Abstract
The Escherichia coli DNA replication machinery must frequently overcome template lesions under normal growth conditions. Yet, the outcome of a collision between the replisome and a leading-strand template lesion remains poorly understood. Here we demonstrate that a single, site-specific, cyclobutane pyrimidine dimer leading-strand template lesion provides only a transient block to fork progression in vitro. The replisome remains stably associated with the fork following collision with the lesion. Leading-strand synthesis is then reinitiated downstream of the damage in a reaction that is dependent on the primase, DnaG, but independent of any of the known replication-restart proteins. These observations reveal that the replisome can tolerate leading-strand template lesions without dissociating by synthesizing the leading strand discontinuously.
Encounters between the DNA replication machinery and obstacles such as DNA damage and RNA polymerase transcription complexes are thought to occur frequently under normal growth conditions (1–4). Following ultraviolet irradiation of E. coli cells, DNA replication continues past multiple lesions generating ssDNA gaps in the newly synthesized DNA (5, 6), implying that replication is reinitiated downstream of lesions in both the leading- and lagging–strand templates. Indeed, the replisome bypasses lagging-strand template damage with little impediment because of the discontinuous nature of lagging-strand synthesis (7–9). Leading-strand lesions may be overcome by reinitiating leading-strand synthesis downstream of damage, as the primase, DnaG, can prime the leading-strand template on a model fork structure following origin-independent replisome assembly (10). This model assumes that the replisome dissociates from the template following collision with the lesion and therefore requires the replication restart proteins to reload the replisome (11). However, the fate of the replisome following a collision with leading-strand template damage has not yet been determined.
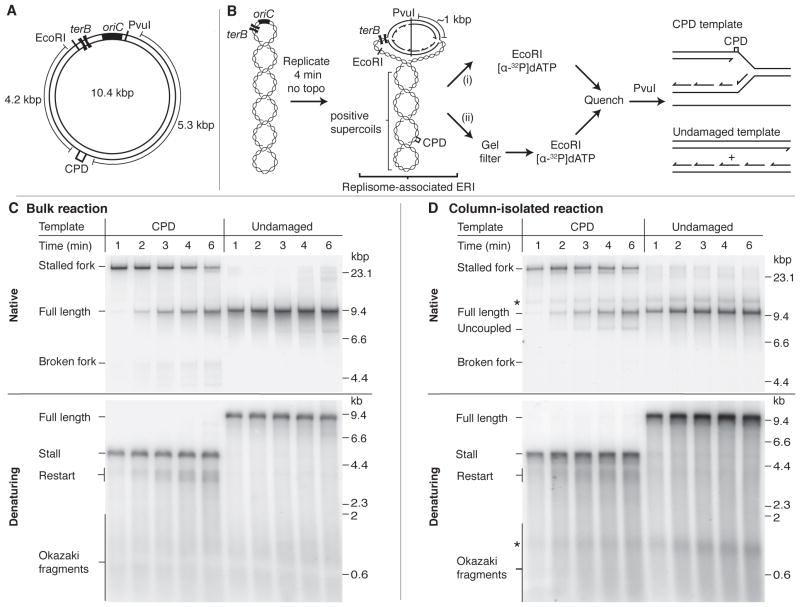
To investigate how the replisome may overcome leading-strand template damage, we constructed a 10.4 kbp plasmid containing the E. coli chromosomal origin of replication, oriC, and a single DNA lesion in the form of a cyclobutane pyrimidine dimer (CPD) (Fig. 1A and fig. S1A). To generate an essentially unidirectional replication system, we inserted two terB sites approximately 400 bp to the counter-clockwise side of oriC. In the presence of the Tus protein, terB blocks replication (12), enabling us to monitor the progression of the clockwise moving fork that will encounter the CPD in the leading-strand template. oriC-dependent replication reactions were initiated in the absence of a topoisomerase to generate stable early-replication intermediates (ERIs) by stalling the clockwise moving replisome approximately 1 kbp from the origin because of the accumulation of positive supercoils (Fig. 1B) (13). Topologically-stalled replisomes were released by EcoRI cleavage (3, 14) (fig. S1b). Replication of an undamaged template in bulk, using only the minimum proteins required to sustain oriC-dependent replication [DnaA, DnaB, DnaC, DnaG, HU, SSB, and the DNA polymerase III holoenzyme (as Pol III* and the β subunit)], generated full-length duplex products of the predicted 9.6 kbp (Fig. 1C, native), consisting of unit-length leading strands and 1 – 2 kb Okazaki fragments (Fig. 1C, denaturing). During the first 2 min of the reaction with the CPD template, virtually all newly synthesized products migrated to a position near the top of the native gel, indicating the presence of a stalled replication fork (Fig. 1C, native). The stalled fork resumed replication to generate full-length duplex DNA (figs. S2 and S5A, B and