Abstract
Introduction
The xenobiotic detoxification system, which protects the human body from external chemicals, comprises drug-metabolizing enzymes and transporters whose expressions are regulated by pregnane X receptor (PXR) and the constitutive androstane receptor (CAR). The progress made in a large number of recent studies calls for a timely review to summarize and highlight these key discoveries.
Areas covered
This review summarizes recent advances in elucidating the roles of PXR and CAR in the xenobiotic detoxification system and highlights the progress in understanding the regulation of PXR and CAR activity at the post-translational levels, as well as the structural basis for the regulation of these two xenobiotic sensors.
Expert opinion
Future efforts are needed to discover novel agonists and antagonists with species and isoform selectivity, to systematically understand the regulation of PXR and CAR at multiple levels (transcriptional, post-transcriptional, and post-translational levels) in response to xenobiotics exposure, and to solve the structures of the full-length receptors, which will be enabled by improved protein expression and purification techniques and approaches. In addition, more efforts will be needed to validate PXR and CAR as disease-related therapeutic targets and thus expand their roles as master xenobiotic sensors.
Keywords: constitutive androstane receptor, drug-drug interactions, pregnane X receptor, xenobiotic detoxification
1. Introduction
In response to the continuous exposure to external chemicals (also known as xenobiotics), the human body has evolved a xenobiotic detoxification system to protect itself from the possible toxicity of these foreign chemicals. The xenobiotic detoxification system is mainly composed of drug-metabolizing enzymes (DMEs) and transporters. Several phases are involved in the process of xenobiotic detoxification and elimination. Phase I cytochromes P450 (CYPs) are enzymes belonging to the monooxygenase superfamily and are highly expressed in the liver and intestine and mainly function to catalyze the first step of detoxification of toxic compounds. By hydroxylation or oxidation reactions, CYPs can convert target compounds into more soluble derivatives that are suitable for excreting from the body [1-3]. Phase II conjugation reactions are catalyzed by a large group of transferases, such as sulfotransferase (SULT), glutathione S-transferase (GST), and UDP-glucuronosyltransferase (UGT) [4], which conjugate polar functional groups onto xenobiotics and endobiotics (i.e., endogenous compounds) to produce water-soluble, inactive metabolites suitable for biliary and urinary excretion [4]. The excretion process is regulated by members of ATP-binding cassette (ABC) transporter family and solute carrier family [5, 6]. These transporters, together with phase I and II enzymes, orchestrate the xenobiotic detoxification process.
The pregnane X receptor (PXR) and the constitutive androstane receptor (CAR), both belong to the nuclear receptor (NR) superfamily, are well-established xenobiotic sensors capable of binding to various structurally diverse chemicals, a characteristic referred to as “ligand promiscuity.” Another ligand-activated transcription factor aryl hydrocarbon receptor (AhR), which is not a member of the NR superfamily, also binds a broad spectrum of xenobiotics, including halogenated aromatic hydrocarbons and polycyclic aromatic hydrocarbons. The roles of AhR in xenobiotic detoxification has been receviewed recently, and will not be discussed here [7]. PXR and CAR are predominantly expressed in the liver. While CAR has little expression in other organs, extrahepatic expression of PXR, such as that in gastrointestinal tract, has been reported [8, 9]. In the absence of xenobiotics, the basal expression level of proteins responsible for xenobiotic detoxification and elimination is low. Upon ligand binding, PXR and CAR are activated to control the expression of a large number of proteins involved in the biotransformation and homeostasis of both exogenous and endogenous substances, including DMEs and transporters, thereby detoxifying xenobiotics. This review summarizes recent advances in elucidating the roles of PXR and CAR in regulating xenobiotic detoxification and highlights the efforts made to understand the regulation of PXR and CAR activity by cellular signaling pathways, as well as the structural basis for the regulation of the receptors.
2. PXR and CAR in Xenobiotic Detoxification
2.1 Species Differences of PXR and CAR in Xenobiotic Detoxification
One critical problem in studying the function of PXR and CAR lies in the ligand selectivity between humans and mice because of the marked species differences in amino acid sequences in their ligand-binding domains (LBDs) [10]. For example, rifampicin and SR12813 are potent agonists for human PXR (hPXR) but not for mouse PXR (mPXR), whereas the potent mPXR agonist 5-pregnen-3β-ol-20-one-16α-carbonitrile (PCN) is a poor agonist for hPXR [9]. On the other hand, 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime (CITCO) is a strong agonist for human CAR (hCAR) but not mouse CAR (mCAR) [11], while 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene,3,3′,5,5′-tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP) is more selective for mCAR than hCAR [12, 13]. To overcome the species differences in ligand recognition and to facilitate functional studies of human receptors in animal models, several humanized mouse models for PXR and CAR have been generated. In the first generation of hPXR or hCAR humanized mouse models, the hPXR or hCAR cDNA was controlled by either the liver-specific albumin promoter [14, 15] or the rat fatty acid–binding protein promoter [16] randomly integrated into the mouse genome where the mPXR or mCAR was deleted. The second-generation hPXR humanized mouse model was developed by using a genomic fragment containing the entire hPXR gene and its promoter randomly integrated into the mouse genome with a Pxr-null background [17]. The latest hPXR or hCAR humanized mouse model was generated through knockin strategies by expressing hPXR or hCAR under the control of its corresponding mouse promoter and a simultaneous interruption of the expression of the corresponding endogenous gene [18]. In the latest humanized mouse model, hPXR or hCAR was expressed in the predicted tissues at physiological levels. In addition, the splice variants of both human receptors were expressed [18]. These new models provided useful means to dissect the species differences of PXR and CAR and predict their roles in xenobiotic detoxification in humans. Noting the significant species differences, the following sections of this review will focus on the function of hPXR and hCAR in the human detoxification system.
2.2 Overlapping Sets of Genes Regulated by PXR and CAR in Xenobiotic Detoxification
PXR and CAR regulate overlapping sets of genes encoding phase I and II DMEs and transporters for xenobiotic detoxification and elimination (Figure 1) and can also be activated by the same sets of compounds, such as phenobarbital and TCPOBOP [3]. Upon activation, both PXR and CAR bind to the promoter of its target gene as a heterodimer with retinoid X receptor (RXR). PXR and CAR regulate gene expression through the PXR responsive element (PXRRE) and the phenobarbital responsive element module (PBREM), respectively, in the promoter regions of target genes [18]. These responsive elements are composed of various repeats of the consensus motif, AG(G/T)TCA, including direct repeats (DR) separated by 3–5 nucleotides (DR3, DR4, or DR5), everted repeats (ER) separated by 6 or 8 nucleotides (ER6 or ER8), inverted repeats (IR) separated by 6 or no nucleotides (IR6 or IR0), or a novel motif named DR-(5n + 4) recently identified through the analysis of the PXR cistrome [19].
Figure 1.
Overlapping and distinctive sets of genes regulated by PXR and CAR in xenobiotic detoxification. Upon activation by xenobiotics, PXR and CAR regulate gene expression through PXR responsive element (PXRRE) and the phenobarbital responsive element module (PBREM) in the promoter regions of target genes encoding phase I and II drug-metabolizing enzymes and drug transporters. “?” indicates genes predicted to be regulated by hPXR and hCAR. “*” indicates genes indentified from rodent experiments that need further validation in human system.
Both hPXR and hCAR can regulate the expression of phase I CYPs, including CYP2B6 [20, 21], CYP2C9 [22, 23], CYP2C19 [24, 25], CYP3A4 [26-28], and CYP3A5 [29]. However, the contribution of CAR and PXR to the regulation of CYPs is different because of the differential preference of binding to different AG(G/T)TCA repeats. The regulation of CYP2B6 and CYP3A4 has been well studied to elucidate the overlapping but differential roles played by PXR and CAR. Whereas PXR strongly binds to the DR4, DR3, or ER6 motif in both CYP2B6 and CYP3A4 promoters [30, 31], CAR only weakly binds to the CYP3A4 proximal ER6 motif but strongly binds to the imperfect DR4 motif in the PBREM of the CYP2B6 gene [31, 32], leading to CAR’s selectivity for CYP2B6 over CYP3A4 in humans. Accordingly, a model highlighting the asymmetrical cross-regulation of CYP2B6 and CYP3A4 by hCAR has been proposed [30, 31, 33]. Genes whose expression is dually regulated by both PXR and CAR continue to be identified. Most recently, the expression of DHCR24, a gene encoding human 24-dehydrocholesterol reductase that catalyzes the last step of cholesterol biosynthesis, was identified as a target gene dually regulated by both hPXR and hCAR through a DR4 motif in its distal promoter region [34].
