Abstract
A diverse T-cell receptor (TCR) repertoire capable of recognizing a broad range of antigenic peptides is thought to be central to effective pathogen-specific immunity by counteracting escape mutations, selecting high-avidity T cells, and providing T-cell specificities with comprehensive functional characteristics. However, evidence that TCR diversity is important for the successful control of human infections is limited. A single-cell strategy for the clonotypic analysis of human CD8+ TCRαβ repertoires was used to probe the diversity and magnitude of individual human cytomegalovirus (CMV)-specific CD8+ T-cells recovered directly ex vivo. We found that CD8+ TCRαβ repertoire diversity, but not the size of the CD8+ T-cell response, was inversely related to circulating CMV-specific antibody levels, a measure that has been correlated epidemiologically with differential mortality risks and found here to be higher in persons with detectable (versus undetectable) CMV viral loads. Overall, our findings indicate that CD8+ T-cell diversity may be more important than T-cell abundance in limiting the negative consequences of CMV persistence, demonstrate high prevalence of both TCRα and β public motif usage, and suggest that a highly diverse TCRαβ repertoire may be an important benchmark and target in the success of immunotherapeutic strategies.
INTRODUCTION
Effective immune responses against human pathogens rely on an armamentarium of T-cell receptors (TCRs) capable of recognizing antigenic epitopes on infectious agents, among other requirements. Although a diverse TCR repertoire would seem to be better adapted to combatting a broad range of human pathogens, direct evidence supporting this fundamental principle is limited (1). The diversity of any pathogen-specific T-cell mediated immune response is defined by the spectrum of TCRαβ heterodimers that bind to introduced, non-self peptides (p) presented in the context of self major histocompatibility complex (MHC) glycoproteins. The range of individual clonotypes bearing TCRs that are specific for a particular pMHC epitope comprise a repertoire that varies substantially in constituent TCR frequency and diversity. Structural TCR variation is primarily associated with the third complementarity determining region (CDR3) that interacts specifically with the MHC-bound peptide (2). These CDR3-region differences are generated by somatic rearrangements within the variable (V) and joining (J) gene segments of the TCRα chain and within the V, D (diversity), and J gene segments of the TCRβ chain (2–4). Although there is some understanding of the importance of epitope-specific T-cell repertoire breadth in the development of effective responses against infections (5), much remains to be learned regarding the selection, maintenance, and functional implications of human TCR diversity (6).
Potential mechanisms by which a diverse TCR repertoire can effect immune protection include the control of viral escape (7, 8), selection of high-avidity cytotoxic T lymphocytes (CTLs) that eliminate infected cells more quickly (9, 10), and the provision of a wide range of T cell structural avidities that fulfill requirements for functional heterogeneity (11, 12). Evidence from a murine model of herpes simplex virus type 1 infection has provided a compelling link between TCR diversity and host resistance in this persistent infection (5). However, the case that TCR diversity is important for the successful CD8+ T cell-mediated control of human infections is far from proven (1). This prompted us to test the impact of TCR repertoire diversity on persistent human cytomegalovirus (CMV) infection.
The ability to quantify a relationship between TCR diversity and microbial defense depends on the sensitivity of the system used to probe the pMHC Class I (pMHCI)-specific immune repertoire. Historically, the approaches used vary greatly in acuity. Flow cytometric analysis with TCR Vβ monoclonal antibodies offers a low-resolution perspective by enumerating the percentages of epitope-specific T cells [detected using pMHCI tetramers (13)] expressing particular Vβ segments (14). Spectratyping offers a more refined TCR repertoire “snapshot” by measuring CDR3 size distributions for DNA amplified by polymerase-chain reaction (PCR) using V- and (for higher resolution) J-segment-specific primers (15). Molecular cloning and sequencing (including pyrosequencing) of TCR CDR3 transcripts is much more precise (16, 17) but, at least for humans, this type of analysis has been limited thus far to determining CDR3α and/or CDR3β repertoires separately, or examining paired CDR3αβ data from a small number of in vitro-expanded CTL clones.
Optimally, the comprehensive assessment of epitope-specific TCRαβ repertoire selection and clonal prevalence requires the identification and enumeration of individual clonotypes. Although recent advances in high-throughput sequencing have begun to shed light on the development and selection of human TCR repertoires by bulk sequencing of lymphocyte populations (17), this approach does not allow the simultaneous characterization of CDR3α and CDR3β sequences for individual T cell clonotypes. Using a single-cell approach, recent analysis in a mouse model system (18) allowed us to characterize CDR3α and CDR3β segments expressed in single, influenza-specific T cells recovered directly from the lung. This protocol has now been adapted to probe the human CMV-specific CD8+ T cell response for individual peripheral blood lymphocytes.
This powerful tool allows the immediate, ex vivo characterization of human epitope-specific T-cell repertoires by single-cell, multiplex clonotypic analysis, without prior knowledge of associated Vα or Vβ gene segment usage. Nested PCR, utilizing comprehensive panels of TCR Vα and Vβ primers paired with Cα and Cβ primers, enables the simultaneous amplification of expressed CDR3α and CDR3β segments from individual, epitope-specific T cells, isolated by pMHCI tetramer-based flow cytometric cell sorting. Nucleotide sequencing of the individual, amplified CDR3 products yields precise CDR3αβ clonotypic data reflective of the in vivo clonal prevalence. The TCR repertoire findings, together with the concurrent measurement of antibody titers and viral loads, have allowed us to reach conclusions concerning the importance of TCRαβ repertoire diversity and clonotype prevalence in the long-term control of persistent CMV infections.
RESULTS
Single-cell human TCR repertoire characterization directly ex vivo
Aiming to view a high resolution “snapshot” of the CMV-specific CD8+ T cell response, which is believed to be important for controlling persistent CMV infections, we developed a strategy that allows the contemporaneous characterization of CDR3αβ repertoires for single epitope-specific CD8+ CTLs recovered directly from peripheral blood. The design and validation of a comprehensive panel of primers specific for all non-pseudogene TCR Vα, Vβ, Cα, and Cβ gene segments is detailed in the Materials and Methods and Supplementary Materials sections (Supplementary Results, Tables S1 and S2, Fig. S1 and S2). All TCR genes are identified using the international ImMunoGeneTics information system (IMGT) nomenclature (19).
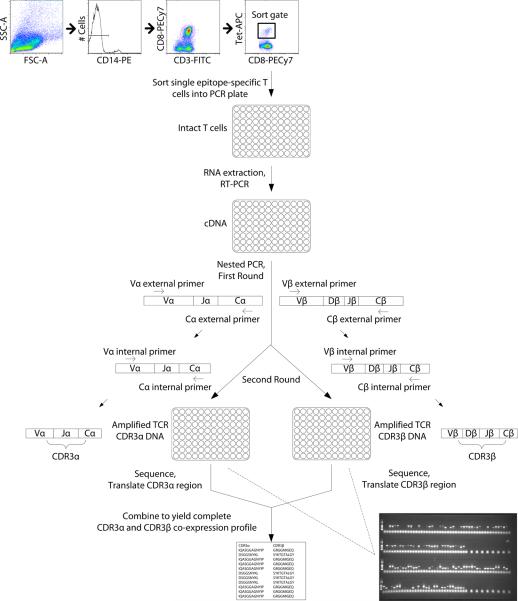
This ex vivo characterization of the epitope-specific T-cell repertoire at the single-cell-level is grounded in the isolation of single CD8+ T cells from unmanipulated human peripheral blood mononuclear cells (PBMCs). The human leukocyte antigen (HLA) A*0201-restricted pp65495–503 NLVPMVATV (CMV-NLV) CTLs constitute the dominant CMV-specific CTL set for individuals expressing that HLA phenotype. Single, tetramer-labeled, CMV-NLV-specific CD8+ T cells were sorted by flow cytometry into the individual wells of a 96-well PCR plate (Fig. 1). A strategy combining reverse-transcription (RT)-PCR and nested PCR then enabled the highly specific amplification of both the CDR3α and CDR3β segments expressed concurrently in each T cell.