C, native,), despite the absence of any of the known origin-independent, replisome-loading proteins. Alkaline gel analysis of the CPD-template reaction (Fig. 1C, denaturing) revealed the presence of a truncated leading strand (stall) and Okazaki fragments. In addition to these anticipated products, a zone of nascent single strands that migrated between 3.5 – 4 kb were generated (restart). The upper length estimate for these products (4 – 4.2 kb) corresponds to the distance (4.2 kb) from the CPD to the end of the template (EcoRI site), and restriction mapping confirmed that they were generated by replication downstream of the CPD (figs. S5B and C, denaturing). Furthermore, the kinetics with which restart products appeared closely mirrored those of the transition from the stalled fork to full-length duplex DNA (compare Fig. 1C native and denaturing). Comparable results were also obtained when the CPD was replaced with an abasic site (fig. S3). These data are consistent with a model whereby leading-strand synthesis is reinitiated downstream of template damage.
Fig. 1.
A single leading-strand template CPD provides only a transient block to replisome progression. (A) Diagram of the CPD-containing plasmid used as template for replication reactions. (B) Illustration showing the staging of typical bulk (i), and column-isolated (ii), oriC-dependent replication reactions using EcoRI to release topologically-stalled forks. The predicted replication products following post-replication PvuI digestion are shown for the CPD and undamaged templates. (C and D) Time courses for replication reactions conducted in bulk (C) or using column-isolated, replisome-associated ERIs (D). Reaction products were digested with PvuI and analyzed by both native and denaturing gel electrophoresis as indicated. (D) The * denotes products formed from ERIs where the leading strand was labeled but not extended following EcoRI cleavage.
To determine if the replisome that initiated replication at oriC was responsible for generating the full-length duplex DNA and restart products observed in the CPD-template reaction, we initiated replication in bulk and isolated ERIs and their associated replisomes free from unbound proteins by gel-filtration chromatography (fig. S4) (15). The proteins that act distributively during replication (16) — DnaG, the β subunit of the Pol III HE, and SSB — were added back to the reactions and forks were released by EcoRI cleavage (Fig. 1D). The products generated in column-isolated reactions, on both the undamaged and CPD templates, were very similar to those observed in bulk (Figs. 1C and D and figs. S5 and 6). One difference was a CPD-template reaction product that migrated faster than full-length DNA on native gels and was resistant to cleavage by restriction enzymes that map downstream of the CPD (figs. S5B and D), indicating that it may be the product of uncoupled replication. The kinetics with which full-length duplex DNA and restart products were formed in both bulk and column-isolated reactions were almost indistinguishable, strongly suggesting that the oriC-loaded replisome can catalyze their formation without the need to recruit additional DNA polymerase or helicase from solution.
Our Pol III* preparation is likely to be composed of complexes that predominantly contain two DNA polymerases, whereas a recent study has suggested that the active replication machinery in E. coli possesses three DNA polymerases (17). We therefore repeated experiments, both in bulk and following column isolation with a Pol III* preparation that should predominantly contain three DNA polymerases (fig. S6 and supplementary methods). No appreciable differences were observed in either the efficiency, or kinetics, with which replication was reinitiated downstream of the CPD.
