Abstract
Alternative splicing (AS) is the primary mechanism by which a limited number of protein coding genes can generate the proteome diversity. We have investigated the role of an alternative splicing factor (ASF), Sfrs1, an arginine/serine (SR) rich-protein family member, during retinal development. Here we report that loss of Sfrs1 function during embryonic retinal development had a profound effect such that it led to a small retina at birth. In addition, the retina underwent further degeneration in the postnatal period. Loss of Sfrs1 function resulted in the death of retinal neurons that were born during early and mid-embryonic development. Ganglion cells, cone photoreceptors, horizontal cells and amacrine cells were produced and initiated differentiation. However, these neurons subsequently underwent cell death through apoptosis. In contrast, Sfrs1 was not required for the survival of the neurons generated later, including later born amacrine cells, rod photoreceptors, bipolar cells and Müller glia. Our results highlight the requirement of Sfrs1-mediated AS for the survival of retinal neurons, with sensitivity defined by the window of time in which the neuron was generated. In all, this is the first description addressing the function of an ASF in vertebrate retinal development.
Introduction
As the genome of different organisms gets sequenced and annotated it is becoming apparent that the complexity of an organism does not depend on the total number of protein coding genes. For example, the current estimate for the number of protein coding genes in the human genome is closer to that observed in mouse (~23,049) (http://www.ensembl.org/Mus_musculus/index.html) which is similar to that observed in Arabidopsis thaliana (Seki et al., 2002). Thus, it has been proposed that the complexity of higher metazoans must arise via the regulation of these genes at the transcriptional and post-transcriptional level. AS is the primary mechanism by which exons of a single gene can be spliced in various combinations to encode a diverse set of proteins. Indeed, 74% of all human genes are known to be alternatively spliced and different tissues exhibit varying degrees of AS (Cheng et al., 2005; Johnson et al., 2003; Kampa et al., 2004). Moreover, when one considers that a single gene can produce multiple isoforms, the number of proteins encoded by the genome will most likely be much higher than the current estimates (Brett et al., 2002). In humans, the genes that are expressed in the central nervous system (CNS) are subjected to the highest degree of AS when compared to other mature tissues (McCullough et al., 2005). However, the role of AS in CNS development is relatively unexplored. Given the complexity of AS combined with that of CNS development, it is important that the system employed to investigate this question should be accessible, well characterized during development and relatively amenable to functional perturbation.
The vertebrate retina is part of the CNS, yet is a relatively simple tissue with six neuronal cell classes (rod photoreceptors, cone photoreceptors, horizontal cells, bipolar cells, amacrine cells and ganglion cells) and one glial type (Müller glia) organized in a stereotypic manner. The birth order of each cell type is conserved, such that ganglion cells, cone photoreceptors and horizontal cells are among the first born cell types, followed by amacrine cells, rod photoreceptors, bipolar cells and Müller glia (1996; Rapaport et al., 2004; Sidman, 1961; Young, 1985a). The production of each postmitotic cell type begins in the central retina and expands from the center to the periphery (Rapaport et al., 2004; Young, 1985a; Young, 1985b). For example, the ganglion cell production begins in the dorsal/central retina at E11.5 followed by ventral/central retina and ends at P0 where the ganglion cells are generated in the periphery (Farah and Easter, 2005; Young, 1985a). A similar pattern of postmitotic cell generation is observed for all other cell types of the retina (Rapaport et al., 2004; Young, 1985b). Retinal development has been the focus of many studies leading to a better understanding of mechanisms that govern cell fate determination and differentiation (Cepko, 1996; Livesey and Cepko, 2001). Indeed, the retina has served as a paradigm for these and other aspects of the developing nervous system. The role of AS in development is very poorly understood and as of yet, no study has focused on the role of ASFs in vertebrate neuronal development. Thus, the current investigation focuses on understanding the role of an ASF called splicing factor arginine/serine-rich 1 (Sfrs1) in retinal development.
