinal manual scores. All automated scores were between −1 and 1 where the 95% confidence point defining the presence of staining was 0. The agreement between automated and manual scores in assigning a ‘positive' or ‘negative' status was assessed using a receiver-operating characteristic (ROC) analysis where the manual score was used as the reference variable, providing a measure of sensitivity, specificity and proportion of cases concordantly classified. Associations with breast cancer-specific survival (BCSS) at 10 years were compared between manual and automated scores using a Cox proportional-hazards model providing a hazard ratio (HR) and 95% confidence interval (95% CI). Known violations of the proportional-hazards assumption (Blows et al, 2010) were accounted for by extending the model to include a coefficient, which was allowed to vary as a function of log time where if the log of the coefficient (T) is <1 hazard falls with time, while if it is >1 hazard increases with time. All statistical analyses were conducted in Intercooled Stata version 11.1 (StataCorp, College Station, TX, USA).
Results
A digital pathology image resource
We used the molecular pathology arm of the large breast study SEARCH for this work (Lesueur et al, 2005; Ali et al, 2011). This is a population based study of women from the east of England with breast cancer. We included 2258 breast tumours and have made digital images for all three markers and reported algorithms freely accessible at: https://www.cri.cam.ac.uk/data/cclab/; username: cclabpub; password: uwzuhq8n.
Objective assessment of signal-to-noise
As part of the nuclear staining analysis, the distribution of all objects (nuclei) for ER was illustrated as a scatter plot according to staining intensity for each TMA slide (Figure 3A and B; Supplementary Figure S1). These plots were inspected in order to identify slides where stained nuclei were not clearly distinguishable from unstained nuclei owing for example, to non-specific staining or excessive counterstain. This in effect provides a visual gauge of signal-to-noise. Although there was considerable variation in signal-to-noise, a population of clearly distinguishable stained objects was identifiable for every TMA slide included in the study; hence, in this instance no slides were excluded on the basis of staining quality. These plots also reflect the overall proportion of stained nuclei. This is illustrated in Figure 3 where plots summarising slides containing substantially different proportions of ER-positive cores as determined by manual scoring, have distinct appearances. The slide summarised in Figure 3A contained 64% ER-positive cores compared with the slide summarised in Figure 3B which contained 79% ER-positive cores. Since the quality of staining was consistently high for ER, we selected TMAs previously stained for the nuclear marker DNA replication licensing factor MCM2 (MCM2) with variable staining quality to demonstrate differences in signal-to-noise detectable by the nuclear algorithm. Figure 3C shows a summary plot with example tissue cores for a TMA slide stained for MCM2 together with examples of tissue cores where an intense counterstain diminishes the signal of positive nuclei. Figure 3D shows a summary plot with example tissue cores for another slide stained for MCM2 with a weak counterstain and background cytoplasmic staining. These plots provide an objective diagnostic of staining quality highlighting slides for further investigation.
Correlation of continuous automated scores with manual ordinal scores
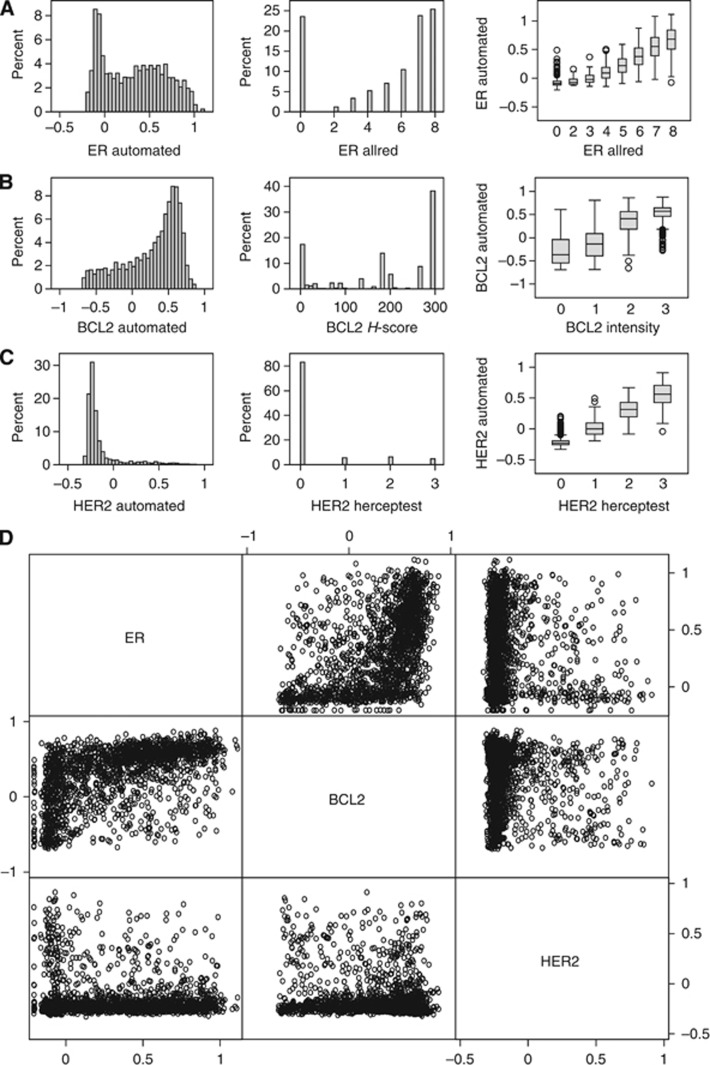