UGT1A1 is the only phase II enzyme whose expression was reported to be dually regulated by hPXR and hCAR. UGT1A1 was initially identified as a CAR target gene when CAR is activated by phenobarbital [35]. Subsequent studies using a humanized PXR mouse model and human cell lines further indicated that hPXR also regulates UGT1A1 expression [25, 36]. GST, another phase II conjugation enzyme, can be induced modestly by rifampicin [37], as well as by phenobarbital [38] and TCPOBOP [39], suggesting that PXR and CAR might regulate the expression of GSTs. Murine GST genes such as Gsta1, Gstm1, and Gstm2 have been shown to be regulated by both mPXR and mCAR [16, 40]. A recent bioinformatic analysis indicated that human GSTM1 and MGST2 contain an ER6 motif, suggesting these two human GST genes might be regulated by hPXR and hCAR [41].
PXR and CAR also regulate the expression of human drug transporters, such as multidrug resistance protein 1 [MDR1; also known as P-glycoprotein (P-gp) or ABCB1], and multi-drug resistance protein 2 (MRP2; also known as ABCC2). Earlier studies showed that agonists of PXR or CAR, such as rifampicin or phenobarbital, can upregulate MDR1 expression in human liver and colon cells [42-44]. A recent study showed that the regulation of MDR1 expression by PXR and CAR also occurs at the human blood–brain barrier [45]. The DR4 motif in the human MDR1 promoter is responsible for its transcriptional regulation by PXR and CAR [46]. Similarly, rifampicin and phenobarbital, the agonists for PXR and CAR, respectively, induce increased mRNA levels of MRP2 in human small intestine cells and human primary hepatocytes [43, 44, 47, 48]. The transcriptional regulation of MRP2 by PXR and CAR is mediated through an ER8 motif in the 5′-flanking region of the MRP2 gene [48]. Other human transporters, such as bile salt export pump and sodium taurocholate cotransporting polypeptide, are also reported to be upregulated by rifampicin and phenobarbital [44], suggesting possible roles of PXR and CAR in regulating the expression of these transporters..
2.3 Distinctive Sets of Genes Regulated by PXR and CAR in Xenobiotic Detoxification
PXR and CAR also regulate distinctive sets of genes (Figure 1). For phase I CYPs, the genes encoding CYP7A1 and CYP4F12 are regulated by PXR. Early studies with a PXR knockout mouse model identified PXR’s contribution to the detoxification process of bile acids by controlling the expression of CYP7A1 [49], a rate-limiting enzyme in bile-acid biosynthesis [50]. Subsequent studies with human hepatocytes confirmed the negative regulation of CYP7A1 expression by PXR. The negative regulation of CYP7A1 gene expression by PXR is achieved by disrupting the interaction between hepatic nuclear factor 4α (HNF4α; also known as NR2A1) and peroxisome proliferator-activated receptor gamma coactivator (PGC-1), which is required for the activation of CYP7A1 gene expression. Rifampicin is required for PXR to interact with HNF4α and PGC-1, leading to reduced interaction between HNF4α and PGC-1 and reduced gene expression of CYP7A1 [51]. CYP4F12 is an enzyme mainly responsible for the metabolism of antihistaminic compounds [52]. A recent study found that PXR regulates the expression of CYP4F12 by binding to the PXRRE in the CYP4F12 gene in a rifampicin-inducible manner in human hepatocytes [53].
The phase I CYPs regulated by CAR are CYP1A1 and CYP1A2, whose expression is increased by phenobarbital treatment. A recent study showed that CAR induces the expression of CYP1A1 and CYP1A2 in human hepatocytes through an ER8 motif in their proximal promoter sequences [54] .
For phase II enzymes, rifampicin-activated PXR was reported to suppress SULT2A1 expression by interfering with the function of HNF4α in human HepG2 cells [55]. A recent study showed that the expression of UGT1A6 was induced with rifampicin treatment in human liver and intestinal precision-cut slices [44, 56]. Whether SULT2A1 and UGT1A6 are direct PXR target genes remains to be validated. Mouse CAR has been shown to regulate the expression of multiple murine UGTs and SULTs [57]. However, whether hCAR can regulate a similar set of genes for phase II enzymes in humans remains unclear, and further evaluation is needed.
Organic anion-transporting polypeptide 1A2 (OATP1A2), a transporter responsible for the uptake of a broad spectrum of substrates [58], was recently identified as a PXR target gene in human breast tissue and cancer [59]. A DR4 motif located 5.7 kb upstream of the transcription initiation site in the human OATP1A2 promoter was identified and confirmed to be a PXR binding site by using chromatin immunoprecipitation. Although activated CAR was found to increase OATP1A2 promoter activity in HepG2 cells, CAR failed to induce OATP1A2 expression in breast cancer cells [59], suggesting that the OATP1A2 is transcriptional target of PXR but not CAR in breast cancers, and tissue selectivity may play a role here. Interestingly, phenobarbital increased but rifampicin decreased the expression of OATP1B3 in human liver cells [59]. Whether OATP1B3 is a CAR target gene has yet to be elucidated.
2.4 PXR and CAR in Drug-Drug Interactions
PXR and CAR regulate gene expression and play major roles in the detoxification of xenobiotics. However, the fact that PXR and CAR induce the expression of CYP3A4, 2B6, 2C9, and 2C19, which are responsible for metabolizing more than 80% of prescription drugs, suggests that the activation of PXR and CAR can lead to undesirable drug-drug interactions (DDIs) [60] [61], cause adverse drug reactions, and contribute to clinical mortality and morbidity.
PXR and CAR contribute to DDIs by increasing drug toxicity and by decreasing therapeutic efficacy. For example, a dual PXR and CAR activator, the antiepileptic drug phenobarbital, can cause acetaminophen-induced hepatotoxicity in a mouse model [15]. Another study showed that pretreatment with the mPXR activator PCN significantly enhanced acetaminophen-induced hepatotoxicity in mice [62]. The PXR- and CAR-mediated hepatotoxicity was linked to the induction of CYP3A and the resulting conversion of acetaminophen to its toxic intermediate metabolite, N-acetyl-p-benzoquinoneimine. Activation of PXR and CAR may also lead to decreased drug efficacy. Phenytoin and carbamazepine, two common antiepileptic drugs, are well-characterized human CAR activators [63, 64]. Combined administration of phenytoin and carbamazepine with other drugs that are metabolized by the CAR-induced CYPs can decrease the drug efficacy or potentiate the toxic effects of these drugs in many clinical applications [65, 66]. Rifampicin and hyperforin, an active component in St. John’s wort, are strong agonists of hPXR. Numerous studies have shown that rifampicin and hyperforin can induce CYP3A expression, resulting in increased clearance and reduced efficacy of many therapeutic drugs during combination therapy, such as anti-HIV protease inhibitors, antihypertensive drugs, and cholesterol-lowering drugs [67, 68]. Furthermore, since PXR can be activated by many anticancer drugs, recent studies support the notion that activation of PXR may compromise the effectiveness of anticancer drugs and contribute to acquired resistance and multi-drug resistance during anticancer chemotherapy [69].
Significant efforts have been made to tackle the PXR- and CAR-mediated DDIs in drug development and clinical therapy. One approach is to identify the potential of lead compounds to activate PXR or CAR during early drug development and chemically modify the drug candidates to minimize their activation of PXR and CAR without compromising their target activity. One such example was shown in a recent study in which the strong PXR transactivation of first-generation IGF-1R inhibitors was minimized by chemical modifications without compromising their IGF-1R inhibition [70]. However, many marketed drugs with PXR and CAR agonistic activity are still in clinical use. In addition, due to the receptors’ ligand promiscuity, minimizing the lead compounds’ potential to activate PXR and CAR requires tremendous chemistry efforts. Such chemical efforts might also alter the pharmacologic profiles of the lead compounds. In light of these considerations, an alternative approach is to discover and develop novel compounds that function as antagonists of PXR or CAR and use them as co-therapy to minimize drug-induced PXR or CAR activation [69, 71, 72]. Recent studies showed that the effectiveness of drugs can be enhanced by antagonizing the inducible activity of PXR with several pharmacologic interventions, such as ecteinascidin-743 [73], ketoconazole [74], FLB-12 [72], sulforaphane [75], A-792611 [76], and coumestrol [77]. Several CAR antagonists or inverse agonists have also been discovered, including meclizine [78], clotrimazole [12], and PK11195 [79]. However, a recent study indicated that meclizine is an hPXR agonist but not an hCAR antagonist or inverse agonist in human hepatocytes [80]. So far, compounds with dual antagonistic effect against both PXR and CAR have not been reported.