Sfrs1 belongs to a highly conserved Serine/Arginine (SR) protein family of RNA processing factors found throughout metazoans and in plants (Zahler, 1999; Zahler et al., 1992). The role of Sfrs1 in splicing has been well documented and only recently has its role in any developmental context been investigated (Xu et al., 2005). In C. elegans, ablation of the Sfrs1 homologue resulted in late embryonic lethality, suggesting that its function is non-redundant in at least one critical stage of development (Kawano et al., 2000; Longman et al., 2001). In this report, we show that Sfrs1 is expressed in the developing mouse retina and is itself regulated by AS. We have identified a new isoform, which is expressed during late embryonic development, and continues to be expressed during postnatal retinal development. The novel isoform lacks the SR domain, which is critical for the nuclear localization of the Sfrs1 protein (Kataoka et al., 1999; Lai et al., 2000; Lai et al., 2001). In mice, the loss of Sfrs1 also resulted in embryonic lethality (Xu et al., 2005). A conditional knockout (cKO) mouse in which the Sfrs1 gene was flanked with lox-P sites (Sfrs1fl/fl) has enabled functional studies of Sfrs1. Xu et al., have shown that loss of Sfrs1 did not cause aberrant proliferation and/or cell death of cardiac progenitors and that embryonic heart development proceeded normally (Xu et al., 2005). However, during postnatal remodeling of the heart, aberrant splicing of specific target genes such as cardiac troponin T (cTnT), the Z-line protein Cypher, and Ca2+/calmodulin-dependent kinase IIδ (CaMKIIδ) resulted in physiological defects in the heart (Xu et al., 2005), demonstrating that Sfrs1 is required for postnatal remodeling of the heart. The present investigation employs the same Sfrs1fl/fl mice to determine the role of Sfrs1 in the developing mouse retina. To specifically ablate Sfrs1 function during retinal development the Sfrs1fl/fl mice were crossed to mice that express Cre recombinase under the regulation of ceh-10 homeo domain containing homologue (Chx10) gene which has been shown to be expressed in retinal progenitor cells (Rowan and Cepko, 2004). The loss of Sfrs1 function resulted in a small eye at birth. We found that loss of Sfrs1 function did not have a significant effect on proliferation. However, neurons generated during early embryonic development underwent apoptosis, which was restricted to this time and stopped after birth. Consequently, most of the neurons that were generated during embryonic development, such as ganglion cells, cone photoreceptors, horizontal cells, and amacrine cells were significantly reduced in the Sfrs1-cKO retina. In contrast, the neuronal cell types that are produced later, such as rod photoreceptors, bipolar cells, late born amacrine cells and Müller glia, differentiated and survived in the Sfrs1-cKO retina. The subset of susceptible neurons were defined primarily by the time of their birth, perhaps due to the temporal heterogeneity in gene expression that has been defined for retinal progenitor cells (Trimarchi 2008).
Materials and Methods
Animal procedures
To generate Sfrs1-cKO retinae, Sfrs1fl/fl mice were crossed to mice expressing Cre under the regulation of Chx10. Mice used for analysis were genotyped for the conditional knock out allele of Sfrs1 by PCR as previously described by Xu et al. (Xu et al., 2005). The presence of the Cre allele was confirmed by PCR (Rivera-Feliciano and Tabin, 2006). The embryos used for characterizing the expression pattern of Sfrs1 were acquired from CD1 time pregnant females that were purchased from Charles River Laboratories, Inc. In addition, when mice were subjected to surgical procedures, all IACUC protocols to minimize pain during and after surgery were observed.
RT-PCR
Retinae from different developmental time points were harvested and total RNA was prepared in Tri-Zol by following the manufacturer’s protocol (Invitrogen, CA). For cDNA synthesis, 5 μg of total RNA from retinae harvested at various time points was used (Kanadia et al., 2006).
In situ hybridization
In situ hybridization on 16 μm cryosections of the retina were performed as described previously (Trimarchi et al., 2007). All the probes used in this report are the same as those published previously (Trimarchi et al., 2008; Trimarchi et al., 2007). For Sfrs1, we employed a 3′UTR probe with sequence that starts at 1620 bp to 2639 bp in the clone (NM_173374) identified in the NCBI database.
Dissociated in situ hybridization (DISH)
Both wild-type and mutant P7 retinae were dissociated by papain digestion and plated on lysine coated slides. Followed by a 10′ fixation in 4% PFA fixation, the cells were subjected to in situ protocol with the same DIG-labeled probes used in section ISH. The presence of RNA was detected by anti-DIG-HRP antibody. Subsequently, the signal was amplified with a tyramide based amplification protocol (PerkinElmer, MA) (Trimarchi et al., 2007).
Immunofluorescence
For immunofluorescence, 16 μm cryosections were first hydrated in phosphate buffered saline (PBS-pH7.4) for 10 minutes at room temperature (RT). This was followed by subjecting the sections to an immunohistochemistry protocol that has been described previously (Kim et al., 2008). The dilution of all the primary antibodies was as follows: chicken anti-GFP (1:2000) (Abcam, MA); mouse anti-Pax6 (1:300) (Covance, CO); rabbit anti-Chx10 (1:300) (Cepko lab); mouse anti-Rhodopsin (4D2) (1:300) (Molday and MacKenzie, 1983); mouse anti-glutamine synthetase (1:300); rabbit anti-red/green opsin (1:300) (Chemicon); and mouse anti-Ki67 (1:250) (BD Pharmigen).