Continuous automated scores and ordinal manual scores were highly correlated. TMAs stained for ER, BCL2 and HER2 had previously been scored by visual inspection of the digital images using standard ordinal scoring systems (Table 2). The distributions of manual and automated scores are illustrated as histograms in Figure 5. Spearman's correlation coefficients for all automated and manual scores are detailed in Table 3. The correlation between the automated score and eight-category Allred ordinal score for ER was the strongest at 0.82, P<0.0001. The histogram of the BCL2 manual H-score shows that although the range of the score is large (0–300) the majority of cases are clustered around the highest and lowest scores while cases with intermediate scores are relatively sparse. This contrasts with the appearance of the histogram for the automated score, which shows a more even distribution of cases through the gradation of staining with a similar cluster of cases at higher scores. This disparity in distribution highlights the ability of automated analysis to distinguish cases with more subtle differences in staining. The BCL2 automated score showed good correlation with the manual modified H-score at 0.73, P<0.0001. Although the distributions of the automated and manual scores for HER2 were the most similar of the three immunostains (Figure 5C), they showed the weakest correlation at 0.64, P<0.0001. This may, in part, be attributable to the relative scarcity of HER2-positive cases (185 cases (11%)). Correlation between automated scores is illustrated as a scatter matrix in Figure 5D. Oestrogen receptor and BCL2 are known to show a strong positive correlation (Dawson et al, 2010). The correlations between the manual and automated scores for ER and BCL2 were very similar at 0.58, P<0.0001 and 0.56, P<0.0001, respectively. Similarly, BCL2 and HER2 showed a negative correlation of −0.24, P<0.0001 by manual scores and −0.009, P<0.0001 by automated scores. Oestrogen receptor and HER2 manual scores were negatively correlated (Spearman's correlation coefficient=−0.19, P<0.0001), but this relationship was not reproduced between the automated scores (Spearman's correlation coefficient=−0.03, P=0.27). However, when restricted to the HER2-positive population as defined by automated analysis, we also find a significant negative correlation with the automated ER score (Spearman's rank correlation −0.27, P<0.0001).
Figure 5.
Distribution of automated and manual scores. Histograms illustrating the distribution of automated scores (left panel), manual scores (centre panel) and boxplots illustrating the distribution of automated scores for each category of the manual score (right panel) for (A) ER, (B) BCL2 and (C) HER2, respectively. (D) Scatter matrix illustrating the relationships between ER, BCL2 and HER2 using automated scores.
Table 3. Correlation between automated and manual scores.
| ER allred | ER automated | BCL2 H-score | BCL2 automated | HER2 Herceptest score | HER2 automated | |
|---|---|---|---|---|---|---|
| ER allred | 1 | |||||
| P-value | ||||||
| ER automated | 0.82 | 1 | ||||
| P-value | <0.0001 | |||||
| BCL2 H-score | 0.58 | 0.54 | 1 | |||
| P-value | <0.0001 | <0.0001 | ||||
| BCL2 automated | 0.46 | 0.56 | 0.73 | 1 | ||
| P-value | <0.0001 | <0.0001 | <0.0001 | |||
| HER2 herceptest score | −0.19 | −0.19 | −0.24 | −0.16 | 1 | |
| P-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| HER2 automated | −0.09 | −0.03 | −0.03 | −0.09 | 0.64 | 1 |
| P-value | 0.7144 | 0.2732 | 0.2621 | 0.0001 | <0.0001 |
Abbreviations: BCL2=B-cell lymphoma protein 2; ER=oestrogen receptor; HER2=human epidermal growth factor receptor 2.