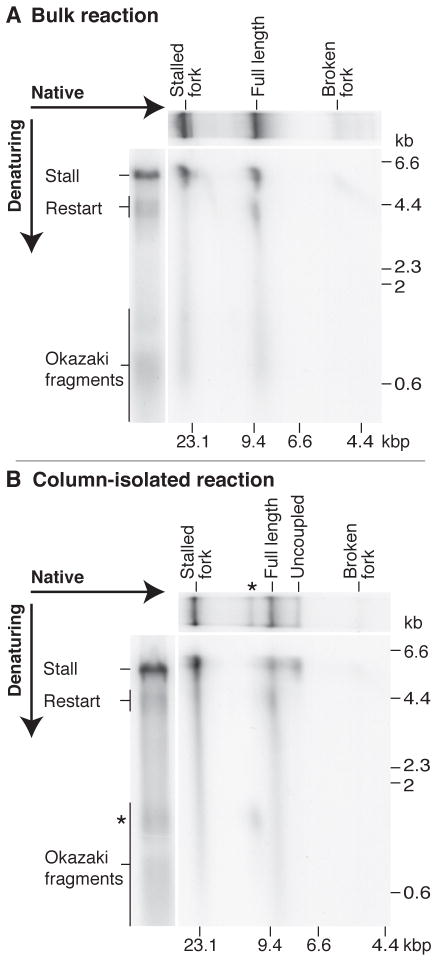
We next analyzed the products of bulk and column-isolated, EcoRI-dependent CPD-template reactions by two-dimensional gel electrophoresis to establish if the restart products detected in denaturing gels were associated with the transition from stalled fork to full-length duplex DNA (Figs. 2A and B). As seen for both reactions, the stalled fork was composed of the 5.3 kb stall product and a smear of Okazaki fragments. Restart fragments were exclusively associated with full-length duplex products, supporting the hypothesis that restart-product formation is coupled to the transition from stalled fork to full-length DNA. Additionally, the putative uncoupled product of the CPD-template reaction was composed solely of leading-strand stall products (Fig. 2B), as would be predicted if, in some cases, template unwinding and lagging-strand synthesis continued to the end of the template but leading-strand reinitiation did not occur (7, 8).
Fig. 2.
Restart products are associated with the production of full-length duplex DNA. (A and B) Two-dimensional gel electrophoretic analysis of the replication products from both bulk (A), and column-isolated (B), EcoRI-dependent reactions using the CPD template. Reactions were incubated for 4 min prior to quenching and PvuI cleavage. Samples of the same reactions were run solely through native and alkaline gels to serve as markers for the major reaction products. The * is as in Fig. 1D.
To investigate whether restart products were the result of leading-strand synthesis, we exploited the manner in which the Tus/ter complex arrests replication by using DNA gyrase to relax positive supercoiling (fig. S7). Under these conditions the clockwise moving fork is replicated to the Tus/ter complex. The leading strand will extend to the first nucleotide of terB, whereas the lagging strand terminates 50–100 bp upstream, generating a region of ssDNA specifically on the lagging-strand template (12). Post-replication EcoRI / PvuI cleavage should therefore only release leading-strand products as full-length duplex DNA (Fig. 3A). Fig. 3B shows that the full-length products of an undamaged-template replication reaction were composed exclusively of leading-strand products. The equivalent result was obtained with the CPD template, in both bulk (Fig. 3C) and column-isolated reactions (fig. S8). However, the full-length duplex products were composed of two nascent chains: the stall and restart, unequivocally showing that restart products were generated by leading-strand synthesis.
Fig. 3.