Immunoblot
Nuclear and cytoplasmic protein extracts were made by subjecting retinae (n=10) from different developmental times to nuclear/cytoplasmic protein extraction protocol from Pierce. Upon fractionation, 30 μg of protein from both cytoplasmic and nuclear extracts was resolved on a 4–20% tris-glycine gradient gel (Invitrogen, CA), followed by transfer of the proteins to a positively charged nylon membrane (Invitrogen, CA), which was then subjected to immunoblot analysis as described previously (Kanadia et al., 2006). The specific primary antibodies used were a mouse monoclonal antibody against Sfrs1 (1:1000) (Lifespan, CA) and a mouse monoclonal antibody against Cugbp1 (1:500) (Abcam, MA).
Electron microscopy
P0 pups were harvested in 0.1 M sodium cacodylate buffer (pH7.4) followed by fixation in 2% paraformaldehyde and 2.5% gluteraldehyde in 0.1 M sodium cacodylate buffer. Retinae were then processed by the Harvard electron microscopy core.
P0 electroporation
P0 pups were electroporated with pCAG-Cre and pCAG-LoxP-Stop-LoxP-GFP plasmids or pCAG-GFP plasmid alone as described previously (Matsuda and Cepko, 2004).
Virus preparation
All viruses were prepared by cotransfection of HEK-293T cells with pQC-H2B-GFP-IRES-Cre plasmid plus a helper plasmid that provides gag, pol and env proteins for packaging along with the VSV-G coat protein plasmid. Virus in the cell culture supernatant media was then concentrated through centrifugation as described previously (Cepko, 1989).
E10 viral infection by ultrasound assisted delivery
Timed pregnant Sfrs1fl/fl and/or Sfrs1wt/wt females were anesthetized under constant isoflurane according to the manufacturer’s recommendations. The pregnant female was secured on a stage in the supine position and an incision was made in the abdomen from the genitalia towards the diaphragm. This incision was then covered with a saline-soaked piece of gauze (37 °C). Through this incision, one uterine horn was carefully extracted from the body cavity and placed on top of the gauze. The embryos were covered with ultrasound gel and imaged live with an ultrasound probe. Upon determining the position of the diencephalon (schematized in Fig. 4) 0.4 μl of virus was injected with a glass needle (custom 0) (Punzo and Cepko, 2008).
Fig. 4. Retinal progenitor cells survive the loss of Sfrs1.
(A) Snapshot of a live ultrasound image of an E10.5 embryo imaged through the uterine wall. Inset shows the newly formed optic vesicles and the diencephalon is traced in white. The next panel shows a glass needle that is introduced into the ventricle where the virus is delivered. (B) P14 retinal sections from Sfrs1wt/wt embryos injected at E10.5 with a virus expressing Cre and nuclear-GFP that were stained for GFP. The white solid arrowhead shows the position of the amacrine cells. (C) P14 retinal sections from Sfrs1fl/fl embryos injected at E10.5 with the same virus injected in wild-type embryos. The white solid arrowhead highlights the absence of the amacrine cells. P7 retinal sections from Sfrs1wt/wt and Sfrs1fl/fl mice that were crossed to RC::PFWE line, which reports the Chx10::Cre excision event by activating nLacZ synthesis (D,E).
P0 Viral infection
P0 pups were first immobilized by hypothermia followed by delivery of 1 × 106 infectious viral particles using a 33 gauge Hamilton needle into the subretinal space (Matsuda and Cepko, 2004).
BrdU-Pulse Labeling
A pregnant female at E16 was first weighed and injected with 3 μg of BrdU/1 gm of body weight. On the seventh day after birth the retinae were harvested and processed for BrdU staining. This was done by first performing antigen retrieval with an antigen unmasking solution (Vector Labs, Inc, CA, USA). The solution was brought to boil in a microwave and the slides were placed in the boiling solution and the power level of the microwave adjusted such that temperature was maintained at around 80 °C for 20 minutes. Then the slides were equilibrated to room temperature (1 hour). Next the slides were fixed in 4% PFA for 20 minutes followed by two 5-minute washes with PBS. Slides were treated with 2N HCL for 30 minutes at room temperature, followed by a brief wash with 0.1 Boric acid (ph 8.5) and IF was performed as described previously (Kim et al., 2008).