Fig. 1.
Overview of single-cell multiplex clonotypic analysis of epitope-specific T cells. Single epitope-specific CD8+ T cells are sorted on a flow cytometric cell sorter into 96-well PCR plates. RT-PCR is performed on the individual cells. The resultant cDNA is subjected to two rounds of nested PCR. In the first round, CDR3α and CDR3β transcript amplification is achieved with the use of a multiplexed, comprehensive panel of external sense Vα and Vβ and antisense Cα and Cβ segment-specific primers. First-round PCR products are subjected to two separate second-round PCRs, incorporating, respectively, a multiplexed panel of external sense Vα and antisense Cα or external sense Vβ and antisense Cβ segment-specific primers. PCR products thus derived are sequenced and translated to yield paired CDR3αβ repertoire data. Inset: Nucleotide products from nested PCR performed on cDNA derived from single CMV-NLV-specific CD8+ T cells from a young adult donor, incorporating TCRα- (top 2 rows) and TCRβ-specific primers (bottom 2 rows).
In our hands, the success rates for obtaining paired CDR3α and CDR3β sequences varied from 22–80% of single cells processed from cryopreserved, human PBMC samples. With maturation of the technique, typically about 50% of sequenced single cells yield paired data. Dual expression of TCRα chains, inadequacy of the mRNA transcripts in individual cells, suboptimal quality of thawed cryopreserved cells, and the loss of what are already small amounts of cellular mRNA due to procedural mechanics, can account for any failures to obtain CDR3αβ co-expression data. Even so, technical constraints that affect the capacity to amplify individual CDR3α and CDR3β segments concurrently may be considered to affect all cells at random and with equal likelihood. Such factors should not, therefore, selectively impact particular CDR3α or CDR3β sequences in ways that bias inferences based on the repertoire structure.
Furthermore, only a single-cell-based repertoire analysis can establish whether or not CDR3α or CDR3β sequence data has been obtained from any particular tetramer+CD8+ T cell. Obviously, approaches based on determining bulk nucleotides from T cell populations cannot provide this insight. Also, unique to our single-cell approach, introducing an additional step that involves subcloning the nested-PCR amplicons in transformed E. coli allows us to identify the productively rearranged CDR3α that can accompany a nonproductively rearranged CDR3α, dual TCRα chains, or other missing CDR3αβ co-expression data. With this additional procedure, we successfully obtained the missing, paired CDR3α or CDR3β sequence in 5 out of 5 CTLs from one donor (Supplementary Results). In addition, earlier analysis has shown that PCR error does not affect sequence validity for the data sets derived from our nested-PCR approach (18).
Determination of CMV-specific CDR3αβ repertoire in immunocompetent adults
Virus-specific CD8+ T cell immunity is thought to play a crucial role in limiting the replication of CMV, which establishes a persistent, lifelong infection in humans (20). Using the strategy for TCR repertoire analysis developed here, we examined the CMV-specific TCR repertoires of unmanipulated, ex vivo peripheral T cells from two young and four older immunocompetent adults with known CMV IgG seropositivity (Table 1, Table S3).
Table 1.
TCR repertoires of CMV-NLV-specific CD8+ T cells from two young and four older adults. TCR V, D, and J nomenclature follows that of IMGT (19).
| Donor | TRAV | CDR3α AA sequence | TRAJ | TRBV | CDR3β AA sequence | TRBD | TRBJ | Prevalence within each donor (%) |
|---|---|---|---|---|---|---|---|---|
| Y1 (n=39) | 1–2 | TDSNYQL | 33 | 6–4 | SSGGGTGEL | 2 | 2–2 | 2.7 |
| 3 | RQDYQL | 33 | 6–5 | GPGTSATDTQ | 2 | 2–3 | 2.7 | |
| 8–1 | CLRGAGSYQL | 28 | 27 | ALGGPNTQ | 1 | 2–3 | 2.7 | |
| 10 | SRRTTGANNL | 36 | 6–6 | SYTLASYEQ | 2 | 2–7 | 2.7 | |
| 14 | RVDGQKLL | 16 | 9 | SPLQGGEL | 1 | 2–2 | 2.7 | |
| 21 | GRSNDYKL | 20 | 9 | SAGGGILATDTQ | 2 | 2–3 | 2.7 | |
| 21 | TGAGYSTL | 11 | 27 | SLLGASLEQ | 1 | 2–7 | 2.7 | |
| 22 | DYGQNF | 26 | 4–2 | SQVPGSYEQ | 2 | 2–7 | 2.7 | |
| 1–2 | GDTGFQKL | 8 | ND | 2.7 | ||||
| 3 | RPRVATGGGNKL | 10 | ND | 2.7 | ||||
| 8–4 | RDLNTGNQF | 49 | ND | 2.7 | ||||
| 19 | SDITGGGNKL | 10 | ND | 2.7 | ||||
| 19 | SEAFRDDKI | 30 | ND | 2.7 | ||||
| 21 | PPQGGSEKL | 57 | ND | 2.7 | ||||
| ND | 6–5 | GPGTSATDTQ | 2 | 2–3 | 13.5 | |||
| ND | 4–2 | TFEGGNEKL | 1 | 1–4 | 2.7 | |||
| ND | 6–1 | RSSYNEQ | 2 | 2–1 | 2.7 | |||
| ND | 6–4 | SSGGGTGEL | 2 | 2–2 | 2.7 | |||
| ND | 6–5 | SFFPGTDTDTQ | 2 | 2–3 | 2.7 | |||
| ND | 6–5 | SISLGFGETQ | 2 | 2–5 | 2.7 | |||
| ND | 6–5 | SYSTNEQ | 2 | 2–1 | 2.7 | |||
| ND | 9 | SAPGTGSHEQ | 1 | 2–7 | 2.7 | |||
| ND | 9 | SDTPTEA | 1 | 1–1 | 2.7 | |||
| ND | 9 | SSGGGTGEL | 2 | 2–2 | 2.7 | |||
| ND | 10–1 | SAPTAPYEQ | 1 | 2–7 | 2.7 | |||
| ND | 13 | SHRNQPQ | 1 | 1–5 | 2.7 | |||
| ND | 14 | SPGQRVNYGY | 1 | 1–2 | 2.7 | |||
| ND | 15 | SRDTSGRARNTQ | 2 | 2–3 | 2.7 | |||
| ND | 19 | GLGSPSTDTQ | 2 | 2–3 | 2.7 | |||
| ND | 27 | SFRGESYGY | 1 | 1–2 | 2.7 | |||
| ND | 28 | SLSAGGPYTGEL | 2 | 2–2 | 2.7 | |||
| ND | 28 | SYPYNEQ | 2–1 | 2.7 | ||||
| ND | 30 | SRGLYGY | 1 | 1–2 | 2.7 | |||
| Y2 (n=69) | 24 | NTGNQF | 49 | 6–5 | SWKTGTGGTGY | 1 | 1–2 | 27.5 |
| 24 | NTGNQF | 49 | 6–5 | SPSTGVTFYGY | 1 | 1–2 | 2.9 | |
| 26–2 | SNNNDM | 43 | 6–5 | SISDLAKNIQ | 2 | 2–4 | 8.7 | |
| 24 | NTGNQF | 49 | ND | 21.7 | ||||
| 26–2 | SNNNDM | 43 | ND | 1.4 | ||||