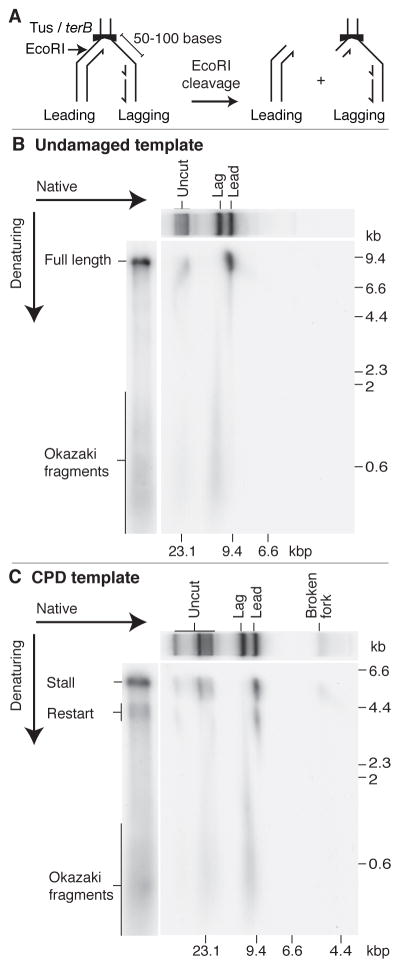
Restart products are generated by leading-strand synthesis. (A) Inhibition of replication by the Tus/ter complex generates a 50–100 base region of ssDNA on the lagging-strand template (12). Post-replication EcoRI cleavage will release leading-strand products as full-length duplex DNA because the restriction site is located 32 bp to the 5′ side of terB. (B and C) Replication reactions were conducted in bulk on undamaged (B) and CPD (C) templates in the presence of DNA gyrase. Following 6-min incubations, products were digested with EcoRI and PvuI prior to analysis by two-dimensional gel electrophoresis, with additional samples run solely through either native or alkaline gels serving as markers. Uncut refers to the positions of products (stalled forks and products of uncoupled replication) that are not released by EcoRI / PvuI cleavage.
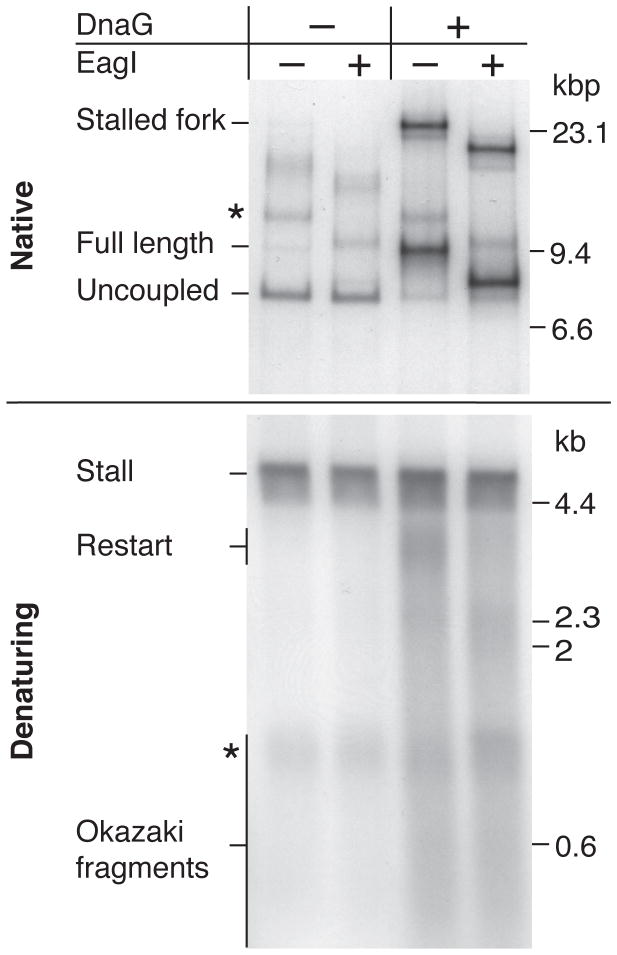
To determine if these products were primase dependent, column-isolated, replisome-associated ERIs were released by EcoRI cleavage in the presence or absence of DnaG. In the absence of DnaG there was virtually no full-length duplex DNA generated (Fig. 4, native). The major reaction product migrated at the position of the uncoupled band and was resistant to EagI cleavage, demonstrating that it was single stranded beyond the CPD. Consistent with these findings, the denaturing gel revealed that restart-product formation was entirely dependent on DnaG, supporting the finding that DnaG can reinitiate leading-strand synthesis by re-priming the leading-strand template (10).