3. Regulation of PXR and CAR Activity
3.1 Subcellular Localization and Protein-Protein Interaction
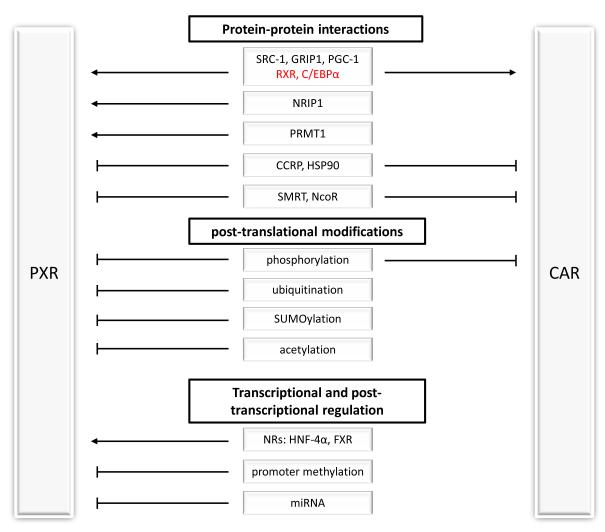
The subcellular localization of PXR to date has been unclear because of the discrepant observation between human and mouse systems. Human PXR appears to be mainly localized in the nucleus, unlike steroid receptors, whose activation involves ligand binding and subsequent cytosol-to-nucleus translocation [81]. As such, PXR is believed to act as a gene silencer in resting states through binding with corepressors such as the silencing mediator of retinoid and thyroid receptors (SMRT) and the nuclear receptor corepressor (NcoR) (Figure 2). These co-repressors can recruit histone deacetylases, thereby stabilizing chromatin and repressing transcription. Ligand binding to PXR in turn results in the dissociation of corepressors and the subsequent association of coactivators, such as steroid receptor coactivator-1 (SRC-1, or NCOA1), SRC-2 (also known as GRIP1), nuclear receptor interacting protein 1 (NRIP1), PGC-1, and forkhead transcription receptor (FHKR/FOXO1) [82]. Structure-based analysis has shown that coactivator binding further promotes the interaction between PXR and its ligand [83]. Recently, PXR was shown to interact with protein arginine methyltransferase 1 (PRMT1), an interaction required for PXR to exert its transcriptional activity [84]. The ligand-induced interaction between PXR and PRMT1 is associated with the concomitant methylation of arginine 3 of histone H4 [84].
Figure 2.
The activity of PXR and CAR is regulated by multiple cellular modulators and cellular processes at multiple levels. Standard arrows indicate an activating effect, and blunt arrows indicate an inhibitory effect.
On the other hand, in vivo studies involving mPXR show a different PXR subcellular localization pattern. Ligand activation of mPXR indeed shows cytoplasmic-to-nuclear translocation in this in vivo system. Mouse PXR was reported to exhibit cytoplasmic localization in resting mouse liver, and upon treatment with PCN, an agonist against mPXR, it shuttles to the nucleus [85]. Cytoplasmic mPXR is found in a complex with Hsp90 and cytoplasmic constitutive active/androstane receptor retention protein (CCRP) [85]. Further studies revealed two putative motifs within mPXR, a nuclear localization signal (NLS) sequence within the DNA-binding domain (DBD) and a xenochemical response signal (XRS) within the LBD, that drive mPXR to the nucleus [86]. However, the overexpression of CCRP does not seem to alter the subcellular localization of hPXR, which remained predominantly localized in the nucleus [81].
The activation of CAR is unique in that there are two mechanisms by which CAR can be activated: direct ligand binding and indirect activation [87]. TCPOBOP was the first selective and potent agonist of mCAR to be described that activates CAR through direct binding to its LBD and the subsequent recruitment of coactivators [13]. The indirect route of CAR activation involves the nuclear translocation of CAR independent of direct ligand binding. Phenobarbital and bilirubin have been shown to activate CAR by driving it into the nucleus without directly binding to the LBD of CAR [88]. In addition, TCPOBOP also drives the nuclear translocation and activation of hCAR without directly binding to the receptor [89]. Similar to PXR, once in the nucleus, CAR recruits coactivators such as PGC-1, SRC-1, and GRIP1 to drive gene transcription [90].
Similar to mPXR, cytoplasmic retention of CAR is modulated by the formation of a complex with CCRP and Hsp90 [91]. However, distinct from mPXR, nuclear accumulation of CAR is driven solely by the XRS, since a single amino acid change within the NLS of CAR abolishes the function of this domain [89]. This would explain a weaker nuclear localization of CAR than with mPXR, which contains 2 functional domains to drive its nuclear localization. A recent study describing a direct interaction between CAR and the membrane-bound PPP1R16A suggested that a subset of CAR exhibits plasma membrane localization [92].
3.2 Post-translational Regulation
PXR activity is also intricately regulated by phosphorylation, although phosphorylation of PXR is mostly correlated with an inhibitory effect (Figure 2) [93]. Treatment of primary rat and human hepatocytes with protein kinase A (PKA) activator leads to the attenuation of Cyp3A1 (rat orthologue of human CYP3A4) and CYP3A4 mRNA levels, respectively. This can be attributed, in part, to the phosphorylation of PXR by PKA [94, 95]. However, a similar treatment of mouse hepatocytes resulted in an increase in Cyp3a11 (mouse orthologue of human CYP3A4) mRNA levels, suggesting species specificity also exist in the regulation of PXR activity by phosphorylation [96].
PXR may also be subjected to phosphorylation by other kinases. In vitro kinase assays with a panel of kinases including protein kinase C [94], 70 kDa ribosomal S6 kinase, glycogen synthase kinase 3, and casein kinase II [95] showed that PXR can be phosphorylated by these kinases in vitro. A systematic approach to mutating serine/threonine residues to aspartic acid revealed that Ser8Asp, Thr57Asp, Ser208Asp, and Thr408Asp resulted in a decrease in PXR transactivation [96]. Work by Pondugula et al. [97] further revealed that the phosphomimetic mutant of PXR, Thr57Asp, also had an altered pattern of subcellular localization. In separate reports by Dong et al. and Lin et al. [98, 99], cyclin-dependent kinase 5 (Cdk5) and Cdk2 were identified as potential kinases that can phosphorylate and inhibit PXR. Findings from these reports suggest that PXR is also regulated by the cell cycle. In all, phosphorylation exerts a negative effect on PXR, in part by altering its pattern of subcellular localization or affecting its interaction with corepressors or coactivators. However, to this end, in vivo phosphorylation of PXR remains undetectable.
The stability of PXR is tightly regulated, although little is known about the mechanism behind its regulation (Figure 2). A semi-quantitative approach determined the half-life of unliganded PXR to be less than 4 hours [100]. Binding of PXR to endocrine-disrupting chemicals increased its half-life, in part due to the disruption of its interaction with suppressor for GAL1, a component of the proteasome [100, 101]. Recent work from Staudinger and colleagues [102] revealed an increase in ubiquitinated PXR after the inhibition of the 26S proteasome with a pharmacologic proteasomal inhibitor, MG132. Proteasomal inhibition also resulted in the inhibition of PXR transactivation, which suggests an interplay between PXR and the ubiquitin pathway [102]. However, it is noteworthy that many coregulators of PXR are also subject to regulation through the proteasomal pathway [103]. More work will need to be done to study PXR’s modulation through ubiquitination.
Other post-translational modifications of PXR involve SUMOylation and lysine acetylation. Similar to ubiquitination, SUMOylation involves the covalent addition of small ubiquitin-like modifier (SUMO) moieties onto its target protein. SUMO modifications on proteins lead to several fates, including nuclear-cytosolic translocation, modulation of protein stability and transcriptional regulation. Ligand-bound SUMOylation of PXR, which occurs mainly through SUMO 2/3 chains, is attenuated by the inflammatory response in the liver [102]. Acetylation of PXR has only recently been found to modulate its activity, and ligand-induced deacetylation of PXR was shown to be modulated by SIRT1 [104].
Similar to PXR, CAR activity is also regulated by phosphorylation. The phosphatase inhibitor okadaic acid prevents nuclear translocation of CAR and blocks phenobarbital-induced gene activation in rodents. Similarly, the overexpression of protein phosphatase 2A (PP2A) enhances TCPOBOP-induced mCAR nuclear translocation in hepatoma cells [87]. Further studies identified Ser202 within the LBD to be the site of phosphorylation [105]. Mutation of Ser202 to Asp abolished its response to phenobarbital and retained CAR in the cytoplasm. In addition, the CAR-Hsp90 complex recruits PP2A after treatment with phenobarbital, suggesting that PP2A is responsible for dephosphorylating Ser202 of CAR [106]. However, the kinases that phosphorylate CAR remain elusive.
Recent evidence also suggests a crosstalk between CAR activation and epidermal growth factor (EGF) receptor, extracellular signal-related kinase (ERK), and adenosine monophosphate-activated protein kinase (AMPK) signaling pathways. EGF was shown to repress phenobarbital-dependent activation of CAR, and the mitogen-activated protein kinase kinase inhibitor U0126 increases phenobarbital-dependent induction of CYP2B in primary rat hepatocytes [107]. Studies using AMPK knockout mice showed that AMPK may be involved in the activation of CAR in the nucleus [108]. Further studies revealed that phenobarbital targets the LKB1 gene to activate AMPK, suggesting that this could be the pathway leading to CAR activation, independent of binding to the LBD of CAR. It remains to be shown whether AMPK or ERK can phosphorylate and modulate CAR’s activity.