| ND | 6–5 | SWKTGTGGTGY | 1 | 1–2 | 18.8 | |||
| ND | 6–5 | SISDLAKNIQ | 2 | 2–4 | 14.5 | |||
| ND | 6–5 | SPSTGVTFYGY | 1 | 1–2 | 2.9 | |||
| ND | 7–9 | SEVTYEQ | 2 | 2–7 | 1.4 | |||
| E1 (n=36) | 5 | IQASGGSYIP | 6 | 15 | GRGGMIGEQ | 2 | 2–7 | 30.6 |
| 5 | IQASGGSYIP | 6 | ND | 11.1 | ||||
| 14 | DSGGSNYKL | 53 | ND | 2.8 | ||||
| ND | 6–5 | SYKTGTALGY | 1 | 1–2 | 2.8 | |||
| ND | 15 | GRGGMIGEQ | 2 | 2–7 | 52.8 | |||
| E2 (n=33) | 25 | KTSYDKV | 50 | 25–1 | EQGMHEQ | 1 | 2–7 | 9.7 |
| 27 | RDNYGQNF | 26 | 12–3 | SWTDLNQPQ | 1 | 1–5 | 6.5 | |
| 24 | STGGANNL | 36 | 6–1 | SEWADTEA | 1 | 1–1 | 6.5 | |
| 6 | GGGYNKL | 4 | 6–2 | SYGRDRENIQ | 1 | 2–4 | 3.2 | |
| 8–6 | RPRNFNKF | 21 | 19 | SLSPSMATYNEQ | 2 | 2–1 | 3.2 | |
| 19 | GNNARL | 31 | 6–5 | SLGTGWEHGY | 1 | 1–2 | 3.2 | |
| 23 | SIGNFGNEKL | 48 | 25–1 | SPARNTEA | 1 | 1–1 | 3.2 | |
| 24 | LSGGKL | 23 | 20–1 | TRTARNQPQ | 1 | 1–5 | 3.2 | |
| 25 | TLSNFGNEKL | 48 | 13 | SLGVASYEQ | 2 | 2–7 | 3.2 | |
| 30 | SIGNFGNEKL | 48 | ND | 6.5 | ||||
| 32 | DNNNDM | 43 | ND | 6.5 | ||||
| 26–1 | RGPSGGSYIP | 6 | ND | 3.2 | ||||
| 27 | SDPNDYKL | 20 | ND | 3.2 | ||||
| 28 | QAEGSQGNL | 42 | ND | 3.2 | ||||
| 29 | QAGYGGSQGNL | 42 | ND | 3.2 | ||||
| 31 | ANNYGQNF | 26 | ND | 3.2 | ||||
| ND | 20–1 | TRTARNQPQ | 1 | 1–5 | 6.5 | |||
| ND | 25–1 | EQGMHEQ | 1 | 2–7 | 6.5 | |||
| ND | 2 | SDPRTRLSGANVL | 2 | 2–6 | 3.2 | |||
| ND | 6–1 | SEWADTEA | 1 | 1–1 | 3.2 | |||
| ND | 7–2 | SPVGSFADTQ | 2 | 2–3 | 3.2 | |||
| ND | 9 | SVAPPSDPTGEL | 2 | 2–2 | 3.2 | |||
| ND | 25–1 | SEPGGEQ | 2 | 2–7 | 3.2 | |||
| E3 (n=72) | 5 | SGSYNTDKL | 34 | 20–1 | RDGLAGHYEQ | 2 | 2–7 | 54.2 |
| 8–3 | AFGNQF | 49 | 27 | SMTSGALYNEQ | 2 | 2–1 | 20.8 | |
| 35 | PMKTSYDKV | 50 | 12–4 | ASANYGY | 1 | 1–2 | 2.8 | |
| 3 | RDTDARL | 31 | ND | 1.4 | ||||
| ND | 27 | SMTSGALYNEQ | 2 | 2–1 | 15.3 | |||
| ND | 20–1 | RDGLAGHYEQ | 2 | 2–7 | 4.2 | |||
| ND | 30 | IPGTGGVEA | 1 | 1–1 | 1.4 | |||
| E4 (n=48) | 24 | PYNNNDM | 43 | 27 | SLEGYTEA | 1 | 1–1 | 22.9 |
| 5 | ITGGGNKL | 10 | 20–1 | TDVTKGQLISGY | 1 | 1–2 | 12.5 | |
| 24 | NTGNQF | 49 | 6–5 | SYQTGTGVYGY | 1 | 1–2 | 2.1 | |
| 24 | NTGNQF | 49 | 6–5 | SYQTGTPYGY | 1 | 1–2 | 2.1 | |
| 20 | QAITGGFKT | 9 | 27 | SLEGYTEA | 1 | 1–1 | 2.1 | |
| 24 | PYNNNDM | 43 | ND | 12.5 | ||||
| 5 | ITGGGNKL | 10 | ND | 6.3 | ||||
| 24 | NTGNQF | 49 | ND | 4.2 | ||||
| 20 | QGTTDSWGKF | 24 | ND | 2.1 | ||||
| 10 | SAENARL | 31 | ND | 2.1 | ||||
| ND | 27 | SLEGYTEA | 1 | 1–1 | 16.7 | |||
| ND | 6–5 | SYQTGTGVYGY | 1 | 1–2 | 8.3 | |||
| ND | 20–1 | SSGALNTEA | 1 | 1–1 | 2.1 | |||
| ND | 6–5 | SYSTGVPYGY | 1 | 1–2 | 2.1 | |||
| ND | 20–1 | TDVTKGQLISGY | 1 | 1–2 | 2.1 |
Nonproductive CDR3α rearrangements are not included in this table. Percentages do not total 100% due to rounding error. The ages of the donors were 34, 23, 90, 88, 71, and 88 years, respectively. Corresponding CDR3 nucleotide sequences are listed in Table S3. Each TCR repertoire was measured in 1 experiment per donor. ND, not determined; AA, amino acid.
Several features of our direct, ex vivo acquired repertoire data are consistent with those reported previously for in vitro cultured and molecularly cloned CMV-NLV-specific CD8+ T cells (21–25). First, sequence-unique clones were observed even among those utilizing one specific Vβ family (e.g., TRBV6−5). Second, CMV-specific CD8+ T cell repertoires were frequently, but not always, dominated by one or two clonotypes, as observed in donors Y2, E1, E3, and E4. Third, TRBV6−5 and TRBV27 were among the most frequently expressed Vβ regions among the T-cell clones characterized for all donors. Fourth, TRBV6−5 was commonly associated with TRBJ1−2 (21–25), as observed in donors Y2, E1, E2, and E4. Fifth, the previously identified public (observed in more than one person) CDR3α sequences, NTGNQF (TRAV24 AJ49) and PYNNNDM (TRAV24 AJ43), were co-expressed with CDR3β sequences that utilized TRBV6−5 and TRBV27, respectively (23), as observed in donors Y2 and E4 (Table 1).
Nonproductively rearranged CDR3α transcripts (Table S4) were found at a low prevalence (6% of all responses) in four donors (Y1, Y2, E3, and E4). This is consistent with data from murine models demonstrating that, although high frequencies of nonproductively rearranged CDR3α transcripts are found after primary infection, such clonotypes are lost with repeated antigenic stimulations (18, 26), as occurs in persistent CMV infection.
High prevalence of public motif usage in CMV-specific CDR3αβ repertoire
The simultaneous amplification of CDR3αβ sequences allowed us to identify several important features of the CMV-NLV TCR repertoire, including the accurate definitions of complete CDR3αβ clonotypes and their in vivo prevalences. We systematically catalogued seven public CDR3α and six public CDR3β motifs, distinct patterns of amino-acid usage (Table 2; a comprehensive list of sequences whence the motifs were identified is provided in Table S5), a broader range than found hitherto (21–25, 27). These motifs, some of which include preferential J region usage, can account for approximately 70% of the epitope-specific response (Fig. 2A). Of all the CMV-NLV-specific responses analyzed here, 53% are characterized by the usage of public CDR3α motifs, 55% by public CDR3β motifs, 71% by either CDR3α or CDR3β motifs, and 38% by the usage of both CDR3α and CDR3β motifs. Public CDR3α motifs commonly (71%) pair with public CDR3β motifs, as do public CDR3β motifs with public CDR3α motifs (68%).