Fig. 4.
DnaG re-primes the leading-strand template downstream of the CPD. Column-isolated CPD-template replication reactions were initiated by EcoRI cleavage in the presence or absence of DnaG for 6 min. Reactions were quenched and digested with either PvuI, or PvuI and EagI, which maps to the region of the template downstream of the CPD (fig. S5B). Samples were analyzed by both native and denaturing gel electrophoresis. The * is as in Fig. 1D.
Our data show that the E. coli replisome can tolerate leading-strand template lesions by remaining associated with the DNA and reinitiating leading-strand synthesis downstream of the damage via DnaG-dependent leading strand re-priming. The reaction is independent of the replication restart proteins, demonstrating that it is an inherent property of the replisome. Bypass of lesions in this manner will generate ssDNA gaps behind the replication fork (6) that are predominantly repaired by the faithful process of RecA-dependent recombination (18). Because the SOS response will not be induced, this mechanism should be particularly advantageous under normal growth conditions when low levels of damage are present, therefore avoiding cell-division arrest and the induction of mutagenic translesion polymerases. However, the leading-strand reinitiation that we observe is not complete and therefore replisome dissociation may occur, the chances of which will be increased when there are multiple lesions in the template, such as under conditions of replication stress. Should the replisome dissociate, origin-independent replisome loading and SOS-inducible systems such as NER and translesion polymerases are likely to become central to cell survival.
Supplementary Material
Acknowledgments
We thank Soon Bahng for establishing the protocol for replicating M13 DNA primed with the CPD oligonucleotide, Carolina Gabbai, Sankalp Gupta, and Stewart Shuman for advice on the manuscript. These studies were supported by NIH grant GM34557.
References
- 1.Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. Nature. 2011;470:554. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox MM, et al. Nature. 2000;404:37. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 3.Guy CP, et al. Mol Cell. 2009;36:654. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGlynn P, Lloyd RG. Nat Rev Mol Cell Biol. 2002;3:859. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 5.Rupp WD, Howard-Flanders P. J Mol Biol. 1968;31:291. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 6.Iyer VN, Rupp WD. Biochem Biophys Acta. 1971;228:117. doi: 10.1016/0005-2787(71)90551-x. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi K, et al. Genes Cells. 2003;8:437. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 8.Pages V, Fuchs RP. Science. 2003;300:1300. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 9.McInerney P, O’Donnell M. J Biol Chem. 2004;279:21543. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 10.Heller RC, Marians KJ. Nature. 2006;439:557. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 11.Heller RC, Marians KJ. Nat Rev Mol Cell Biol. 2006;7:932. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 12.Hill TM, Marians KJ. Proc Natl Acad Sci USA. 1990;87:2481. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiasa H, Marians KJ. J Biol Chem. 1994;269:16371. [PubMed] [Google Scholar]
- 14.Marians KJ, Hiasa H, Kim DR, McHenry CS. J Biol Chem. 1998;273:2452. doi: 10.1074/jbc.273.4.2452. [DOI] [PubMed] [Google Scholar]
- 15.Carr KM, Kaguni JM. J Biol Chem. 2001;276:44919. doi: 10.1074/jbc.M107463200. [DOI] [PubMed] [Google Scholar]
- 16.Wu CA, Zechner EL, Marians KJ. J Biol Chem. 1992;267:4030. [PubMed] [Google Scholar]
- 17.Reyes-Lamothe R, Sherratt DJ, Leake MC. Science. 2010 Apr 23;328:498. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berdichevsky A, Izhar L, Livneh Z. Mol Cell. 2002;10:917. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.