Unlike PXR, little has been studied on other post-translational modifications of CAR, such as ubiquitination and acetylation. To date, only Ser202 has been found to be phosphorylated, which results in the nuclear translocation of CAR, as described above. Based on CAR’s similarity to PXR and vitamin D receptor (VDR), both of which undergo ubiquitination, it is possible that CAR is also subject to ubiquitination. More efforts will be needed to explore the post-translational regulation on CAR.
4. Structural Basis of PXR and CAR on Ligand Binding
PXR and CAR are modular proteins, sharing common structural features that are characteristic of NRs [109]. A conserved N-terminal DBD encompasses zinc-finger motifs that bind to distinct target genes at the hormone response elements. The LBD is connected to the DBD through a hinge region, which is an important element for the function of NRs [110]. The LBDs of PXR and CAR include large, flexible binding pockets with broad specificity for ligands, and they are involved in the interactions of these NRs with coregulatory proteins and heterodimerization with RXR.
Several crystal structures of the LBD have been reported, which depict a compact α-helix sandwich fold (Figure 3). The LBD of PXR consists of three sets of α-helices: α1/α3, α4/α5/α8/α9, and α7/α10. In addition, a layer of five stranded anti-parallel β-sheets includes two novel β-strands (β1 and β1′) not observed in other NRs. PXR contains an insert of approximately 60 amino acids between helices α1 and α3, which contribute to the formation of β1, β1′, and the novel helix α2 [111]. The CAR LBD comprises three β-strands and 11 α-helices [112, 113]. Moreover, two 310 helices (α2 and α2′) were observed as rigid regions that can contribute to the hydrophobic character of the binding pocket, and it was speculated that they may contribute to the formation of a gate for ligand access [114].
Figure 3.
Crystal structure of the hCAR LBD (PDB code 1XV9) and hPXR LBD (PDB code 1ILH). The LBD of PXR (dark blue) and CAR (light blue) were aligned, displaying their respective AF-2 region (pink) and coactivator SRC-1 peptide (green). The β1 and β1′ strands unique in PXR are shown in yellow, and the helix x (H-x) seen in the CAR structure is depicted in red. The complex of CAR LBD with the RXRα LBD (salmon) illustrates a large dimerization interface.
PXR and CAR interact with coactivators such as SRC-1 and corepressors like SMRT through the ligand-dependent activating function 2 (AF-2) helix within the LBD, which recognizes the Leu-Xxx-Xxx-Leu-Leu motifs (Xxx = any other amino acid) in coactivators and the sequence Ile/Leu-Xxx-Xxx-Ile/Val-Ile in corepressors [90]. The interaction of the agonist with the LBD leads to the exposure of a hydrophobic surface for coactivator binding, which in turn is stabilized by a “charge clamp.” Based on crystal structures, the SRC-1 peptide appeared to be buried in a groove on the surface of the PXR LBD composed of AF-2, α3, and α4 [115]. Crystal structure analyses of the CAR LBD have uncovered key elements of the constitutive nature of CAR. A prominent feature is the short and rigid AF-2 helix, which lacks the C-terminal extension present in other NRs [112-114]. This allows for the interaction between the free carboxylate and Lys195 (hCAR), leading to further stabilization of the active AF-2 conformation. A short “helix x” replaces the extended loop linking α10 and AF-2. The helix x is composed of only 4-6 residues (depending on the species), with a single amino acid separating it from the AF-2 helix, thereby restricting and favoring the AF-2 in the active orientation. However, it was argued that helix x might not be a major contributor to the basal activity level, because other NRs have a comparable helix without being constitutively active [116].
RXR forms complexes with several other NRs, including PXR and CAR. Association with RXR increases the affinity of CAR for ligands and coactivators [112]. The CAR-RXR structure indicates the involvement of helices α7, α9, and α10 and the loop connecting α8 and α9 [112, 113]. A hydrophobic region was notably observed, surrounded by polar contacts. It was suggested that the large heterodimerization surface may contribute to the constitutive nature of CAR by stabilizing the active conformation [117]. Although there are similarities in the secondary structure of RXRα in the CAR/RXRα and the PPARγ/RXRα complexes, differences were discerned at the dimer interface [112, 113, 118]. The AF-2 helix for both CAR and RXRα were seen in their active orientation, with the latter being further stabilized by the interaction of its C-terminus extension with the CAR LBD [113].
Biochemical and crystallographic evidence indicates that PXR homodimerizes where the terminal β1′ strands from each monomer interlock to form the dimer interface [119]. Corresponding Trp223 and Tyr225 residues intertwine to form a novel tryptophan zipper, and a network of six intermolecular hydrogen bonds provides added stability to the interface. A Trp223Ala–Tyr225Ala double mutant prevents homodimerization without affecting protein folding while retaining the ability to bind ligands and DNA. As the mutants exhibited a marked reduction in CYP3A4 induction in response to PXR agonists SR12813 and rifampicin, it was postulated that coactivator recruitment was disrupted. The homodimerization mode of PXR may represent a distinct dimeric organization not observed in other NRs [120].
The LBD is depicted as a flexible and dynamic pocket. The highly promiscuous nature of PXR becomes evident by analyzing the substantially large binding cavity, with a volume of more than 1100 Å3 [111], which can accommodate a wide range of chemicals of varying sizes and structures. On the other hand, the LBD of CAR has a smaller volume in the range of 525 Å3 - 675 Å3 [112-114], which partly explains its more limited repertoire of agonists in contrast to PXR’s. The ligand pocket for both NRs is formed predominantly by nonpolar residues, creating a mostly hydrophobic and uncharged cavity. Of 28 amino acids forming the PXR binding pocket, eight are polar or charged residues [111]. Approximately three quarters of the 27 residues in the CAR cavity are nonpolar [114]. In the hPXR crystal structure, a salt bridge was observed between Glu321 and Arg410 that reduced the charge effect [111]. Similarly, the charge on the Asp238 was seen to be neutralized by Arg156 in the murine CAR structure [114]. A second salt bridge in the vicinity of the hPXR binding pocket further reduces the charge environment (between Asp205 and Arg413), while the side chains of Glu225 and Lys235 in murine CAR were directed away from the pocket. Based on cell-based reporter assays using PXR mutants, the amino acid residues participating in these electrostatic interactions were believed to be involved in the basal activity of PXR [111].
Three-dimensional structure analyses of PXR in both the apo (unliganded) and holo (liganded) forms have provided a wealth of information about the conformational changes that follow ligand binding. The first PXR LBD structure was reported as a complex with the cholesterol-lowering drug SR12813, which was discerned in three distinct positions within the cavity [111]. Subsequently, other structures involving PXR-SR12813 in complex with an SRC-1 coactivator peptide revealed a single agonist binding mode [115]. Thus, it was hypothesized that the PXR binding pocket can accommodate ligands in multiple positions, but a single active orientation is stabilized upon coactivator binding. The PXR-ligand interaction appears to be a dynamic process, leading to structural changes that might affect the interaction between PXR and its protein partners. Binding of hyperforin changes the pocket shape and increases its volume from 1294 to 1544 Å3. The agonists, colupulone and rifampicin, increase the thermal motion in various sections of the PXR LBD, including the AF-2 region and areas formed by β1 and β1′. The structure of CAR has been in complex with the ligands 5β-pregnanedione, CITCO [113], and TCPOBOP [112], which amplifies its transcriptional activity above basal levels. In the complex with hCAR, 5β-pregnanedione and CITCO were not seen to form direct contacts with the AF-2 region, which was obstructed from binding cavity by a barrier comprising the residues Phe161, Asn165, Phe234, and Tyr326 [113]. In contrast, the high potency of the mCAR agonist TCPOBOP was attributed to its ability to penetrate this barrier [112]. Androstenol represses the constitutive activity of CAR, and the 3D model of this inverse agonist bound to mCAR provides structural explanations for an interesting mode of modulating the activity of an NR that differs from traditionally viewed NR antagonists [114]. In the presence of androstenol, CAR displays α10 and α11 as two distinct helices separated by a “kink,” rather than as a single continuous helix seen in the active conformation. The researchers suggested that the ligand-induced “kink” unfolds a series of events that results in the destabilization of the active conformation of the AF-2 helix [114].
5. Conclusion
As xenobiotic sensors, PXR and CAR play pivotal roles in xenobiotic detoxification. Different from other NRs, PXR and CAR share some unique features, including ligand promiscuity, species differences, and regulation of the expression of a large number of overlapping or distinctive sets of genes responsible for xenobiotic detoxification (Figure 1). On one hand, these features enable PXR and CAR to physiologically exert the detoxifying function and protect the body against the toxic effect of environmental pollutants. On the other hand, PXR and CAR can also be activated pharmacologically by prescription drugs. The undesirable DDIs mediated by PXR and CAR are among the most important causes of adverse drug reactions. To deal with this “double-edged sword”, several strategies have been implemented to minimize the undesirable activation of PXR and CAR in drug development and clinical therapy. Moreover, the ligand promiscuity and structural flexibility of PXR and CAR set the stage for precise control of their activation through transcriptional and post-translational regulation. The need for both NRs and coregulators to modulate gene transcription links the regulation of PXR and CAR to a vast network of signaling pathways, and displays the complex roles that PXR and CAR play in the control of xenobiotic detoxification. Structural studies of PXR and CAR enable the understanding of the overall architecture of their LBDs, which differ in some aspects from the canonical view of the LBD seen in other NRs. The 3D models provide vital clues to the promiscuity of PXR and the constitutive nature of CAR and lay the foundation for predicting potential agonists and the development of antagonists to be used as co-therapeutics.