Concordance of dichotomisation for automated vs manual scores
In order to assign patients to ‘positive' or ‘negative' categories using the automated score, the population was divided at the level of the 95% confidence point that there was staining present as defined against the scatter of unstained objects; notably this is an unsupervised method and was not influenced by the dichotomous manual score. There was excellent concordance between the automated and manual scores in assigning cases to ‘positive' and ‘negative' categories. Receiver-operating characteristic analysis is detailed in Table 4. Cross-tabulations of dichotomous scores by marker are shown in Table 5. HER2 showed the best agreement between manual and automated dichotomised scores with 96% of cases classified concordantly at a sensitivity of 98.4% and a specificity of 95.7%. Dichotomisation of automated scores for ER also performed well with 93.2% of cases classified concordantly. The assignment of cases as BCL2+ or BCL2− using the automated method concordantly classified 87.3%. This unsupervised assignment of cases to ‘positive' and ‘negative' categories highlights the potential for our automated analysis to act as an unbiased classifier avoiding many of the pitfalls associated with manual scoring.
Table 4. ROC analysis of dichotomous automated score vs dichotomous manual score.
| Automated score | N | Sensitivity, % | Specificity, % | Concordant classification, % | AUC (95% CI) |
|---|---|---|---|---|---|
| ER | 1664 | 94.4 | 89.4 | 93.2 | 0.92 (0.90–0.94) |
| BCL2 | 1679 | 89.1 | 79.3 | 87.3 | 0.84 (0.82–0.87) |
| HER2 | 1647 | 98.4 | 95.7 | 96.0 | 0.97 (0.96–0.98) |
Abbreviations: AUC=area under curve, BCL2=B-cell lymphoma protein 2; CI=confidence interval; HER2=human epidermal growth factor receptor 2; ER=oestrogen receptor; ROC=receiver-operating characteristic.
Table 5. Cross-tabulation of automated vs manual dichotomous scores.
|
ER manual (%) |
BCL2 manual (%) |
HER2 manual (%) |
||||
|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | |
|
ER automated (%) | ||||||
| Negative | 354 (89) | 71 (6) | ||||
| Positive |
42 (11) |
1197 (94) |
|
|
|
|
|
BCL2 automated (%) | ||||||
| Negative | 238 (79) | 151 (11) | ||||
| Positive |
|
|
62 (21) |
1228 (89) |
|
|
|
HER2 automated (%) | ||||||
| Negative | 1399 (96) | 3 (2) | ||||
| Positive | 63 (4) | 182 (98) | ||||
Abbreviation: ER=oestrogen receptor.
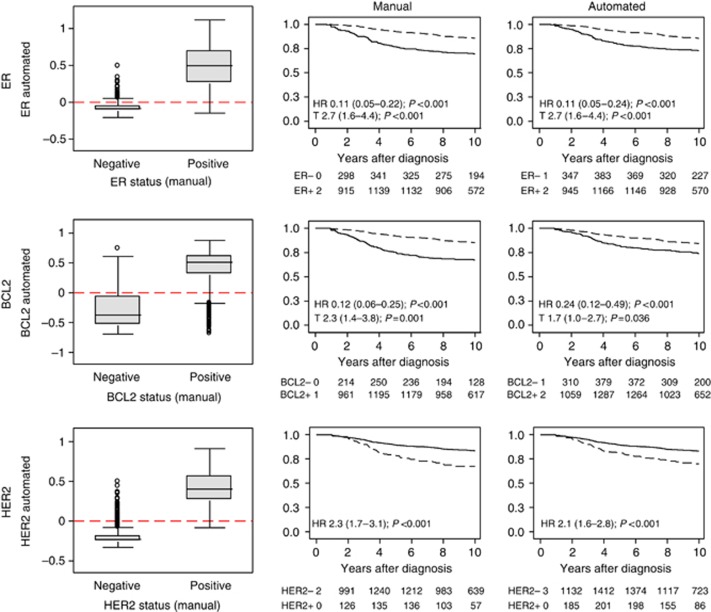
These patterns of concordance between dichotomous manual and automated scores were reflected in estimates of association with BCSS (Table 6; Figure 6). While both ER and HER2 showed near identical estimates between manual and automated scores, estimates for BCL2 manual (HR, 0.12; 95% CI, 0.06–0.25; P<0.001; T, 2.3 (1.4–3.8); P=0.001) and automated (HR, 0.24; 95% CI, 0.12–0.49; P<0.001; T, 1.7; 95% CI, 1.0–2.7; P=0.036) scores were slightly different. This disparity in survival prediction is consistent with observations that the method for analysis of cytoplasmic stains performed least well in terms of concordance with manual scores.