Table 2.
Public motifs that emerged among donors in this and previous reports of CMV-NLV-specific TCR repertoire.
| Motif No. | Public CDR3α motif | Donors and previous reports in which public motif was found | Prevalence among all responses in this study (%) |
|---|---|---|---|
| 1 | *nGNQF | Y2, E3, E4, Trautmann, et al. (23), Day, et al. (25) | 14.1 |
| 2 | *nYGQNF | Y1, E2, Trautmann, et al. (23), Day, et al. (25) | 2.1 |
| 3 | *nNNNDM | Y2, E2, E4, Trautmann, et al. (23) | 10.5 |
| 4 | *nNFGNEKL | E2, Trautmann, et al. (23) | 2.1 |
| 5 | *nTGANNL | Y1, E2, Trautmann, et al. (23) | 1.6 |
| 6 | *nTGGGNKL | Y1, E4 | 5.1 |
| 7 | *nTSYDKV | E2, Trautmann, et al. (23) | 1.6 |
|
| |||
| Public CDR3β motif | |||
|
| |||
| 8 | S*nTG*nGY | Y2, E1, E2, E4, Weekes, et al. (21), Trautmann, et al. (23), Price, et al. (24), Day, et al. (25), Khan, et al. (22) | 13.0 |
| 9 | S*nTEA | Y1, E2, E4, Weekes, et al. (21), Trautmann, et al. (23), Price, et al. (24), Day, et al. (25), Khan, et al. (22), Venturi, et al. (27) | 11.7 |
| 10 | *nTDTQ | Y1, Trautmann, et al. (23), Price, et al. (24), Day, et al. (25) | 4.3 |
| 11 | *nRD*nG*YEQ | E3, Day, et al. (25), Khan, et al. (22) | 9.7 |
| 12 | *nSYEQ | Y1, E2, Weekes, et al. (21), Price, et al. (24), Khan, et al. (22) | 1.5 |
| 13 | *nTSGALYNEQ | E3, Trautmann, et al. (23) | 6.1 |
*n denotes a variable region of the public motif consisting of n amino acids, where n can range from 1 to 10. Percentages in this table are derived from all responses (N = 297 TCR sequences), regardless of whether TCRα and β co-expression data are available.
Fig. 2.
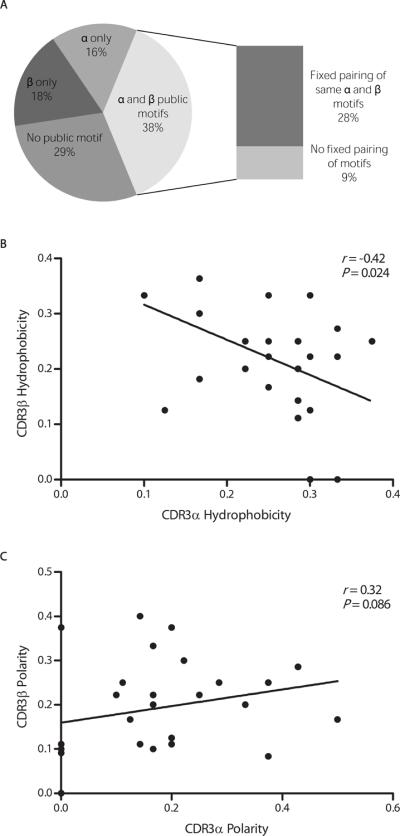
Relationships between paired CDR3α and CDR3β sequences in CMV-NLV-specific CD8 T-cell repertoire. (A) Prevalence of CDR3α and CDR3β public motif usage among all responses. Percentages were derived from paired CDR3αβ data (N = 135 TCR sequences). (B) and (C) Relationships between hydrophobicity (B) or polarity (C) values within paired CDR3α and CDR3β amino acid sequences. Hydrophobicity and polarity are defined in the text. In (B) and (C), each symbol represents one CDR3αβ pair. Correlation was assessed using the Spearman's rank correlation coefficient. Percentages do not total 100% due to rounding error.
Only a contemporaneous TCRαβ analysis allows the identification of TCRαβ pairing rules. Thus far, we have considered the prevalence of pairing for any public CDR3α with any public CDR3β. However, we also observed consistently repeated instances of the same public CDR3α pairing with the same public CDR3β, a profile we tentatively characterize as a “dual public clonotype.” For instance, the public CDR3α sequence, NTGNQF, is always paired with the public β motif, S*nTG*nGY (where n represents a variable number of amino acids), among all donors in the present and in previous studies (23, 25) (Table S5). Another public CDR3α sequence, AFGNQF, was consistently associated with the same public CDR3β motif, *nTSGALYNEQ, in this and a previous study (23). The public CDR3α sequence, PYNNNDM, paired repeatedly with the public CDR3β motif, S*nTEA, in this and a previous study (23). In all, approximately 28% of the CMV-NLV-specific responses are characterized by a dual public CDR3αβ motif (Fig. 2A). Taken together, these data indicate equal contributions by the CDR3α and CDR3β in shaping this CMV-NLV-specific T-cell response.
Beyond these TCRαβ public motif pairings, further insights into pairing rules were probed at the ultrastructural level. We systematically enumerated polar and hydrophobic (relative to glycine) amino acids in all of the paired CDR3αβ sequences in this study and generated “normalized polarity” and “hydrophobicity” values for each sequence by dividing by the total number of amino acids in each respective sequence. What we found was that the hydrophobicities of the CDR3α and CDR3β sequences were negatively correlated to a moderate extent (Spearman r = −0.42, P = 0.024; Fig. 2B). That is, a lower CDR3α hydrophobicity was balanced by a higher CDR3β hydrophobicity, and vice versa. In contrast, such an inverse relationship was not definitively observed for CDR3αβ amino acid polarity values, which were comparable within CDR3αβ pairs (Fig. 2C). Both hydrogen bonds and van der Waals forces contribute to the interaction between the TCR and the CMV-NLV epitope (28). Taken together, our data suggest that the totality of hydrophobic contacts within the CDR3αβ region is a determinant of optimal CDRαβ pairing for this epitope-specific response that is more tightly regulated than polar contacts.
Estimating TCR repertoire diversity
Our single-cell analysis of the CMV-NLV-specific TCRαβ repertoire thus far has provided insights into the selection and pairing of CDR3αβ sequences in a persistent human infection. Although there are suggestions that TCR repertoire diversity is important for human anti-microbial defense (29), direct evidence supporting this hypothesis has been both limited and hard to generate. Therefore, we next focused on determining a useful, quantitative measure of pathogen-specific TCRαβ repertoire diversity for this continuing, in vivo CTL response.
Simpson's Diversity Index (SDI), a summary statistic often applied to repertoire analysis, provides a relative measure of the richness and evenness of a population. Borrowed from ecology, the SDI is both less sample-size-dependent than other measures and more applicable to the comparison of diversities in samples with smaller sample-to-total population size ratios (30). These properties are particularly important when it comes to the assessment of human TCR repertoires, where only a very small fraction can be sampled from the total PBMC and tissue-located CTL pool. Thus far, the use of SDI to characterize the constitution of antigen-specific TCR repertoires has focused mainly on probing CDR3β profiles (31).
With other pathogens, the experience so far has been that either the TCRα or the TCRβ chain can undergo more restrictive selection, whereas the other chain is less constrained (32). A key determinant of such a biased selection is thought to be the dominance of one TCR chain over the other in making contacts with the foreign peptide (33). It is thus important to probe both CDR3α and CDR3β repertoires concurrently for single TCRαβ pairs. The present analysis allows us to characterize repertoire diversity of TCRαβ clonotypes as they occur in nature. In addition, we are able to ask whether similar diversities are found between the CDR3α and CDR3β profiles, or whether one compartment is more restricted than the other.