6. Expert opinion
Over the past decade, the discovery that PXR and CAR regulate the expression of genes encoding DMEs and transporters has greatly contributed to our knowledge of the mechanisms of xenobiotic detoxification and elimination. Although PXR and CAR primarily regulate the expression of the phase I DMEs CYPs, recent mouse model studies have revealed that PXR and CAR may also regulate non-CYP genes involved in phase I drug metabolism, such as CES1/2 [121] and Aldo-keto reductases [122]. While these studies potentially expand the scope of genes in phase I drug metabolism regulated by PXR and CAR, due to the species differences of PXR and CAR, future studies in a human system are needed. To continue to investigate the species differences, a recent study profiled 2816 clinically used drugs for their ability to activate hPXR and rat PXR. The results from this study further highlight the species specificity of PXR activation [123]. In addition to species specificity, in human, 3 isoforms of PXR and 15 isoforms of CAR have been identified with different molecular features, which potentially associate with distinct biological activity (reviewed in [90]). Arecent study discovered several PXR and CAR isoform-specific ligands [124]. The discovery of PXR and CAR ligands with species and isoforms selectivity will provide valuable tools to dissect their differential roles in the detoxification system. Similar to their roles in regulating xenobiotic detoxification, PXR and CAR also play critical roles in endobiotic metabolism governing the homeostasis of endogenous substances (reviewed in [125]), suggesting that PXR and CAR can be potential therapeutic targets for diseases such as hepatic steatosis, bone disorders, inflammatory bowel disease, and cancer [126]. We expect to see more efforts to establish PXR and CAR as disease-related therapeutic targets, thus expanding their classical roles as master xenobiotic sensors. Indeed, their roles as master xenobiotic sensors actually increase their significance as potential therapeutic targets.
Apart from post-translational regulation, PXR and CAR are also subject to transcriptional and post-transcriptional regulation. The expression of PXR and CAR is tissue-specific and is regulated by other NRs, including HNF-4α [127], glucocorticoid receptor [128] and farnesoid X receptor [129]. The expression of PXR is also regulated by methylation [130, 131], and by miRNA [132]. Little is known of the regulation of CAR expression by methylation and miRNA. The levels of PXR and CAR directly contribute to their activity. Since transcriptional, post-transcriptional, and post-translational regulation of PXR and CAR all contribute to their activity levels and these regulations can be affected by signaling pathways induced by xenobiotics, we expect to see more studies that systematically investigate the activation of PXR and CAR in response to xenobiotics at multiple levels.
There is considerable interest in the prediction of potential PXR or CAR activators early in the drug discovery process to filter out compounds that can potentially lead to DDIs [133]. The existing LBD crystal structures of both NRs start to shed light on the mechanisms that govern ligand binding and NR activation, and the increasing collection of available x-ray structures with structurally diverse ligands will facilitate the refining of in silico approaches. However, considerable roadblocks still exist, taking into account the dynamic and promiscuous nature of the ligand-binding pocket of these two NRs. The ligand-binding cavity of PXR can adjust its volume to accommodate compounds of different sizes, and docking molecules to proteins whose pocket can “breathe” will remain a major challenge. Moreover, ligands have been shown to bind in multiple orientations, such as the case of SR12813 in PXR [111] and CITCO in CAR [113].
A great majority of the structural studies covering members of the NR family involve either the LBD or DBD as isolated domains, but investigations encompassing full-length proteins are lacking. Structural information on intact NRs is slowly emerging, providing glimpses into the complex network among ligands, DNA, and interacting proteins. The first crystal structure of an intact NR was reported for the PPARγ/RXRα duplex, which allows the visualization of the interactions among domains [134]. The structure of the RXR/VDR/DNA complex revealed by cryo-electron microscopy underscores the hinge region as an important element in NR function [135], regulating the orientation of the LBD relative to the DNA. Structural studies of full-length VDR/RXR by hydrogen-deuterium exchange mass spectrometry revealed a long-range allosteric effect involving DNA/ligand binding and coactivator interaction with the NR complex [136]. The valuable information obtained from these crystal and solution-based structures reveal an intricate inter- and intra-domain communication, and the role of NRs as scaffold proteins can be altered by the binding of ligands, DNA, and interacting proteins [110]. In the same manner, the elucidation of the intact PXR and CAR structures may expose novel domain-domain interactions, especially considering that they both have unique structural features in the LBD that are not observed in other NRs.
Although current knowledge is based on the most common isoforms of PXR and CAR, it is noteworthy that studies on the other isoforms from the structural point of view have been overlooked. The constitutive activity of CAR and its ligand-mediated activity vary across isoforms. Differential activation of PXR between isoforms has also been noted. Structural investigations might also provide valuable insights into the basis for species-dependent ligand preference, which is necessary to understand drug-drug interactions in animals that need to be extrapolated to humans. We anticipate seeing more efforts to investigate the structures of full-length NRs, including those for various isoforms of PXR and CAR. Improved protein expression and purification techniques and approaches will greatly facilitate these efforts.
Article highlights.
As xenobiotic sensors, PXR and CAR play critical roles in xenobiotic detoxification to protect the human body from the toxicity of external chemicals.
PXR and CAR share some unique features: ligand promiscuity, species differences, and the ability to regulate a large number of overlapping or distinctive sets of genes involved in xenobiotic detoxification.
PXR and CAR can be activated by a number of marketed drugs, leading to undesirable drug-drug interactions that are the major cause of adverse drug reactions.
The subcellular localization of PXR and CAR and their interactions with cofactors can affect their transcriptional activity.
The activity of PXR and CAR can be modulated at the post-translational level, including phosphorylation, ubiquitination, SUMOylation, and lysine acetylation.
Structural analysis provides insight into the promiscuity of PXR and the constitutive activity of CAR.
Acknowledgements
This work was supported in part by the National Institute of General Medical Sciences [Grant GM086415], National Institutes of Health National Cancer Institute [Grant P30-CA21765], the American Lebanese Syrian Associated Charities (ALSAC) and St. Jude Children’s Research Hospital. The authors thank members of the Chen group for their valuable discussions and David Galloway for editing the manuscript.
Reference List
- 1.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–50. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 2.Nebert DW, Gonzalez FJ. P450 genes: structure, evolution, and regulation. Annu Rev Biochem. 1987;56:945–93. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- 3.Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–66. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 4.McCarver DG, Hines RN. The ontogeny of human drug-metabolizing enzymes: phase II conjugation enzymes and regulatory mechanisms. J Pharmacol Exp Ther. 2002;300:361–6. doi: 10.1124/jpet.300.2.361. [DOI] [PubMed] [Google Scholar]
- 5.Ayrton A, Morgan P. Role of transport proteins in drug discovery and development: a pharmaceutical perspective. Xenobiotica. 2008;38:676–708. doi: 10.1080/00498250801923855. [DOI] [PubMed] [Google Scholar]
- 6.El-Sheikh AA, Masereeuw R, Russel FG. Mechanisms of renal anionic drug transport. Eur J Pharmacol. 2008;585:245–55. doi: 10.1016/j.ejphar.2008.02.085. [DOI] [PubMed] [Google Scholar]
- 7.Tolson AH, Wang H. Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv Drug Deliv Rev. 2010;62:1238–49. doi: 10.1016/j.addr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baes M, Gulick T, Choi HS, et al. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–52. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kliewer SA, Moore JT, Wade L, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. •• This is the first report of the identification of PXR.
- 10.Ekins S, Mirny L, Schuetz EG. A ligand-based approach to understanding selectivity of nuclear hormone receptors PXR, CAR, FXR, LXRalpha, and LXRbeta. Pharm Res. 2002;19:1788–800. doi: 10.1023/a:1021429105173. [DOI] [PubMed] [Google Scholar]
- 11.Maglich JM, Parks DJ, Moore LB, et al. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–83. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 12.Moore LB, Parks DJ, Jones SA, et al. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275:15122–7. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- 13.Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–8. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie W, Barwick JL, Downes M, et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–9. doi: 10.1038/35019116. •• This is the first report of the humanized PXR mouse model.
- 15.Zhang J, Huang W, Chua SS, et al. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298:422–4. doi: 10.1126/science.1073502. •• This is the first report of the humanized CAR mouse model.