Table 6. Comparison of estimates of association with 10-year BCSS between manual and automated scores.
| Marker | N (events) | HR (95% CI) | P-value | T (95% CI) | P-value |
|---|---|---|---|---|---|
| ER manual | 1663 (249) | 0.11 (0.05–0.22) | <0.001 | 2.7 (1.6–4.4) | <0.001 |
| ER automated | 1663 (249) | 0.11 (0.05–0.24) | <0.001 | 2.7 (1.6–4.4) | <0.001 |
| BCL2 manual | 1678 (246) | 0.12 (0.06–0.25) | <0.001 | 2.3 (1.4–3.8) | 0.001 |
| BCL2 automated | 1678 (246) | 0.24 (0.12–0.49) | <0.001 | 1.7 (1.0–2.7) | 0.036 |
| HER2 manual | 1646 (243) | 2.3 (1.7–3.1) | <0.001 | NA | NA |
| HER2 automated | 1646 (243) | 2.1 (1.6–2.8) | <0.001 | NA | NA |
Abbreviations: CI=confidence interval; HR=hazard ratio; BCSS=breast cancer specific survival.
ER and BCL2 violate the proportional hazards assumption, so the Cox model was fitted in which the natural logarithm of the hazard ratio (β) varies linearly with the natural logarithm of time. Thus, the HR at time t=exp(ln(HR)+t.ln(T)).
Figure 6.
Concordance between automated and manual scores. Boxplots illustrating the distribution of the automated continuous score by manual ‘positive' or ‘negative' category, where the automated score was divided at ‘0' (red dashed line) to generate the equivalent dichotomous score (first panel) for (A) ER, (B) BCL2 and (C) HER2 respectively. Kaplan-Meier survival plots comparing manual (second panel) and automated (third panel) dichotomous scores for (A) ER, (B) BCL2 and (C) HER2, where the solid and dashed lines represent negative and positive cases respectively.
In order to investigate the reasons for discordance of dichotomous scores between automated and manual assessment, discordant cases were reviewed by two pathologists (HRA and BM-A). Each case was re-assigned as ‘positive' or ‘negative' according to a consensus decision and cases were also scored for the number of tumour cells present (more or less than 50 cells), presence of contaminating normal breast epithelium (absent or present) and lymphocytic infiltration (absent, sparse, marked). The results are detailed in Supplementary Table S2. Of the 184 cases stained for ER and discordantly scored between methods, 15 were reassigned following review to concordant categories. Similarly, 21 cases stained for BCL2 were reassigned to concordant categories following review, of 213 originally discordant cases. Notably, a large proportion of cases classified as ‘positive' by the automated method and ‘negative' by manual assessment (63 (48%)) contained an inflammatory infiltrate which is a probable cause of misclassification since B-lymphocytes express BCL2. Review of discordant cases stained for HER2 resulted in the reclassification of four cases to concordant categories of a total of 66 discordant cases. The reasons for discordance between methods for ER and HER2 arise as a result of the different thresholds used for positivity since the cutpoint at which the automated score was dichotomised was not optimised against the manual dichotomous score. For example, of cases classified as ER negative by automated analysis and ER positive by manual assessment, 56 (79%) were attributed an Allred score of 3 or 4 with just 2 (3%) with scores of 7 and 8.
Discussion
The utility of IHC in assaying expression and localisation of proteins in tissues has led to its integration in both cancer research and clinical practice. However the subjective and semi-quantitative nature of IHC continues to limit its utility. For the first time, our approach to the problem of objectively interpreting complex microscopic images takes full advantage of existing robust, validated algorithms in the field of astronomy. We have described methods for the automated analysis of immunostains encompassing all three subcellular compartments. These algorithms produce objective continuous data which is highly correlated with manual scores produced by visual inspection. Moreover, we described an unsupervised method for assigning a cutpoint in order to classify ‘positive' and ‘negative' cases. This method showed excellent concordance with classification according to manual scores and very similar associations with survival.