Results
Sfrs1 was expressed during retinal development and was itself regulated by AS
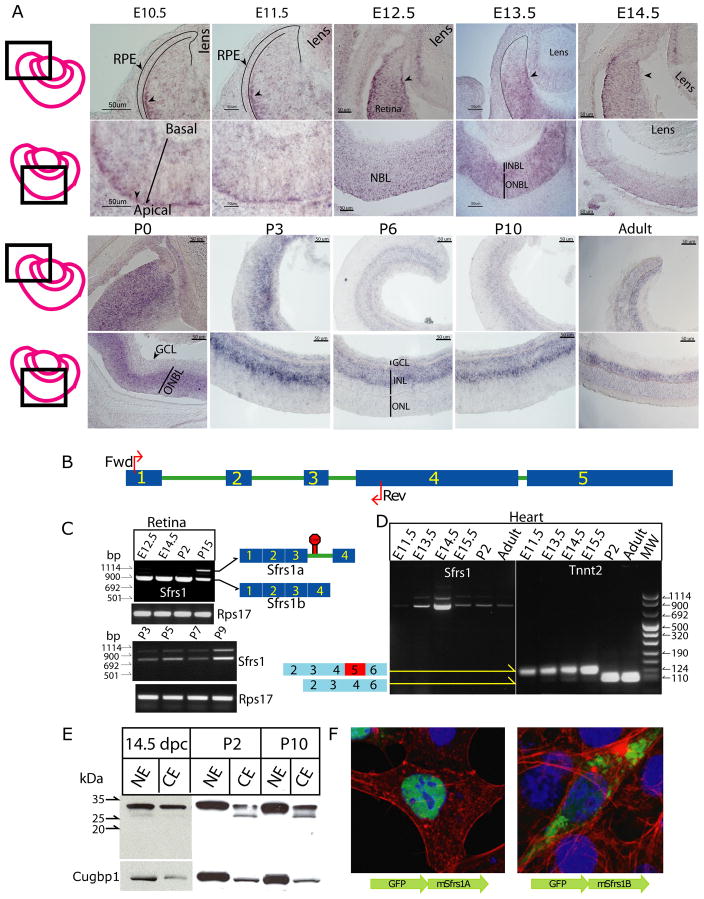
To understand the role of Sfrs1 during retinal development, we first characterized its expression pattern. RNA in situ hybridization (ISH) was employed on retinal sections from embryonic day (E) 10.5 to postnatal day 30 (P30) (Fig. 1A). At E10.5 and E11.5, expression of Sfrs1 was detected in progenitor cells in both the center and in the peripheral retina. However, in the peripheral retina, Sfrs1 expression did not extend to the edge, a portion of the retina that is known to give rise to the ciliary body (Davis-Silberman and Ashery-Padan, 2008). In addition, the ISH signal for Sfrs1 was enriched in cells at the apical end of the retina (Fig. 1A, black arrowhead), where the retinal progenitor cells undergo M phase of their cell cycle (Sidman, 1961; Young, 1985b). Similar asymmetric enrichment of the ISH signal for Sfrs1 was observed elsewhere in the brain, such as in the olfactory epithelium (Supplemental Fig. 1). ISH for cell division cycle 20 (Cdc20), a known marker of the G2/M phase of the cell cycle, and IF for another known marker of the M phase, phosphorylated-histone H3 (PH3) were employed (Supplementary Fig. 1) (Trimarchi et al., 2008; Weinstein, 1997) to determine if the region of Sfrs1 expression in the olfactory epithelium was also the region of M phase (Kawauchi et al., 2005). As predicted, both, the ISH for Cdc20 and IF for PH3 confirmed that the mRNA for Sfrs1 was enriched in areas expressing these genes, thereby suggesting the involvement of Sfrs1 in cells in the G2/M phase of the cell cycle. At E12.5, the expression was expanded to the entire outer neuroblastic layer (ONBL) (Fig. 1A) and was also observed in the lens epithelium. Specifically, the signal was stronger in the ONBL as compared to the postmitotic cells that are found in the newly forming inner neuroblastic layer (INBL). In addition, at E13.5, expression of Sfrs1 was transiently enriched in cells lining the vitreal side of the peripheral retina (Fig. 1A–E 13.5, arrowhead), but this expression pattern was no longer observed by E14.5 at which time the expression of Sfrs1 in the ONBL persisted. Again, the expression of Sfrs1 at P0 was in the area of progenitor cells and not in the INBL where postmitotic cells reside (Fig. 1A). During postnatal development, the expression of Sfrs1 was restricted in the newly forming inner nuclear layer (INL) in the central retina, while in the periphery the expression of Sfrs1 was observed in area of progenitor cells. At P6, Sfrs1 expression became restricted to the lower half of the INL, where mostly amacrine and displaced ganglion cells are found, and in the ganglion cell layer (GCL), where ganglion and displaced amacrine cells are found (Fig. 1A). This pattern was also observed at P10 and at P30 (Fig. 1A).
Fig. 1. Sfrs1 expression during retinal development.