- 16.Gong H, Singh SV, Singh SP, et al. Orphan nuclear receptor pregnane X receptor sensitizes oxidative stress responses in transgenic mice and cancerous cells. Mol Endocrinol. 2006;20:279–90. doi: 10.1210/me.2005-0205. [DOI] [PubMed] [Google Scholar]
- 17.Ma X, Idle JR, Gonzalez FJ. The pregnane X receptor: from bench to bedside. Expert Opin Drug Metab Toxicol. 2008;4:895–908. doi: 10.1517/17425255.4.7.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheer N, Ross J, Rode A, et al. A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest. 2008;118:3228–39. doi: 10.1172/JCI35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui JY, Gunewardena SS, Rockwell CE, Klaassen CD. ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res. 2010;38:7943–63. doi: 10.1093/nar/gkq654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin B, Moore LB, Stoltz CM, et al. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60:427–31. [PubMed] [Google Scholar]
- 21.Sueyoshi T, Kawamoto T, Zelko I, et al. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson SS, LeCluyse EL, Negishi M, Goldstein JA. Regulation of human CYP2C9 by the constitutive androstane receptor: discovery of a new distal binding site. Mol Pharmacol. 2002;62:737–46. doi: 10.1124/mol.62.3.737. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Ferguson SS, Negishi M, Goldstein JA. Induction of human CYP2C9 by rifampicin, hyperforin, and phenobarbital is mediated by the pregnane X receptor. J Pharmacol Exp Ther. 2004;308:495–501. doi: 10.1124/jpet.103.058818. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Ferguson SS, Negishi M, Goldstein JA. Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol Pharmacol. 2003;64:316–24. doi: 10.1124/mol.64.2.316. [DOI] [PubMed] [Google Scholar]
- 25.Gardner-Stephen D, Heydel JM, Goyal A, et al. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab Dispos. 2004;32:340–7. doi: 10.1124/dmd.32.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodwin B, Hodgson E, D’Costa DJ, et al. Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol Pharmacol. 2002;62:359–65. doi: 10.1124/mol.62.2.359. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann JM, McKee DD, Watson MA, et al. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blumberg B, Sabbagh W, Jr., Juguilon H, et al. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burk O, Koch I, Raucy J, et al. The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR) J Biol Chem. 2004;279:38379–85. doi: 10.1074/jbc.M404949200. [DOI] [PubMed] [Google Scholar]
- 30.Faucette SR, Sueyoshi T, Smith CM, et al. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther. 2006;317:1200–9. doi: 10.1124/jpet.105.098160. • An improved model on asymmetrical cross-regulation by hCAR but not hPXR was proposed.
- 31.Xie W, Barwick JL, Simon CM, et al. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000;14:3014–23. doi: 10.1101/gad.846800. • This is the first report on cross-regulation of xenobiotic response by PXR and CAR.
- 32.Kakizaki S, Yamamoto Y, Ueda A, et al. Phenobarbital induction of drug/steroid-metabolizing enzymes and nuclear receptor CAR. Biochim Biophys Acta. 2003;1619:239–42. doi: 10.1016/s0304-4165(02)00482-8. [DOI] [PubMed] [Google Scholar]
- 33.Faucette SR, Zhang TC, Moore R, et al. Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther. 2007;320:72–80. doi: 10.1124/jpet.106.112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshinari K, Ohno H, Benoki S, Yamazoe Y. Constitutive androstane receptor transactivates the hepatic expression of mouse Dhcr24 and human DHCR24 encoding a cholesterogenic enzyme 24-dehydrocholesterol reductase. Toxicol Lett. 2012;208:185–91. doi: 10.1016/j.toxlet.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Sugatani J, Kojima H, Ueda A, et al. The phenobarbital response enhancer module in the human bilirubin UDP-glucuronosyltransferase UGT1A1 gene and regulation by the nuclear receptor CAR. Hepatology. 2001;33:1232–8. doi: 10.1053/jhep.2001.24172. [DOI] [PubMed] [Google Scholar]
- 36.Xie W, Yeuh MF, Radominska-Pandya A, et al. Control of steroid, heme, and carcinogen metabolism by nuclear pregnane X receptor and constitutive androstane receptor. Proc Natl Acad Sci U S A. 2003;100:4150–5. doi: 10.1073/pnas.0438010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Younes M, Schlichting R, Siegers CP. Glutathione S-transferase activities in rat liver: effect of some factors influencing the metabolism of xenobiotics. Pharmacol Res Commun. 1980;12:115–29. doi: 10.1016/s0031-6989(80)80069-5. [DOI] [PubMed] [Google Scholar]
- 38.Pickett CB, Telakowski-Hopkins CA, Ding GJ, et al. Rat liver glutathione S-transferases. Complete nucleotide sequence of a glutathione S-transferase mRNA and the regulation of the Ya, Yb, and Yc mRNAs by 3-methylcholanthrene and phenobarbital. J Biol Chem. 1984;259:5182–8. [PubMed] [Google Scholar]
- 39.Nims RW, Sinclair PR, Sinclair JF, et al. Dose-response relationships for the induction of P450 2B by 1,4-bis[2-(3,5-dichloropyridyloxy)]benzne (TCPOBOP) in rat and cultured rat hepatocytes. Xenobiotica. 1993;23:1411–26. doi: 10.3109/00498259309059450. [DOI] [PubMed] [Google Scholar]
- 40.Maglich JM, Stoltz CM, Goodwin B, et al. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–46. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 41.Higgins LG, Hayes JD. Mechanisms of induction of cytosolic and microsomal glutathione transferase (GST) genes by xenobiotics and pro-inflammatory agents. Drug Metab Rev. 2011;43:92–137. doi: 10.3109/03602532.2011.567391. [DOI] [PubMed] [Google Scholar]
- 42.Burk O, Arnold KA, Geick A, et al. A role for constitutive androstane receptor in the regulation of human intestinal MDR1 expression. Biol Chem. 2005;386:503–13. doi: 10.1515/BC.2005.060. [DOI] [PubMed] [Google Scholar]
- 43.Jigorel E, Le VM, Boursier-Neyret C, et al. Differential regulation of sinusoidal and canalicular hepatic drug transporter expression by xenobiotics activating drug-sensing receptors in primary human hepatocytes. Drug Metab Dispos. 2006;34:1756–63. doi: 10.1124/dmd.106.010033. [DOI] [PubMed] [Google Scholar]
- 44.Olinga P, Elferink MG, Draaisma AL, et al. Coordinated induction of drug transporters and phase I and II metabolism in human liver slices. Eur J Pharm Sci. 2008;33:380–9. doi: 10.1016/j.ejps.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Chan GN, Hoque MT, Cummins CL, Bendayan R. Regulation of P-glycoprotein by orphan nuclear receptors in human brain microvessel endothelial cells. J Neurochem. 2011;118:163–75. doi: 10.1111/j.1471-4159.2011.07288.x. [DOI] [PubMed] [Google Scholar]
- 46.Geick A, Eichelbaum M, Burk O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem. 2001;276:14581–7. doi: 10.1074/jbc.M010173200. [DOI] [PubMed] [Google Scholar]
- 47.Fromm MF, Kauffmann HM, Fritz P, et al. The effect of rifampin treatment on intestinal expression of human MRP transporters. Am J Pathol. 2000;157:1575–80. doi: 10.1016/S0002-9440(10)64794-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kast HR, Goodwin B, Tarr PT, et al. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–15. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- 49.Staudinger J, Liu Y, Madan A, et al. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab Dispos. 2001;29:1467–72. [PubMed] [Google Scholar]
- 50.Shinkyo R, Guengerich FP. Cytochrome P450 7A1 cholesterol 7alpha-hydroxylation: individual reaction steps in the catalytic cycle and rate-limiting ferric iron reduction. J Biol Chem. 2011;286:4632–43. doi: 10.1074/jbc.M110.193409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, Chiang JY. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–G84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 52.Hashizume T, Imaoka S, Mise M, et al. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J Pharmacol Exp Ther. 2002;300:298–304. doi: 10.1124/jpet.300.1.298. [DOI] [PubMed] [Google Scholar]
- 53.Hariparsad N, Chu X, Yabut J, et al. Identification of pregnane-X receptor target genes and coactivator and corepressor binding to promoter elements in human hepatocytes. Nucleic Acids Res. 2009;37:1160–73. doi: 10.1093/nar/gkn1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshinari K, Yoda N, Toriyabe T, Yamazoe Y. Constitutive androstane receptor transcriptionally activates human CYP1A1 and CYP1A2 genes through a common regulatory element in the 5′-flanking region. Biochem Pharmacol. 2010;79:261–9. doi: 10.1016/j.bcp.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 55.Fang HL, Strom SC, Ellis E, et al. Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther. 2007;323:586–98. doi: 10.1124/jpet.107.124610. [DOI] [PubMed] [Google Scholar]
- 56.van de Kerkhof EG, de Graaf IA, Ungell AL, Groothuis GM. Induction of metabolism and transport in human intestine: validation of precision-cut slices as a tool to study induction of drug metabolism in human intestine in vitro. Drug Metab Dispos. 2008;36:604–13. doi: 10.1124/dmd.107.018820. [DOI] [PubMed] [Google Scholar]
- 57.Shelby MK, Klaassen CD. Induction of rat UDP-glucuronosyltransferases in liver and duodenum by microsomal enzyme inducers that activate various transcriptional pathways. Drug Metab Dispos. 2006;34:1772–8. doi: 10.1124/dmd.106.010397. [DOI] [PubMed] [Google Scholar]