Methods for the automated analysis of in situ protein expression have been previously described and are commercially available (Camp et al, 2002; Cordon-Cardo et al, 2007; Donovan et al, 2008; Rexhepaj et al, 2008; Turbin et al, 2008; Faratian et al, 2009; Turashvili et al, 2009; Bolton et al, 2010; Tuominen et al, 2010; Brugmann et al, 2012). These methods use different assays and different techniques for image analysis. Quantitative immunofluorescence offers the advantage of a larger dynamic range than IHC, however the detection of protein expression in different subcellular compartments is reliant on the simultaneous detection of a protein known to localise to the compartment of interest (Camp et al, 2002). This can limit the potential flexibility of the assay since multiple reactions are conducted on the same tissue section, necessitating the same antigen retrieval conditions for all proteins of interest and antibodies raised in different species in order to avoid cross-reaction (Camp et al, 2002). Techniques previously described for the automated analysis of IHC have been shown to perform well; however, these tend to be limited to stains localising to the nucleus for which commercial methods have also been shown to produce results concordant with manual scores (Rexhepaj et al, 2008; Turbin et al, 2008; Faratian et al, 2009; Tuominen et al, 2010, 2012). Unlike some other methods, our algorithm accounts for staining variability by adjusting for the differences between stained and unstained nuclei. This adjustment is especially important for cases with a more intense counterstain which can otherwise obscure weakly stained nuclei. In addition, by inspecting plots depicting the scatter of stained and unstained nuclei for each slide (Figure 3A–D; Supplementary Figure S1), our method enables the identification of slides with potentially poor-quality or artefactual staining for further consideration.
The phenomenon of bimodality in manual score distribution has been discussed previously with respect to ER (Rimm et al, 2007; Schnitt, 2006). Here, we corroborate the contention that a bimodal distribution of scores is an artefact of human interpretation of subtly different images rather than a true distribution. The histograms presented in Figure 5 illustrate relatively bimodal distributions for BCL2 and ER staining compared with the automated scores which show a more continuous pattern. This illustrates the potential for automated analysis of IHC to better reflect true differences in protein abundance between tumours, hence facilitating improved outcome prediction.
These methods have some limitations. First, the performance of the methods for each subcellular compartment differed significantly. Overall, the cytoplasmic method performed least well of the three in terms of concordantly classified cases and survival prediction compared to manual methods. This is in large part attributable to the misclassification of BCL2-expressing lymphocytes as tumour cells. This represents an area for on-going development and highlights the need for enduring collaboration. It also demonstrates the extent to which particular phenomena may be stain-specific and the advantage of making methodological adjustments as the need arises. The adaptation of existing astronomical algorithms makes this iterative process more flexible and efficient. Second, our attention has focussed on the use of these high-throughput methods for use with TMAs as part of large translational studies. It is in the context of research that these methods are most likely to make an impact. Their potential clinical utility including application to whole-tissue sections has not been evaluated. Indeed, the proportion of cases discordantly classified is greater than would be acceptable in a clinical context. However, for research purposes these techniques have substantial advantages over manual methods including the provision of quantitative information which may uncover novel associations which ultimately influence clinical practice.
Digital pathology represents an important adjunct to genomic data by enabling us to link data across platforms accounting for the cellular heterogeneity of tumours. The progress of genomic research has been substantially facilitated by the existence of public repositories of genomic data. In the same vein, we have made all TMA images and associated algorithms available for public access. We hope that this resource will enable other researchers to contribute to the development of digital pathology and to learn from our experience thus far.
Conclusion
In summary, we have developed a series of algorithms adapted from astronomy for the automated assessment of immunostains localising to the nucleus, cytoplasm and membrane. We find that automated scores show excellent correlation with scores based on visual inspection and can effectively divide the population into ‘positive' and ‘negative' groups significantly associated with outcome in an unsupervised manner. These methods constitute a high-throughput pipeline for the generation of objective, reproducible and continuous IHC data (Walton et al, 2010). This study takes advantage of a unique digital pathology resource by bringing together the expertise of researchers from diverse disciplines in order to develop a true systems pathology approach to cancer medicine.
Acknowledgments
We are very grateful to the participants of the SEARCH breast study who permitted the use of their tissue for research and to the many individuals who have made this work possible. We acknowledge the SEARCH team, the Eastern Cancer Registration and Information Centre, and the Histopathology Core Facility at the CRUK Cambridge Research Institute for immunohistochemical staining and digital image acquisition.