Summarized in Table 3, the SDIs for each donor confirm preliminary interpretations from a visual survey of the spectrum of repertoire diversity (Table 1): The highest diversity was found in the TCR repertoire of donor Y1, which comprised many clonotypes of approximately equal frequency, whereas the lowest diversity was that found for donor E1, who had one dominant clonotype and a rare clonotype for both the CDR3α and CDR3β. In general, there was good concordance in the extent of CDR3α and CDR3β diversity within individuals. With two individuals, donor Y2 and E4, the spectrum of CDR3α and CDR3β diversities showed evidence of greater discordance, but there was no consistency for greater diversity in one or the other of TCRα and TCRβ chains. These data suggest that neither CDR3α nor CDR3β is more selectively restricted in the human CMV-NLV response.
Table 3.
Simpson's Diversity Index (SDI) for each donor's CMV-NLV-specific CD8+ TCR repertoire.
| Simpsons's Diversity Index | |||
|---|---|---|---|
|
|
|||
| Donor | CDR3αβ | CDR3α | CDR3β |
| Y1 | 1 | 0.98 | 0.97 |
| Y2 | 0.47 | 0.28 | 0.55 |
| E1 | 0 | 0.14 | 0.065 |
| E2 | 0.94 | 0.97 | 0.93 |
| E3 | 0.45 | 0.46 | 0.53 |
| E4 | 0.63 | 0.81 | 0.33 |
, where c is the number of different clonoptypes in each TCR repertoire sample, ni is the clone size of the ith clonotype, and n is the total number of TCR sequences sampled. The SDI ranges from 0 to 1, representing the minimal and maximal diversity, respectively.
TCR repertoire diversity, CTL response magnitude, and circulating antibody levels
Circulating CMV-specific antibody levels correlate with viral loads in cancer patients receiving chemotherapy (34) and are associated with differential mortality risks in immunocompetent older adults (35–37). Therefore, we next examined the relationship between pathogen-specific TCR repertoire diversity and the circulating CMV-specific IgG concentration, using contemporaneously obtained blood specimens. Strikingly, a strong inverse correlation was found between CMV-NLV-specific CDR3αβ repertoire diversity and the serum anti-CMV IgG concentration (Spearman r = −1.0, P = 0.0028; Fig. 3A). Similar, but less robust, trends were observed when CDR3α and CDR3β repertoire diversities were examined separately, relative to the anti-CMV IgG concentration (Spearman r = −0.94, P = 0.017 and r = −0.83, P = 0.058, respectively; Fig. 3B).
Fig. 3.
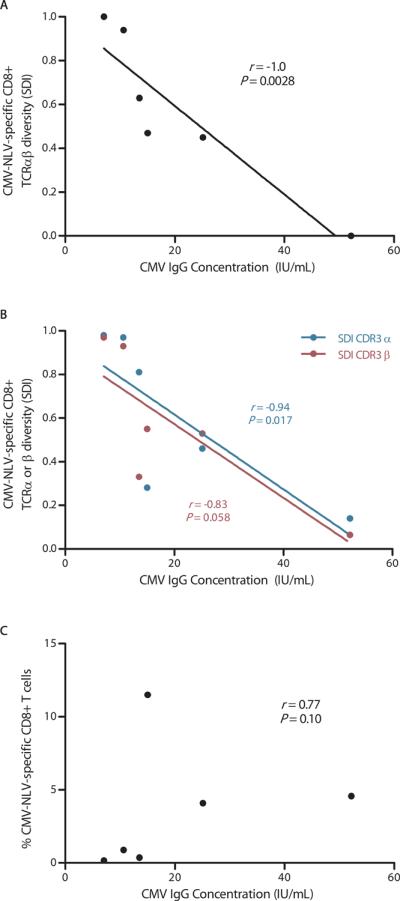
Relationship between CMV-NLV-specific CTL response and serum anti-CMV IgG concentration. (A) Relationship between CMV-NLV-specific CD8+ TCRαβ repertoire diversity, expressed as the Simpson's Diversity Index (SDI; see text and footnote to Table 2), and CMV IgG concentration. (B) Examination of the CMV-NLV-specific CD8+ TCRα and TCRβ repertoire diversities, as separate variables, in relationship with the CMV IgG concentration. (C) Relationship between the magnitude of the CTL response, measured as the frequency of CMV-NLV pMCHI tetramer+ CD8+ T cells (as percentage of all CD8+ T cells), and the CMV IgG concentration. Correlation was assessed using the Spearman's rank correlation coefficient. Each TCR repertoire was measured in 1 experiment per donor. CMV IgG concentration was measured in duplicates.
Because of this inverse association between CMV-specific TCRαβ repertoire diversity and the virus-specific IgG level, we next asked whether the CTL response magnitude showed any obvious relationship to the virus-specific antibody response. Among all donors, we found that the size of the CMV-NLV-specific CD8+ CTL response, determined by the percentage of CD8+ T cells binding the CMV-NLV pMHCI tetramers, bore no obvious relationship to serum anti-CMV IgG concentrations (Fig. 3C). Thus, if we regard CMV-specific antibody titers as a surrogate for virus reactivation, the numbers of circulating epitope-specific CTLs do not obviously correlate in any way with variations in CMV antigen load. It is well known from studies in experimental systems (1) that there is no necessary relationship between TCR diversity and CD8 T cell response magnitude.
Although circulating CMV viral load is routinely quantifiable in immunocompromised persons as a measure of virus reactivation (38), CMV reactivation is less readily detectable in immunocompetent individuals (39). Using quantitative PCR, we measured CMV DNA in plasma contemporaneous with serum CMV-specific IgG in our study participants. Although no evidence of virus presence was found for the three donors with the lowest anti-CMV IgG levels, the CMV viral load was > 300 copies/mL in the three donors with the highest antibody levels (Fig. S3A). To further probe this dichotomy, we measured plasma CMV loads in 24 immunocompetent adults and found that serum CMV IgG concentrations were generally higher in virus carriers with detectable CMV than in those where we found no CMV DNA (P = 0.011; Fig. S3B). Taken together, our data suggest that CMV-specific CD8+ TCR repertoire diversity is more important than CD8+ T cell response magnitude for the control of persistent CMV infection.
DISCUSSION
Although CMV is known to reactivate under circumstances of immunodeficiency or immunosuppression, as when a concurrent HIV infection progresses to AIDS or in post-transplant settings (40), evidence of any such reactivation is, as confirmed here, seldom detectable in immunocompetent carriers (39). Thus, an alternative measure of CMV antigen load is needed for monitoring clinically normal, persistently infected virus carriers. We and others have recently found a direct correlation between CMV-specific antibody levels and differential morbidity and mortality risks in older adults (35–37). It thus seems reasonable that heightened levels of CMV-specific circulating antibodies may reflect suboptimal CTL-mediated immune control, with IgG titers functioning, in effect, as a surrogate measure of virus reactivation (34).
In general, a varied TCR repertoire is believed to mediate the optimal control of pathogens by increasing the probability that microbial escape mutations might still be recognized by one or other TCRαβ pair (7, 8). Greater diversity is also thought to facilitate the selection of high-avidity CTLs that are more effective players in immune defense (5, 10), although this association between TCR avidity and repertoire diversity is not a consistent finding (41). Because the CMV-NLV pp65 epitope is relatively stable, with rare mutations found in clinical samples (42), any advantage conferred by greater breadth of the CMV-NLV-specific TCR repertoire is unlikely to reflect protection from viral escape. Although a diverse TCR repertoire can in principle enhance functional heterogeneity and thus pathogen control, detailed analysis in mice infected with readily eliminated influenza A virus has not established any clear association between TCR repertoire diversity and various measures of the cognate pMHC responsiveness, including cytotoxicity and cytokine production (41, 43). Indeed, functional heterogeneity can be generated from a single antigen-specific T cell clone in a process that appears to be stochastic, and thus not related to the involvement of a spectrum of TCRs (44).