- 58.Franke RM, Scherkenbach LA, Sparreboom A. Pharmacogenetics of the organic anion transporting polypeptide 1A2. Pharmacogenomics. 2009;10:339–44. doi: 10.2217/14622416.10.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer zu Schwabedissen HE, Tirona RG, Yip CS, et al. Interplay between the nuclear receptor pregnane X receptor and the uptake transporter organic anion transporter polypeptide 1A2 selectively enhances estrogen effects in breast cancer. Cancer Res. 2008;68:9338–47. doi: 10.1158/0008-5472.CAN-08-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veith H, Southall N, Huang R, et al. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nat Biotechnol. 2009;27:1050–5. doi: 10.1038/nbt.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wienkers LC, Heath TG. Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discov. 2005;4:825–33. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 62.Guo GL, Moffit JS, Nicol CJ, et al. Enhanced acetaminophen toxicity by activation of the pregnane X receptor. Toxicol Sci. 2004;82:374–80. doi: 10.1093/toxsci/kfh286. [DOI] [PubMed] [Google Scholar]
- 63.Wang H, Faucette S, Moore R, et al. Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem. 2004;279:29295–301. doi: 10.1074/jbc.M400580200. [DOI] [PubMed] [Google Scholar]
- 64.Faucette SR, Zhang TC, Moore R, et al. Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther. 2007;320:72–80. doi: 10.1124/jpet.106.112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haidar C, Jeha S. Drug interactions in childhood cancer. Lancet Oncol. 2011;12:92–9. doi: 10.1016/S1470-2045(10)70105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jankovic SM, Dostic M. Choice of antiepileptic drugs for the elderly: possible drug interactions and adverse effects. Expert Opin Drug Metab Toxicol. 2012;8:81–91. doi: 10.1517/17425255.2012.645535. [DOI] [PubMed] [Google Scholar]
- 67.Sousa M, Pozniak A, Boffito M. Pharmacokinetics and pharmacodynamics of drug interactions involving rifampicin, rifabutin and antimalarial drugs. J Antimicrob Chemother. 2008;62:872–8. doi: 10.1093/jac/dkn330. [DOI] [PubMed] [Google Scholar]
- 68.Mai I, Bauer S, Perloff ES, et al. Hyperforin content determines the magnitude of the St John’s wort-cyclosporine drug interaction. Clin Pharmacol Ther. 2004;76:330–40. doi: 10.1016/j.clpt.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 69.Chen T. Overcoming drug resistance by regulating nuclear receptors. Adv Drug Deliv Rev. 2010;62:1257–64. doi: 10.1016/j.addr.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zimmermann K, Wittman MD, Saulnier MG, et al. SAR of PXR transactivation in benzimidazole-based IGF-1R kinase inhibitors. Bioorg Med Chem Lett. 2010;20:1744–8. doi: 10.1016/j.bmcl.2010.01.087. [DOI] [PubMed] [Google Scholar]
- 71.Chen T. Nuclear receptor drug discovery. Curr Opin Chem Biol. 2008;12:418–26. doi: 10.1016/j.cbpa.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 72.Venkatesh M, Wang H, Cayer J, et al. In vivo and in vitro characterization of a first-in-class novel azole analog that targets pregnane X receptor activation. Mol Pharmacol. 2011;80:124–35. doi: 10.1124/mol.111.071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–90. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 74.Huang H, Wang H, Sinz M, et al. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26:258–68. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- 75.Zhou C, Poulton EJ, Grun F, et al. The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol Pharmacol. 2007;71:220–9. doi: 10.1124/mol.106.029264. [DOI] [PubMed] [Google Scholar]
- 76.Healan-Greenberg C, Waring JF, Kempf DJ, et al. A human immunodeficiency virus protease inhibitor is a novel functional inhibitor of human pregnane X receptor. Drug Metab Dispos. 2008;36:500–7. doi: 10.1124/dmd.107.019547. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Li H, Moore LB, et al. The phytoestrogen coumestrol is a naturally occurring antagonist of the human pregnane X receptor. Mol Endocrinol. 2008;22:838–57. doi: 10.1210/me.2007-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang W, Zhang J, Wei P, et al. Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol Endocrinol. 2004;18:2402–8. doi: 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- 79.Li L, Chen T, Stanton JD, et al. The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol. 2008;74:443–53. doi: 10.1124/mol.108.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lau AJ, Yang G, Rajaraman G, et al. Differential effect of meclizine on the activity of human pregnane X receptor and constitutive androstane receptor. J Pharmacol Exp Ther. 2011;336:816–26. doi: 10.1124/jpet.110.175927. [DOI] [PubMed] [Google Scholar]
- 81.Saradhi M, Sengupta A, Mukhopadhyay G, Tyagi RK. Pregnane and Xenobiotic Receptor (PXR/SXR) resides predominantly in the nuclear compartment of the interphase cell and associates with the condensed chromosomes during mitosis. Biochim Biophys Acta. 2005;1746:85–94. doi: 10.1016/j.bbamcr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 82.Moore DD, Kato S, Xie W, et al. International Union of Pharmacology. LXII. The NR1H and NR1I receptors: constitutive androstane receptor, pregnene X receptor, farnesoid X receptor alpha, farnesoid X receptor beta, liver X receptor alpha, liver X receptor beta, and vitamin D receptor. Pharmacol Rev. 2006;58:742–59. doi: 10.1124/pr.58.4.6. [DOI] [PubMed] [Google Scholar]
- 83.Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. Journal of molecular biology. 2003;331:815–28. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 84.Xie Y, Ke S, Ouyang N, et al. Epigenetic regulation of transcriptional activity of pregnane X receptor by protein arginine methyltransferase 1. J Biol Chem. 2009;284:9199–205. doi: 10.1074/jbc.M806193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Squires EJ, Sueyoshi T, Negishi M. Cytoplasmic localization of pregnane X receptor and ligand-dependent nuclear translocation in mouse liver. J Biol Chem. 2004;279:49307–14. doi: 10.1074/jbc.M407281200. [DOI] [PubMed] [Google Scholar]
- 86.Kawana K, Ikuta T, Kobayashi Y, et al. Molecular mechanism of nuclear translocation of an orphan nuclear receptor, SXR. Mol Pharmacol. 2003;63:524–31. doi: 10.1124/mol.63.3.524. [DOI] [PubMed] [Google Scholar]
- 87.Li H, Wang H. Activation of xenobiotic receptors: driving into the nucleus. Expert Opin Drug Metab Toxicol. 2010;6:409–26. doi: 10.1517/17425251003598886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang W, Zhang J, Chua SS, et al. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci U S A. 2003;100:4156–61. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zelko I, Sueyoshi T, Kawamoto T, et al. The peptide near the C terminus regulates receptor CAR nuclear translocation induced by xenochemicals in mouse liver. Mol Cell Biol. 2001;21:2838–46. doi: 10.1128/MCB.21.8.2838-2846.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.di Masi A, De Marinis E, Ascenzi P, Marino M. Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Mol Aspects Med. 2009;30:297–343. doi: 10.1016/j.mam.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 91.Kobayashi K, Sueyoshi T, Inoue K, et al. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol Pharmacol. 2003;64:1069–75. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- 92.Sueyoshi T, Moore R, Sugatani J, et al. PPP1R16A, the membrane subunit of protein phosphatase 1beta, signals nuclear translocation of the nuclear receptor constitutive active/androstane receptor. Mol Pharmacol. 2008;73:1113–21. doi: 10.1124/mol.107.042960. [DOI] [PubMed] [Google Scholar]
- 93.Pondugula SR, Dong H, Chen T. Phosphorylation and protein-protein interactions in PXR-mediated CYP3A repression. Expert Opin Drug Metab Toxicol. 2009;5:861–73. doi: 10.1517/17425250903012360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ding X, Staudinger JL. Induction of drug metabolism by forskolin: the role of the pregnane X receptor and the protein kinase a signal transduction pathway. J Pharmacol Exp Ther. 2005;312:849–56. doi: 10.1124/jpet.104.076331. [DOI] [PubMed] [Google Scholar]
- 95.Lichti-Kaiser K, Xu C, Staudinger JL. Cyclic AMP-dependent protein kinase signaling modulates pregnane x receptor activity in a species-specific manner. J Biol Chem. 2009;284:6639–49. doi: 10.1074/jbc.M807426200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lichti-Kaiser K, Brobst D, Xu C, Staudinger JL. A systematic analysis of predicted phosphorylation sites within the human pregnane X receptor protein. J Pharmacol Exp Ther. 2009;331:65–76. doi: 10.1124/jpet.109.157180. • This paper describes a systematic analysis of putative PXR phosphorylation sites (serine or threonine)
- 97.Pondugula SR, Brimer-Cline C, Wu J, et al. A phosphomimetic mutation at threonine-57 abolishes transactivation activity and alters nuclear localization pattern of human pregnane x receptor. Drug Metab Dispos. 2009;37:719–30. doi: 10.1124/dmd.108.024695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong H, Lin W, Wu J, Chen T. Flavonoids activate pregnane x receptor-mediated CYP3A4 gene expression by inhibiting cyclin-dependent kinases in HepG2 liver carcinoma cells. BMC Biochem. 2010;11:23. doi: 10.1186/1471-2091-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin W, Wu J, Dong H, et al. Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem. 2008;283:30650–7. doi: 10.1074/jbc.M806132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masuyama H, Inoshita H, Hiramatsu Y, Kudo T. Ligands have various potential effects on the degradation of pregnane X receptor by proteasome. Endocrinology. 2002;143:55–61. doi: 10.1210/endo.143.1.8578. [DOI] [PubMed] [Google Scholar]
- 101.Masuyama H, Suwaki N, Tateishi Y, et al. The pregnane X receptor regulates gene expression in a ligand- and promoter-selective fashion. Mol Endocrinol. 2005;19:1170–80. doi: 10.1210/me.2004-0434. [DOI] [PubMed] [Google Scholar]
- 102.Staudinger JL, Xu C, Biswas A, Mani S. Post-translational modification of pregnane x receptor. Pharmacol Res. 2011;64:4–10. doi: 10.1016/j.phrs.2011.02.011. • First report of the regulation of PXR stability through ubiquitination.