(A) In situ hybridization with DIG-labeled anti-sense RNA probe detecting Sfrs1 RNA during retinal development from E10.5 to P30. The top panel shows the expression of Sfrs1 in the peripheral retina, while the bottom panel shows the central retina. A schematic on the far left demonstrates the position of the retina shown in the subsequent panels. (B) A schematic representation of the Sfrs1 gene with all its exons (
 ) and introns (
) and introns (
 ). The red arrows mark the position of the primers used to investigate the AS status of Sfrs1 by RT-PCR analysis. (C) Two RT-PCR products of Sfrs1 coding sequence with the time points denoted on the top. The bottom panel shows PCR products for Rps17, which was used to check the quality of the cDNA preparation. The identity of each isoform is schematized on the right, which shows that the top band retains intron 3 while the lower band is the canonical isoform with all four exons. (D) RT-PCR products of Sfrs1 coding sequence from heart cDNA from different developmental time points are shown and the quality of the cDNA was assessed by amplifying exon-2 to exon-6 of Tnnt2, a heart specific gene. (E) Immunoblot analysis (nuclear and cytoplasmic retinal extracts) to detect the production of the new isoform of Sfrs1 at E14.5, P2 and P10. Fractionation and equal protein loading was assayed by probing for Cugbp1. (F) Localization of the two isoforms of Sfrs1 was done by fusing Sfrs1A and Sfrs1B with GFP followed by transfection of NIH3T3 cells that were counterstained with phalloidin (red) and DAPI (blue) prior to acquiring the images.
). The red arrows mark the position of the primers used to investigate the AS status of Sfrs1 by RT-PCR analysis. (C) Two RT-PCR products of Sfrs1 coding sequence with the time points denoted on the top. The bottom panel shows PCR products for Rps17, which was used to check the quality of the cDNA preparation. The identity of each isoform is schematized on the right, which shows that the top band retains intron 3 while the lower band is the canonical isoform with all four exons. (D) RT-PCR products of Sfrs1 coding sequence from heart cDNA from different developmental time points are shown and the quality of the cDNA was assessed by amplifying exon-2 to exon-6 of Tnnt2, a heart specific gene. (E) Immunoblot analysis (nuclear and cytoplasmic retinal extracts) to detect the production of the new isoform of Sfrs1 at E14.5, P2 and P10. Fractionation and equal protein loading was assayed by probing for Cugbp1. (F) Localization of the two isoforms of Sfrs1 was done by fusing Sfrs1A and Sfrs1B with GFP followed by transfection of NIH3T3 cells that were counterstained with phalloidin (red) and DAPI (blue) prior to acquiring the images.
Next, we characterized the expression of Sfrs1 via RT-PCR analysis. A recent report based upon bioinformatic analysis indicated that the protein level of most of the SR-protein family members is regulated by AS (Lareau et al., 2007; Ni et al., 2007). Thus, we investigated whether AS might modulate the protein levels of Sfrs1 during retinal development. To determine the AS status of Sfrs1, which has 5 exons (Fig. 1B), a forward primer in exon-1 and reverse primer in exon-4 (red arrows, Fig. 1B) were utilized. The results show that Sfrs1 was regulated via AS in a temporal manner such that two isoforms of Sfrs1 were observed postnatally and only one isoform (Sfrs1a) was observed embryonically (Fig. 1C). During postnatal development, expression of Sfrs1a isoform continued, but an additional isoform of Sfrs1 (Sfrs1b) at a higher molecular weight was also expressed (Fig. 1C). The temporal regulation of Sfrs1 via AS led us to investigate whether a similar form of regulation was also employed during the development of other tissues. We chose the heart, since the role of Sfrs1 in heart development has been investigated (Xu et al., 2005). RT-PCR analysis with the same primers showed that during embryonic heart development, both the isoforms of Sfrs1 were detected, while during postnatal time points, only the Sfrs1a isoform was observed (Fig. 1D).
Sequence analysis of the two isoforms from the retina and the heart showed that the embryonic isoform of Sfrs1 was the canonical isoform with its two RNA recognition motifs and the RS domain. The other isoform (Sfrs1b) showed that it retained intron 3, thereby shifting the reading frame of the coding sequence. Consequently, the Sfrs1b isoform lacked the RS domain. This was an interesting observation, since Sfrs1 protein is one of the few SR-proteins that shuttles between the cytoplasm and the nucleus and the phosphorylation of the RS domain is required for its nuclear localization (Caceres et al., 1998; Ma et al., 2008). Thus, during retinal development, the AS of Sfrs1 produces an isoform that would most likely fail to translocate into the nucleus. In agreement with the RT-PCR analysis, the immunoblot analysis showed two immunoreactive bands in the cytoplasmic fraction from the postnatal, but not the embryonic retina (Fig. 1E). In addition, the lower molecular weight isoform (Sfrs1b) of Sfrs1 was at a significantly higher level in the cytoplasmic fraction and at a much lower level in the nuclear fraction (Fig. 1E). Since the new Sfrs1b isoform lacks the RS domain, which is required for the nuclear translocation, the low level expression of Sfrs1b in the nuclear fraction might be due to cross contamination between the two fractions. The inability of the Sfrs1b isoform to shuttle into the nucleus was further investigated by fusing the coding sequence of Sfrs1a and Sfrs1b isoforms to GFP, followed by transfection of each plasmid into NIH-3T3 cells. As predicted, the canonical isoform, Sfrs1a, was predominantly in the nucleus while the Sfrs1b isoform was not observed in the nucleus (Fig. 1F). In summary, Sfrs1 was expressed across retinal development and was found to be regulated by AS in a temporal manner.