Author contributions
HRA, MI and CC designed the study. CC, JDB, NW and PDP provided study oversight. MI generated and conducted all image analysis algorithms and contributed to the manuscript. HRA provided histopathological support during algorithm generation, scored TMAs, conducted all statistical analyses comparing methods and wrote the manuscript with CC. LM provided infrastructural and database support. SJD and EP scored TMAs from the SEARCH study. FMB constructed TMAs and managed the clinical database for the SEARCH study. BM-A and HRA conducted histopathological review of cases discordant between automated and manual methods. JDB and NAW conceived of the collaboration and provided strategic advice during its progress. PDP and CC are the project leaders for molecular pathology studies in SEARCH. All authors read and approved the final manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005;116 (3:340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- Ali HR, Dawson SJ, Blows FM, Provenzano E, Pharoah PD, Caldas C. Cancer stem cell markers in breast cancer: pathological, clinical and prognostic significance. Breast Cancer Res. 2011;13 (6:R118. doi: 10.1186/bcr3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blows F, Driver K, Schmidt M, Broeks A, van Leeuwen F, Wesseling J, Cheang M, Gelmon K, Nielsen T, Blomqvist C, Heikkilä P, Heikkinen T, Nevanlinna H, Akslen L, Bégin L, Foulkes W, Couch F, Wang X, Cafourek V, Olson J, Baglietto L, Giles G, Severi G, McLean C, Southey M, Rakha E, Green A, Ellis I, Sherman M, Lissowska J, Anderson W, Cox A, Cross S, Reed M, Provenzano E, Dawson S, Dunning A, Humphreys M, Easton D, García-Closas M, Caldas C, Pharoah P, Huntsman D. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7 (5:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton KL, Garcia-Closas M, Pfeiffer RM, Duggan MA, Howat WJ, Hewitt SM, Yang XR, Cornelison R, Anzick SL, Meltzer P, Davis S, Lenz P, Figueroa JD, Pharoah PD, Sherman ME. Assessment of automated image analysis of breast cancer tissue microarrays for epidemiologic studies. Cancer Epidemiol Biomarkers Prev. 2010;19 (4:992–999. doi: 10.1158/1055-9965.EPI-09-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann A, Eld M, Lelkaitis G, Nielsen S, Grunkin M, Hansen JD, Foged NT, Vyberg M. Digital image analysis of membrane connectivity is a robust measure of HER2 immunostains. Breast Cancer Res Treat. 2012;132 (1:41–49. doi: 10.1007/s10549-011-1514-2. [DOI] [PubMed] [Google Scholar]
- Callagy G, Cattaneo E, Daigo Y, Happerfield L, Bobrow LG, Pharoah PD, Caldas C. Molecular classification of breast carcinomas using tissue microarrays. Diagn Mol Pathol. 2003;12 (1:27–34. doi: 10.1097/00019606-200303000-00004. [DOI] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8 (11:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C, Kotsianti A, Verbel DA, Teverovskiy M, Capodieci P, Hamann S, Jeffers Y, Clayton M, Elkhettabi F, Khan FM, Sapir M, Bayer-Zubek V, Vengrenyuk Y, Fogarsi S, Saidi O, Reuter VE, Scher HI, Kattan MW, Bianco FJ, Wheeler TM, Ayala GE, Scardino PT, Donovan MJ. Improved prediction of prostate cancer recurrence through systems pathology. J Clin Invest. 2007;117 (7:1876–1883. doi: 10.1172/JCI31399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, Le Quesne J, Baglietto L, Severi G, Giles GG, McLean CA, Callagy G, Green AR, Ellis I, Gelmon K, Turashvili G, Leung S, Aparicio S, Huntsman D, Caldas C, Pharoah P. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer. 2010;103 (5:668–675. doi: 10.1038/sj.bjc.6605736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan MJ, Hamann S, Clayton M, Khan FM, Sapir M, Bayer-Zubek V, Fernandez G, Mesa-Tejada R, Teverovskiy M, Reuter VE, Scardino PT, Cordon-Cardo C. Systems pathology approach for the prediction of prostate cancer progression after radical prostatectomy. J Clin Oncol. 2008;26 (24:3923–3929. doi: 10.1200/JCO.2007.15.3155. [DOI] [PubMed] [Google Scholar]