- 103.Lonard DM, O’Malley BW. Chapter 4 emerging roles of the ubiquitin proteasome system in nuclear hormone receptor signaling. Prog Mol Biol Transl Sci. 2009;87:117–35. doi: 10.1016/S1877-1173(09)87004-X. [DOI] [PubMed] [Google Scholar]
- 104.Biswas A, Pasquel D, Tyagi RK, Mani S. Acetylation of pregnane X receptor protein determines selective function independent of ligand activation. Biochem Biophys Res Commun. 2011;406:371–6. doi: 10.1016/j.bbrc.2011.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hosseinpour F, Moore R, Negishi M, Sueyoshi T. Serine 202 regulates the nuclear translocation of constitutive active/androstane receptor. Mol Pharmacol. 2006;69:1095–102. doi: 10.1124/mol.105.019505. [DOI] [PubMed] [Google Scholar]
- 106.Yoshinari K, Kobayashi K, Moore R, et al. Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett. 2003;548:17–20. doi: 10.1016/s0014-5793(03)00720-8. [DOI] [PubMed] [Google Scholar]
- 107.Joannard F, Rissel M, Gilot D, et al. Role for mitogen-activated protein kinases in phenobarbital-induced expression of cytochrome P450 2B in primary cultures of rat hepatocytes. Toxicol Lett. 2006;161:61–72. doi: 10.1016/j.toxlet.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 108.Rencurel F, Foretz M, Kaufmann MR, et al. Stimulation of AMP-activated protein kinase is essential for the induction of drug metabolizing enzymes by phenobarbital in human and mouse liver. Mol Pharmacol. 2006;70:1925–34. doi: 10.1124/mol.106.029421. [DOI] [PubMed] [Google Scholar]
- 109.Ingraham HA, Redinbo MR. Orphan nuclear receptors adopted by crystallography. Curr Opin Struct Biol. 2005;15:708–15. doi: 10.1016/j.sbi.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 110.Nwachukwu JC, Nettles KW. The nuclear receptor signalling scaffold: insights from full-length structures. The EMBO journal. 2012;31:251–3. doi: 10.1038/emboj.2011.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Watkins RE, Wisely GB, Moore LB, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–33. doi: 10.1126/science.1060762. • First reported crystal structure of the PXR LBD.
- 112.Suino K, Peng L, Reynolds R, et al. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. • One of the first to report the crystal structure of the CAR LBD.
- 113.Xu RX, Lambert MH, Wisely BB, et al. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell. 2004;16:919–28. doi: 10.1016/j.molcel.2004.11.042. • One of the first to report the crystal structure of the CAR LBD.
- 114.Shan L, Vincent J, Brunzelle JS, et al. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol Cell. 2004;16:907–17. doi: 10.1016/j.molcel.2004.11.037. • One of the first to report the crystal structure of the CAR LBD.
- 115.Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. Journal of molecular biology. 2003;331:815–28. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 116.Windshugel B, Jyrkkarinne J, Vanamo J, et al. Comparison of homology models and X-ray structures of the nuclear receptor CAR: assessing the structural basis of constitutive activity. Journal of molecular graphics & modelling. 2007;25:644–57. doi: 10.1016/j.jmgm.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 117.Moore DD. CAR: three new models for a problem child. Cell metabolism. 2005;1:6–8. doi: 10.1016/j.cmet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 118.Gampe RT, Jr., Montana VG, Lambert MH, et al. Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000;5:545–55. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 119.Noble SM, Carnahan VE, Moore LB, et al. Human PXR forms a tryptophan zipper-mediated homodimer. Biochemistry. 2006;45:8579–89. doi: 10.1021/bi0602821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Calgaro MR, Neto Mde O, Figueira AC, et al. Orphan nuclear receptor NGFI-B forms dimers with nonclassical interface. Protein science : a publication of the Protein Society. 2007;16:1762–72. doi: 10.1110/ps.062692207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Staudinger JL, Xu C, Cui YJ, Klaassen CD. Nuclear receptor-mediated regulation of carboxylesterase expression and activity. Expert Opin Drug Metab Toxicol. 2010;6:261–71. doi: 10.1517/17425250903483215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu MJ, Takahashi Y, Wada T, et al. The aldo-keto reductase Akr1b7 gene is a common transcriptional target of xenobiotic receptors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2009;76:604–11. doi: 10.1124/mol.109.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shukla SJ, Sakamuru S, Huang R, et al. Identification of clinically used drugs that activate pregnane X receptors. Drug Metab Dispos. 2011;39:151–9. doi: 10.1124/dmd.110.035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dring AM, Anderson LE, Qamar S, Stoner MA. Rational quantitative structure-activity relationship (RQSAR) screen for PXR and CAR isoform-specific nuclear receptor ligands. Chem Biol Interact. 2010;188:512–25. doi: 10.1016/j.cbi.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ihunnah CA, Jiang M, Xie W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim Biophys Acta. 2011;1812:956–63. doi: 10.1016/j.bbadis.2011.01.014. • A comprehensive review on PXR’s functions focusing on endobiotic metabolism.
- 126.Ma X, Idle JR, Gonzalez FJ. The pregnane X receptor: from bench to bedside. Expert Opin Drug Metab Toxicol. 2008;4:895–908. doi: 10.1517/17425255.4.7.895. • A comprehensive review on PXR’s roles in human diseases.
- 127.Tirona RG, Lee W, Leake BF, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med. 2003;9:220–4. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 128.Shi D, Yang D, Yan B. Dexamethasone transcriptionally increases the expression of the pregnane X receptor and synergistically enhances pyrethroid esfenvalerate in the induction of cytochrome P450 3A23. Biochem Pharmacol. 2010;80:1274–83. doi: 10.1016/j.bcp.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jung D, Mangelsdorf DJ, Meyer UA. Pregnane X receptor is a target of farnesoid X receptor. J Biol Chem. 2006;281:19081–91. doi: 10.1074/jbc.M600116200. [DOI] [PubMed] [Google Scholar]
- 130.Misawa A, Inoue J, Sugino Y, et al. Methylation-associated silencing of the nuclear receptor 1I2 gene in advanced-type neuroblastomas, identified by bacterial artificial chromosome array-based methylated CpG island amplification. Cancer Res. 2005;65:10233–42. doi: 10.1158/0008-5472.CAN-05-1073. [DOI] [PubMed] [Google Scholar]
- 131.Habano W, Gamo T, Terashima J, et al. Involvement of promoter methylation in the regulation of Pregnane X receptor in colon cancer cells. BMC Cancer. 2011;11:81. doi: 10.1186/1471-2407-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283:9674–80. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 133.Jyrkkarinne J, Windshugel B, Ronkko T, et al. Insights into ligand-elicited activation of human constitutive androstane receptor based on novel agonists and three-dimensional quantitative structure-activity relationship. Journal of medicinal chemistry. 2008;51:7181–92. doi: 10.1021/jm800731b. [DOI] [PubMed] [Google Scholar]
- 134.Chandra V, Huang P, Hamuro Y, et al. Structure of the intact PPAR-gamma-RXR-nuclear receptor complex on DNA. Nature. 2008;456:350–6. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Orlov I, Rochel N, Moras D, Klaholz BP. Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. The EMBO journal. 2011;31:291–300. doi: 10.1038/emboj.2011.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang J, Chalmers MJ, Stayrook KR, et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nature structural & molecular biology. 2011;18:556–63. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]