Sfrs1-cKO mice had small eyes at birth
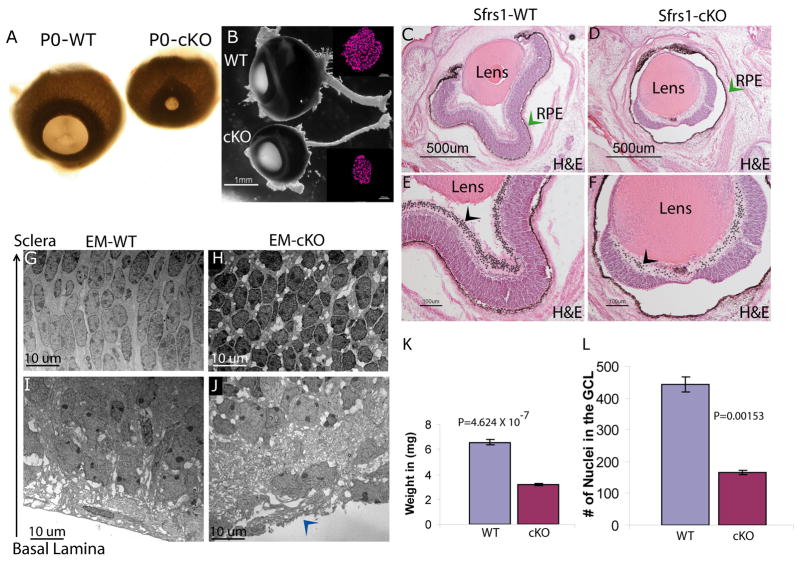
The role of Sfrs1 during retinal development was investigated by genetic ablation of Sfrs1 function by crossing the Sfrs1fl/fl mouse to a Chx10::Cre-recombinase transgenic mouse, which express Cre in retinal progenitor cells as early as E10.5 (Rowan and Cepko, 2004). As a result, the majority of the developing retina should lack Sfrs1 function. In contrast to the normal embryonic development of the Sfrs1 null hearts (Xu et al., 2005), the Sfrs1-cKO retinae underwent aberrant embryonic development which was inferred from the observation that Sfrs1-cKOmice were born with small eyes (Fig. 2A). Gross morphological analysis of the Sfrs1-null cKO eyes also revealed a thin optic nerve and total eye weight comparison showed that the mutant eyes weighed approximately half of the weight of the eyes from the wild-type littermate controls (Fig. 2B,2K). The eye was sectioned and stained with hematoxylin and eosin (H&E), which revealed that the there was a drastic decrease in the size of the retina, but not in the retinal pigmented epithelium (RPE) or the lens (Fig. 2C and 2D). The retina was detached from the RPE (Fig. 2D, green arrow) while abutting the lens and exhibited a drastic decrease in the cellularity of the GCL along with a perturbed inner limiting membrane (ILM) (Fig. 2F, arrowhead). Quantification of the number of nuclei in the GCL that were observed in the H&E stained retinal sections (Fig. 2E, 2F, arrowhead) confirmed the drastic decrease in the cellularity of the GCL (Fig. 2L), which in turn was reflected in a thin optic nerve (Figure 2B, inset). Ultrastructural analysis showed that cells in the mutant retina were highly disorganized with vacuolar inclusions, unlike the wild-type retina, which had elongated nuclei with a defined polarity (Fig. 2G–J). The absence of the ILM that was detected in the H&E stained sections was further confirmed by ultrastructural analysis (Fig. 2J, arrowhead). The mutant retina exhibited a dysmorphic fibrous layer instead of the well organized ILM that was observed in the wild-type control retina (Fig. 2I). In summary, loss of Sfrs1 function had a profound effect on embryonic development of the retina and possibly that of the ciliary body, which together might have caused the microphthalmia observed at birth.
Fig. 2. Sfrs1-cKO mice have small eyes at P0.
(A) Gross morphological analysis of the Sfrs1 cKO eye at birth. The image on the left shows the frontal view of Sfrs1 wild-type and Sfrs1 cKO eyes. (B) A lateral view of the eyes with the optic nerve and the inset shows the crossection of both the wild-type and the mutant optic nerves that are stained with DAPI and pseudo colored in magenta. Hematoxylin and eosin (H&E) stain of P0 wild-type and mutant retina, with a green arrowhead showing the retinal pigmented epithelium (C, D). Higher magnification of the same section as seen in D&E, with each ganglion cell nuclei marked with a dot (black arrowhead) (F,G). Ultrastructural analysis of wild-type and mutant retina (G–J), with the basal lamina indicated in the mutant retina (J-blue arrowhead). (K) Total weight comparison of the wild-type (n=4) and mutant (n=4) eyes. (L) Quantification of the ganglion cell layer cellularity in the wild-type (n=3) and mutant (n=3) retinae.