- Faratian D, Kay C, Robson T, Campbell FM, Grant M, Rea D, Bartlett JM. Automated image analysis for high-throughput quantitative detection of ER and PR expression levels in large-scale clinical studies: the TEAM Trial Experience. Histopathology. 2009;55 (5:587–593. doi: 10.1111/j.1365-2559.2009.03419.x. [DOI] [PubMed] [Google Scholar]
- Hoaglin DC, Mosteller F, Tukey JW.1983Understanding Robust and Exploratory Data AnalysisWiley series in probablility and mathematical statistics
- Irwin M. Automatic analysis of crowded fields. MNRAS. 1985;214:575. [Google Scholar]
- Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, Adelaide J, Koki A, Houvenaeghel G, Hassoun J, Maraninchi D, Viens P, Birnbaum D, Bertucci F. Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res. 2005;65 (3:767–779. [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch M, Sauter G, Kallioniemi O. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4 (7:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- Lesueur F, Pharoah P, Laing S, Ahmed S, Jordan C, Smith P, Luben R, Wareham N, Easton D, Dunning A, Ponder B. Allelic association of the human homologue of the mouse modifier Ptprj with breast cancer. Hum Mol Genet. 2005;14 (16:2349–2356. doi: 10.1093/hmg/ddi237. [DOI] [PubMed] [Google Scholar]
- Makretsov NA, Huntsman DG, Nielsen TO, Yorida E, Peacock M, Cheang MC, Dunn SE, Hayes M, van de Rijn M, Bajdik C, Gilks CB. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin Cancer Res. 2004;10 (18 Part 1:6143–6151. doi: 10.1158/1078-0432.CCR-04-0429. [DOI] [PubMed] [Google Scholar]
- Rexhepaj E, Brennan DJ, Holloway P, Kay EW, McCann AH, Landberg G, Duffy MJ, Jirstrom K, Gallagher WM. Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res. 2008;10 (5:R89. doi: 10.1186/bcr2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm DL, Giltnane JM, Moeder C, Harigopal M, Chung GG, Camp RL, Burtness B. Bimodal population or pathologist artifact. J Clin Oncol. 2007;25 (17:2487–2488. doi: 10.1200/JCO.2006.07.7537. [DOI] [PubMed] [Google Scholar]
- Schnitt SJ. Estrogen receptor testing of breast cancer in current clinical practice: what's the question. J Clin Oncol. 2006;24 (12:1797–1799. doi: 10.1200/JCO.2005.05.0666. [DOI] [PubMed] [Google Scholar]
- Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12 (4:R56. doi: 10.1186/bcr2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen VJ, Tolonen TT, Isola J. ImmunoMembrane: a publicly available web application for digital image analysis of HER2 immunohistochemistry. Histopathology. 2012;60 (5:758–767. doi: 10.1111/j.1365-2559.2011.04142.x. [DOI] [PubMed] [Google Scholar]
- Turashvili G, Leung S, Turbin D, Montgomery K, Gilks B, West R, Carrier M, Huntsman D, Aparicio S. Inter-observer reproducibility of HER2 immunohistochemical assessment and concordance with fluorescent in situ hybridization (FISH): pathologist assessment compared to quantitative image analysis. BMC Cancer. 2009;9:165. doi: 10.1186/1471-2407-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbin DA, Leung S, Cheang MC, Kennecke HA, Montgomery KD, McKinney S, Treaba DO, Boyd N, Goldstein LC, Badve S, Gown AM, van de Rijn M, Nielsen TO, Gilks CB, Huntsman DG. Automated quantitative analysis of estrogen receptor expression in breast carcinoma does not differ from expert pathologist scoring: a tissue microarray study of 3,484 cases. Breast Cancer Res Treat. 2008;110 (3:417–426. doi: 10.1007/s10549-007-9736-z. [DOI] [PubMed] [Google Scholar]
- Walton NA, Brenton JD, Caldas C, Irwin MJ, Akram A, Gonzalez-Solares E, Lewis JR, Maccallum PH, Morris LJ, Rixon GT. PathGrid: a service-orientated architecture for microscopy image analysis. Philos Transact A Math Phys Eng Sci. 2010;368 (1925:3937–3952. doi: 10.1098/rsta.2010.0158. [DOI] [PubMed] [Google Scholar]
- Wells DC, Greisen EW, Harten RH. FITS – a flexible image transport system. A&AS. 1981;44:363. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.