The situation with larger and more complex persistent viruses may, however, be very different. Using a single-cell, direct ex vivo approach to characterize the spectrum of TCRαβ repertoires of human CMV-specific CD8+ T cells, we found that virus-specific CTL TCRαβ repertoire diversity correlates inversely with CMV-specific antibody levels, an observation consistent with the idea that CMV reactivation events (and thus antigen load) are more effectively limited by CTLs expressing a diversity of TCRαβ pairs. This observation is especially noteworthy in view of the lack of association between response magnitude for CMV-NLV-specific CTLs and anti-CMV IgG level. Although CMV has evolved molecular mechanisms for subverting class I MHC antigen presentation (45), this immune evasion strategy does not function to prevent the generation of a diverse CTL response in natural infections (46). Presumably this reflects the need to maintain a balance, with CTLs helping to control persistent CMV (47) while allowing the host to survive and transmit the infection.
The protection afforded by the human CMV-specific CD8+ T cell response has been formally demonstrated by the adoptive transfer of in vitro-expanded CTLs into immunosuppressed, transplant recipients who failed antiviral chemotherapy (20). Although indications from studies involving immunosuppressed patients suggest that virus clearance may depend on achieving threshold numbers of antigen-specific CD8+ T cells (47), variations in CTL response magnitude beyond that baseline may be less important than TCR diversity when it comes to pathogen control, particularly in immunocompetent humans. Our data suggest that a potential key, but as yet unmeasured, ingredient predictive of success in such immunotherapeutic strategies may be the diversity of the infused TCR repertoire (48), an issue that would seem to merit future analysis. Also, although high TCR/pMHCI avidity profiles are thought by some to be optimal for virus control, the molecular pathways that elicit cytotoxic effector function can evidently be triggered via a weaker TCR signal than that required for cytokine production (12). Again TCR diversity is likely to contribute importantly, with TCR/pMHCI avidity alone not being the best predictor of CTL-mediated effector function.
An alternative interpretation of our data is that chronic antigen load leads to a narrowing of the TCR repertoire, as it concurrently increases the antibody level. This is suggested by earlier findings (derived from cloned T-cell lines), indicating that the CMV-specific TCR repertoire is less diverse in immunosuppressed patients (23). However, long-term immunosuppression could be associated with a contraction of the TCR repertoire (49). Indeed, in immunocompetent individuals, despite a tendency for limited clonotype dominance after CMV primary infection (25), the CMV-specific TCRβ repertoire appears to be stable at least over a two-year follow-up (50). This stability suggests that the repertoire structure is maintained despite potential fluctuations in viral load. Thus, it seems more likely that the repertoire profile is the predictor, rather than the outcome, variable in our data. Still, it would be worthwhile to examine the stability of CMV repertoire profiles in the same individuals over an extended time scale (>10 years) and spanning the period when potential CMV-related morbidities may arise (>65 years of age). Pairing detailed repertoire analysis with changes in antigen-specific T cell activation phenotypes might also contribute to our understanding of the relationship between TCR diversity and viral control.
Careful analysis of the CMV-specific CD8+ TCRαβ repertoire structure provided additional insights into the selection of this antigen-specific CTL response to a persistent, ubiquitous pathogen. We observed a high prevalence of public CDR3αβ motifs, together with a substantial minority of consistently paired public CDR3α and CDR3β motifs, and an inverse correlation in the hydrophobicity of the paired TCRα and β chains. Indeed, to our knowledge, the present analysis provides the first human epitope-specific CD8+ TCR CDR3αβ repertoire that reflects in vivo clonotype prevalence and diversity, for any pathogen-specific (or other) T cell response. In contrast to the discordant usage of TCRα and TCRβ chains observed in other instances of epitope-specific responses (32), we found comparable repertoire metrics between CDR3α and CDR3β for these CMV-NLV-specific CTLs. This observation, together with the relatively considerable percentage of dual public clonotypes, suggests that both CDR3 counterparts contribute equally to pMHC contact.
To date, this strategy for human antigen-specific TCRαβ repertoire analysis has only been applied to peripheral blood lymphocyte populations, though it has potential application for the analysis of both naïve and immune T cell sets recovered in samples as diverse as cord blood and cancer biopsies. Assaying T cell populations directly ex vivo in this way will, for example, allow us to ask just how representative the clonally expanded (in culture) CTLs used in immunotherapy protocols are when compared with the natural host response in T cell donors. Such single-cell analysis also facilitates the precise, longitudinal tracking of human antigen-specific T-cell clonotypes from the acute response into long-term memory. This allows us to monitor profiles of clonotype fate and involvement for T-cell responses to both persistent and acute infections, and in autoimmune disease (51), and also permits a much more defined analysis of how therapeutic interventions may modify cell-mediated immunity.
In conclusion, the single-cell-based approach to TCR repertoire analysis described here enables detailed characterization of epitope-specific CD8+ T-cell clonotypes as they persist in vivo and offers important insights into the role, and relative importance of pathogen-specific TCR repertoire diversity in microbial defense. The analysis also provides insight into the selection of TCRαβ public motifs in a persistent viral infection, and gives baseline information for the further structural analysis of human TCRαβ pairing. Furthermore, the availability of this analytical system will facilitate much more defined assessments of the ways that T-cell repertoire diversity determines both immune competence and immunodominance, while enabling the tracking of individual T cells during immunotherapy applications (including vaccination strategies), in the pathogenesis of disease, and in long-term memory.
MATERIALS AND METHODS
Subjects and PBMC samples
The study protocols were approved by the institutional review boards of Johns Hopkins University, Baltimore, MD, and St. Jude Children's Research Hospital, Memphis, TN. Written informed consent was obtained from all study participants. Heparinized venous whole blood was obtained peripherally from immunocompetent individuals not taking immunomodulatory pharmaceutical agents. PBMCs were isolated via density gradient centrifugation (GE Healthcare Ficoll-Paque PLUS) and cryopreserved in liquid nitrogen until the time of analysis. HLA typing was performed by the Johns Hopkins University Immunogenetics Laboratory (Supplementary Methods).
Design and synthesis of TCR segment-specific primers
Nucleotide sequences of TCR Vα (TRAV), Cα (TRAC), Vβ (TRBV), and Cα (TRBC) genes were retrieved from IMGT®, the international ImMunoGeneTics information system®, http://www.imgt.org (19). A total of 47 human TRAV, 1 TRAC, 54 TRBV, and 2 TRBC functional and open-reading-frame genes were identified from the IMGT database. TRAC and TRBC genes were separately grouped into related families by phylogenetic analysis. Primers of 17–23 bp length were designed to target each family of related V genes and the TRAC and TRBC genes, allowing for up to three degenerate base pairs. Primers were synthesized by standard phosphoramidite chemistry on Applied Biosystems 3900 DNA synthesizers using Applied Biosytems reagents. A total of 40 external/internal pairs of sense TRAV, 27 sense TRBV, and 1 each of antisense TRAC and TRBC gene segment-specific primers were generated (Table S1). For inclusion in nested PCRs, TRAV and TRBV primers were multiplexed to a concentration of 5 μM for each primer, and TRAC and TRBC primers were reconstituted to a concentration of 20 μM. Primer validation is described in Supplementary Methods.
Isolation of single CMV-NLV-specific CD8+ T cells
After staining with fluorochrome-conjugated monoclonal antibodies (Supplementary Methods), single CD14-CD3+CD8+tetramer+ cells were sorted directly into a 96-well PCR plate (Eppendorf) using a MoFlo flow cytometer, fitted with a Cyclone single-cell deposition unit. Negative controls (no actual cell sorted) were interspersed between the samples (1 in 10).