Sfrs1 loss of function caused cell death during embryonic retinal development
To determine the cause of the small retina in the Sfrs1-cKO mice at P0, we investigated whether the mutant retina had a proliferation defect during embryonic development. Ki-67 antibody, a well characterized proliferation marker (Gerdes et al., 1991) was employed on retinal sections from E12.5, E13.5, E18.0 and P0. Interestingly, proliferation of retinal progenitors in the Sfrs1-cKO retina did not appear to be altered in comparison to its littermate wild-type controls (Fig. 3A–3H). This result was further validated by PH3 staining in the Sfrs1-cKO retina at E12.5 (Supplemental Fig. 2). Moreover, quantification of the PH3+ cells did not show a significant change in the number of proliferating cells (Supplemental Fig. 2). Therefore, we investigated cell death as a cause for the small eye in the Sfrs1-cKOmice. Terminal dUTP nick end labeling (TUNEL) assay revealed a significant increase in the TUNEL+ cells at E12.5, E13.5, and E18.0 in the mutant retina (Fig. 3I–N). TUNEL+ cells observed in Sfrs1-cKO retina at E12.5 and E13.5 were predominantly in the central retina (Fig. 3J, 3L), while the majority of the TUNEL+ cells at E18.0 were observed in the periphery (Fig. 3N). While at P0, very few TUNEL+ cells were observed (Fig. 3P). Furthermore, quantification of the TUNEL+ cells in the mutant retina at each stage revealed that the cell death peaked around E13.5 followed by a decrease in the number of TUNEL+ cells at E18 and a few TNUEL+ cells at P0 (Fig. 3Q–T). In summary, loss of Sfrs1 function during embryonic development caused cell death and did not cause an observable reduction in proliferation. However, a subtle reduction in proliferation could not be ruled out.
Fig. 3. Proliferation and cell death.
(A–H) Proliferation as determined by Ki67 immunofluorescence in Sfrs1-WT and cKO retina at E12.5, E13.5, E18.0 and P0. (I–Q) Cell death as determined by TUNEL analysis in Sfrs1-WT and cKO retina at E12.5, E13.5, E18.0 and P0. (Q–T) Quantification of the number of TUNEL+ cells per section from different animals observed in the mutant retina as compared to the wild-type retina at E12.5 (n=4 for Sfrs1-WT and cKO), E13.5 (n=3), E18 (n=3) and P0 (n=5) (R–U).
Sfrs1 loss of function did not result in death of retinal progenitor cells
There are three different scenarios in which dying cells might explain the small eye phenotype. The dying cells might be progenitor cells, they might be postmitotic cells, or they might be both types of cells. Given that Sfrs1 is expressed in progenitor cells during embryonic development, progenitor cell death was investigated following the loss of Sfrs1. This was achieved by ablating Sfrs1 function in retinal progenitor cells utilizing a retrovirus expressing Cre along with nuclear GFP (Histone-2B-GFP fusion). The premise underlying this experiment was that this type of retrovirus can only infect mitotic cells, thereby targeting only the retinal progenitor cells. Moreover, the incorporation of the viral genome in to the progenitor cell allows the marking of all its progeny. This retrovirus was delivered to E10.5 Sfrs1fl/fl embryos with the aid of an ultrasound guided injection device, followed by detection of GFP immunofluorescence at P14 (Fig. 4B, 4C). Since one of the possibilities subsequent to the loss of Sfrs1 function was death of the progenitor cell, which would result in the absence of GFP+ cells at P14, we controlled for this possibility by co-injecting a virus that expressed membrane-GFP (mGFP). An additional control was to repeat the same experiment in the Sfrs1wt/wt mice. Analysis at P14 showed several clones of H2B-GFP+ cells in the retinae of infected Sfrs1fl/fl mice (Fig. 4C), indicating the survival of the initially infected progenitor cells. However, there was a difference in the cell type composition of the clones in the Sfrs1fl/fl mice relative to those in the Sfrs1wt/wt mice. Based on the position of the GFP+ cells, the majority of the GFP+ cells in the Sfrs1fl/fl mice were determined to be either rod photoreceptors, bipolar cells, or Müller glia (Fig. 4C). In contrast, the cell types observed in the Sfrs1wt/wt mice as determined by the position of the nuclei, included rod photoreceptors, horizontal cells, bipolar cells, Müller glia, amacrine cells and ganglion cells (Fig. 4B). The Cre negative mGFP+ clones were similar in composition to those of the wild-type (data not shown). In all, the loss of Sfrs1 most likely did not result in the death of retinal progenitor cells.