RT PCR, multiplex nested PCR, and sequencing
After sorting, plates were stored at −80 °C until downstream processing. cDNA was synthesized from single cells in PCR plates using the iScript cDNA Synthesis Kit (Bio-Rad) in 2.5-μL reaction mixes, each containing 0.5 μL of 5× iScript reaction mix, 0.5 μL of iScript reverse transcriptase, and 0.1% Triton X-100 (Sigma) that were incubated at 25 °C for 5 min, 42 °C for 30 min, and 80 °C for 5 min. TCR transcripts from each cell were amplified by multiplex nested PCR in 25-μL reaction mixes containing 2.5 μL of cDNA. The first-round PCR was performed with 0.75 units of Taq DNA polymerase (Invitrogen), 2.5 μL of 10x PCR Buffer (Invitrogen) (containing KCl, (NH4)2SO4, and 15 mM MgCl2), 0.5 μL of 10 mM dNTP (Invitrogen), 2.5 pmol each of the external sense TRAV and TRBV primers, and 10 pmol each of the external antisense TRAC and TRBC primers listed in Table S1. Instead of 10× PCR buffer, the second-round PCR utilized CoralLoad PCR Buffer (Invitrogen) containing 2 marker dyes for gel loading. 2.5-μL aliquots of the first-round PCR products served as templates for 2 separate second-round PCRs that incorporated, respectively, either (i) internal sense TRAV primers and internal antisense
TRAC primer or (ii) internal sense TRBV primers and internal antisense TRBC primer, listed in Table S1. The PCR conditions were 95 °C for 2 min, followed by 35 cycles of 95 °C for 20 sec, 52 °C for 20 sec, and 72 °C for 45 sec, followed by 1 cycle of 72 °C for 7 min. PCR products were purified and sequenced with the relevant TRAC or TRBC primer (Supplementary Methods).
CMV IgG and viral load measurement
CMV IgG concentrations were determined in stored (−70° C) serum, using a commercial enzyme-linked immunosorbent assay (ELISA) kit (GenWay Biotech, San Diego, CA) (Supplementary Methods). Each specimen was tested in duplicate and the optical density was calibrated against four standard reference specimens to give the quantitative IgG concentration. The intra-assay coefficient of variation was 2.8%. All assays were performed in a masked fashion. CMV viral load was determined using quantitative PCR (Supplementary Methods).
Statistical analysis
Spearman's rank correlation coefficients were calculated using Prism 5 software. Two-group comparison of antibody levels was done using the nonparametric Wilcoxon rank-sum test. All P values were derived from two-tailed tests; a value of less than 0.05 was considered significant. Prevalences of TCR sequences and motifs among all responses were derived from TCR clone sizes that were normalized within each study participant (expressed as percentages that totaled 100% within each individual).
Supplementary Material
Acknowledgments
We thank M. Leffell for assistance with HLA typing, and A. Valsamakis and M. Forman for technical guidance in CMV viral load measurement.
Funding: Supported by NIH-National Institute of Allergy and Infectious Diseases (NIAID) grants AI70251 (P.C.D.) and AI077714 (P.G.T.); the American Syrian Lebanese Associated Charities (P.C.D., P.G.T.); NIH-National Institute on Aging grants AG033113 (G.C.W.) and AG021334, through the Johns Hopkins Older Americans Independence Center (G.C.W.); Atlantic Philanthropies, American Geriatrics Society, the John A. Hartford Foundation, and the Association of Subspecialty Professors (T. Franklin Williams Research Scholars Award, G.C.W.); and the Johns Hopkins Biology of Healthy Aging Program (G.C.W.).
Footnotes
Author contributions: G.C.W., P.D., J.A.M., P.C.D., and P.G.T. designed the study; G.C.W. did the experiments and analyzed the data, with supervision from P.D., J.A.M., and P.G.T.; G.C.W. and P.C.D. wrote the manuscript, with all authors contributing, reviewing, and approving the final version.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Turner SJ, La Gruta NL, Kedzierska K, Thomas PG, Doherty PC. Functional implications of T cell receptor diversity. Curr Opin Immunol. 2009;21:286–290. doi: 10.1016/j.coi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen JL, Esser U, Fazekas de St Groth B, Reay PA, Davis MM. Mapping T-cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 3.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell TK, Alt FW. Molecular characterization of the lymphoid V(D)J recombination activity. J Biol Chem. 1989;264:10327–10330. [PubMed] [Google Scholar]
- 5.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 6.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 7.Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, Selin LK. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116:1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derby M, Alexander-Miller M, Tse R, Berzofsky J. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J Immunol. 2001;166:1690–1697. doi: 10.4049/jimmunol.166.3.1690. [DOI] [PubMed] [Google Scholar]
- 10.Kedzierska K, La Gruta NL, Davenport MP, Turner SJ, Doherty PC. Contribution of T cell receptor affinity to overall avidity for virus-specific CD8+ T cell responses. Proc Natl Acad Sci U S A. 2005;102:11432–11437. doi: 10.1073/pnas.0504851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valitutti S, Muller S, Dessing M, Lanzavecchia A. Different responses are elicited in cytotoxic T lymphocytes by different levels of T cell receptor occupancy. J Exp Med. 1996;183:1917–1921. doi: 10.1084/jem.183.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faroudi M, Utzny C, Salio M, Cerundolo V, Guiraud M, Muller S, Valitutti S. Lytic versus stimulatory synapse in cytotoxic T lymphocyte/target cell interaction: manifestation of a dual activation threshold. Proc Natl Acad Sci U S A. 2003;100:14145–14150. doi: 10.1073/pnas.2334336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 14.Valmori D, Dutoit V, Lienard D, Lejeune F, Speiser D, Rimoldi D, Cerundolo V, Dietrich PY, Cerottini JC, Romero P. Tetramer-guided analysis of TCR beta-chain usage reveals a large repertoire of melan-A-specific CD8+ T cells in melanoma patients. J Immunol. 2000;165:533–538. doi: 10.4049/jimmunol.165.1.533. [DOI] [PubMed] [Google Scholar]
- 15.Pannetier C, Even J, Kourilsky P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 16.Bousso P, Casrouge A, Altman JD, Haury M, Kanellopoulos J, Abastado JP, Kourilsky P. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity. 1998;9:169–178. doi: 10.1016/s1074-7613(00)80599-3. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr., Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr., Davis RW, Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dash P, McClaren JL, Oguin TH, 3rd, Rothwell W, Todd B, Morris MY, Becksfort J, Reynolds C, Brown SA, Doherty PC, Thomas PG. Paired analysis of TCRalpha and TCRbeta chains at the single-cell level in mice. J Clin Invest. 2011;121:288–295. doi: 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Einsele H, Roosnek E, Rufer N, Sinzger C, Riegler S, Loffler J, Grigoleit U, Moris A, Rammensee HG, Kanz L, Kleihauer A, Frank F, Jahn G, Hebart H. Infusion of cytomegalovirus (CMV)-specific T cells for the treatment of CMV infection not responding to antiviral chemotherapy. Blood. 2002;99:3916–3922. doi: 10.1182/blood.v99.11.3916. [DOI] [PubMed] [Google Scholar]
- 21.Weekes MP, Wills MR, Mynard K, Carmichael AJ, Sissons JG. The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J Virol. 1999;73:2099–2108. doi: 10.1128/jvi.73.3.2099-2108.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PA. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol. 2002;169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 23.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, Bonneville M. Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 24.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day EK, Carmichael AJ, ten Berge IJ, Waller EC, Sissons JG, Wills MR. Rapid CD8+ T cell repertoire focusing and selection of high-affinity clones into memory following primary infection with a persistent human virus: human cytomegalovirus. J Immunol. 2007;179:3203–3213. doi: 10.4049/jimmunol.179.5.3203. [DOI] [PubMed] [Google Scholar]
- 26. In unpublished data, we observed low frequencies of murine epitope-specific T cells expressing a nonproductively rearranged TCRα transcript with repeated antigenic stimulations.
- 27.Venturi V, Chin HY, Asher TE, Ladell K, Scheinberg P, Bornstein E, van Bockel D, Kelleher AD, Douek DC, Price DA, Davenport MP. TCR beta-chain sharing in human CD8+ T cell responses to cytomegalovirus and EBV. J Immunol. 2008;181:7853–7862. doi: 10.4049/jimmunol.181.11.7853. [DOI] [PubMed] [Google Scholar]
- 28.Gras S, Saulquin X, Reiser JB, Debeaupuis E, Echasserieau K, Kissenpfennig A, Legoux F, Chouquet A, Le Gorrec M, Machillot P, Neveu B, Thielens N, Malissen B, Bonneville M, Housset D. Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope. J Immunol. 2009;183:430–437. doi: 10.4049/jimmunol.0900556. [DOI] [PubMed] [Google Scholar]
- 29.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 30.Magurran AE. Measuring biological diversity. Blackwell Science; Oxford: 2004. [Google Scholar]
- 31.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321:182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Zhong W, Dixit SB, Mallis RJ, Arthanari H, Lugovskoy AA, Beveridge DL, Wagner G, Reinherz EL. CTL recognition of a protective immunodominant influenza A virus nucleoprotein epitope utilizes a highly restricted Vbeta but diverse Valpha repertoire: functional and structural implications. J Mol Biol. 2007;372:535–548. doi: 10.1016/j.jmb.2007.06.057. [DOI] [PubMed] [Google Scholar]
- 33.Day EB, Guillonneau C, Gras S, La Gruta NL, Vignali DA, Doherty PC, Purcell AW, Rossjohn J, Turner SJ. Structural basis for enabling T-cell receptor diversity within biased virus-specific CD8+ T-cell responses. Proc Natl Acad Sci U S A. 2011;108:9536–9541. doi: 10.1073/pnas.1106851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo CP, Wu CL, Ho HT, Chen CG, Liu SI, Lu YT. Detection of cytomegalovirus reactivation in cancer patients receiving chemotherapy. Clin Microbiol Infect. 2008;14:221–227. doi: 10.1111/j.1469-0691.2007.01895.x. [DOI] [PubMed] [Google Scholar]
- 35.Strandberg TE, Pitkala KH, Tilvis RS. Cytomegalovirus antibody level and mortality among community-dwelling older adults with stable cardiovascular disease. Jama. 2009;301:380–382. doi: 10.1001/jama.2009.4. [DOI] [PubMed] [Google Scholar]
- 36.Wang GC, Kao WH, Murakami P, Xue QL, Chiou RB, Detrick B, McDyer JF, Semba RD, Casolaro V, Walston JD, Fried LP. Cytomegalovirus infection and the risk of mortality and frailty in older women: a prospective observational cohort study. Am J Epidemiol. 2010;171:1144–1152. doi: 10.1093/aje/kwq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts ET, Haan MN, Dowd JB, Aiello AE. Cytomegalovirus Antibody Levels, Inflammation, and Mortality Among Elderly Latinos Over 9 Years of Follow-up. Am J Epidemiol. 2010;172:363–371. doi: 10.1093/aje/kwq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Einsele H, Ehninger G, Hebart H, Wittkowski KM, Schuler U, Jahn G, Mackes P, Herter M, Klingebiel T, Loffler J, Wagner S, Muller CA. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood. 1995;86:2815–2820. [PubMed] [Google Scholar]
- 39.Ling PD, Lednicky JA, Keitel WA, Poston DG, White ZS, Peng R, Liu Z, Mehta SK, Pierson DL, Rooney CM, Vilchez RA, Smith EO, Butel JS. The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: a 14-month longitudinal study. J Infect Dis. 2003;187:1571–1580. doi: 10.1086/374739. [DOI] [PubMed] [Google Scholar]
- 40.Gandhi MK, Khanna R. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. The Lancet infectious diseases. 2004;4:725–738. doi: 10.1016/S1473-3099(04)01202-2. [DOI] [PubMed] [Google Scholar]
- 41.La Gruta NL, Thomas PG, Webb AI, Dunstone MA, Cukalac T, Doherty PC, Purcell AW, Rossjohn J, Turner SJ. Epitope-specific TCRbeta repertoire diversity imparts no functional advantage on the CD8+ T cell response to cognate viral peptides. Proc Natl Acad Sci U S A. 2008;105:2034–2039. doi: 10.1073/pnas.0711682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaia JA, Gallez-Hawkins G, Li X, Yao ZQ, Lomeli N, Molinder K, La Rosa C, Diamond DJ. Infrequent occurrence of natural mutations in the pp65(495–503) epitope sequence presented by the HLA A*0201 allele among human cytomegalovirus isolates. J Virol. 2001;75:2472–2474. doi: 10.1128/JVI.75.5.2472-2474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moffat JM, Handel A, Doherty PC, Turner SJ, Thomas PG, La Gruta NL. Influenza epitope-specific CD8+ T cell avidity, but not cytokine polyfunctionality, can be determined by TCRbeta clonotype. J Immunol. 2010;185:6850–6856. doi: 10.4049/jimmunol.1002025. [DOI] [PubMed] [Google Scholar]
- 44.Stemberger C, Huster KM, Koffler M, Anderl F, Schiemann M, Wagner H, Busch DH. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Ahn K, Gruhler A, Galocha B, Jones TR, Wiertz EJ, Ploegh HL, Peterson PA, Yang Y, Fruh K. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity. 1997;6:613–621. doi: 10.1016/s1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 46.Manley TJ, Luy L, Jones T, Boeckh M, Mutimer H, Riddell SR. Immune evasion proteins of human cytomegalovirus do not prevent a diverse CD8+ cytotoxic T-cell response in natural infection. Blood. 2004;104:1075–1082. doi: 10.1182/blood-2003-06-1937. [DOI] [PubMed] [Google Scholar]
- 47.Cwynarski K, Ainsworth J, Cobbold M, Wagner S, Mahendra P, Apperley J, Goldman J, Craddock C, Moss PA. Direct visualization of cytomegalovirus-specific T-cell reconstitution after allogeneic stem cell transplantation. Blood. 2001;97:1232–1240. doi: 10.1182/blood.v97.5.1232. [DOI] [PubMed] [Google Scholar]
- 48.Moss P, Rickinson A. Cellular immunotherapy for viral infection after HSC transplantation. Nat Rev Immunol. 2005;5:9–20. doi: 10.1038/nri1526. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez CM, Opelz G, Giraldo MC, Pelzl S, Renner F, Weimer R, Schmidt J, Arbelaez M, Garcia LF, Susal C. Evaluation of T-cell receptor repertoires in patients with long-term renal allograft survival. Am J Transplant. 2005;5:746–756. doi: 10.1111/j.1600-6143.2005.00756.x. [DOI] [PubMed] [Google Scholar]
- 50.Hadrup SR, Strindhall J, Kollgaard T, Seremet T, Johansson B, Pawelec G, thor Straten P, Wikby A. Longitudinal studies of clonally expanded CD8 T cells reveal a repertoire shrinkage predicting mortality and an increased number of dysfunctional cytomegalovirus-specific T cells in the very elderly. J Immunol. 2006;176:2645–2653. doi: 10.4049/jimmunol.176.4.2645. [DOI] [PubMed] [Google Scholar]
- 51.Menezes JS, van den Elzen P, Thornes J, Huffman D, Droin NM, Maverakis E, Sercarz EE. A public T cell clonotype within a heterogeneous autoreactive repertoire is dominant in driving EAE. J Clin Invest. 2007;117:2176–2185. doi: 10.1172/JCI28277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez JL, Storch GA. Multiplex, quantitative, real-time PCR assay for cytomegalovirus and human DNA. J Clin Microbiol. 2002;40:2381–2386. doi: 10.1128/JCM.40.7.2381-2386.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.