The aforementioned inference was further bolstered by the genetic ablation strategy that employed the Chx10-Cre line along with a reporter line called RC::PFWE in the Sfrs1fl/fl background. The RC::PFWE strain is a Cre-reporter line which expresses nuclear-β-galactosidase after a Cre-mediated recombination event (Farago et al., 2006). Thus, every progenitor cell that expresses Cre should not only activate the transcription of nLacZ, but should also excise the Sfrs1 gene. The results of this experiment showed that some retinal progenitor cells did not die, because there were nLacZ+ neurons in the P7 mutant retina (Fig. 4E). These neurons, as determined by their position, were mostly photoreceptors, bipolars and Müller glia (Fig. 4E). In contrast, the wild-type littermate retina showed many cells expressing nLacZ in the ONL (rod photoreceptors, cone receptors), INL (bipolar cells, horizontal cells, amacrine cells and Muller Glia) and in the GCL (ganglion cells) (Fig. 4D). Like in the mutant retina, the wild type retina had cells that were negative for nLacZ, which was due to the mosaic expression of Chx10::Cre (Rowan and Cepko, 2004). In summary, the data gathered from the two experiments described above suggests that loss of Sfrs1 function did not cause progenitor cell death.
Postmitotic cells were generated in the Sfrs1-cKO retina during early embryonic development
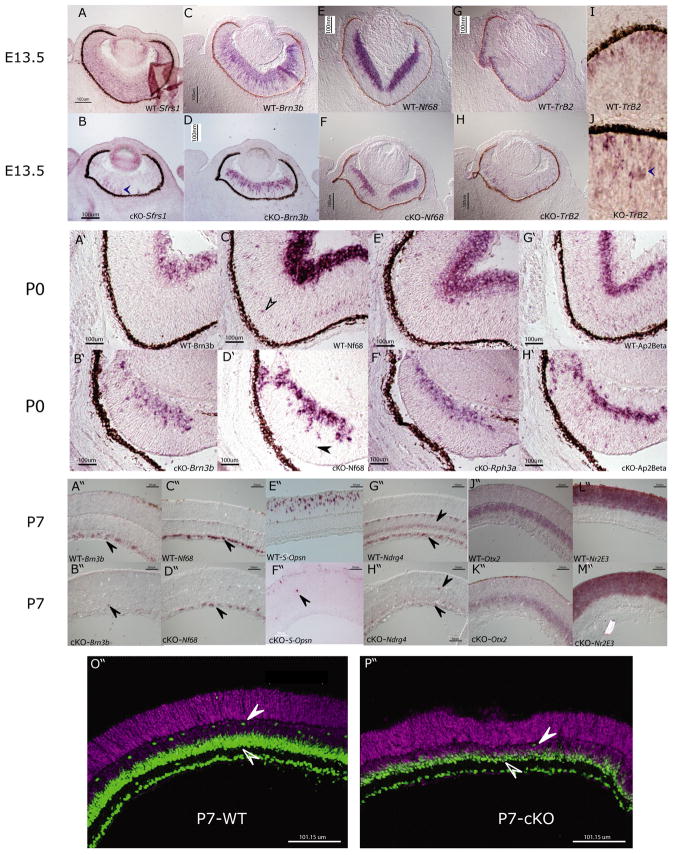
To examine whether the postmitotic cells were dying in the Sfrs1 null retinae, we first determined whether postmitotic cells were produced during embryonic development. For this, RNA ISH with probes that label differentiated neurons was employed on E13.5 retinal sections. First, the loss of the Sfrs1 transcript was assayed in the Sfrs1 cKO retina and was confirmed (Fig. 5B). While the majority of the retina lacked Sfrs1, a few cells (Fig. 5B, arrowhead) were positive for Sfrs1, reflecting the mosaic expression of Cre (Rowan and Cepko, 2004). Subsequently, ISH analysis on serial sections with probes, such as neurofilament like light chain-68 (Nf68), brain 3b (Brn3b) and thyroid hormone receptor beta 2 (Trβ2), that mark postmitotic cells at this stage was performed. Indeed, ganglion cells were produced in the mutant retina, as seen by the strong ISH signal for Nf68 and Brn3b (Fig. 5D, 5F). Since at this stage there is increased cell death in the Sfrs1-cKO retina, it cannot be ruled out that some of these ganglion cells die immediately after they were produced and some did not. Similarly, cones were produced, as determined by the ISH signal for Trβ2 in the Sfrs1-cKO retina (Fig. 5H). However, Trβ2+ cone photoreceptors in the mutant retina were located throughout the ONBL (Fig. 5J, arrowhead), rather than abutting the RPE, as was seen in the wild-type littermate (Fig. 5I). In summary, these data suggest that at least some neurons can be produced and can initiate differentiation, with cones showing aberrant localization.
Fig. 5. Cell type composition in the Sfrs1-cKO retina.
In situ hybridization with DIG-labeled anti-sense RNA probes detecting Sfrs1 (A, B); Brn3b (C, D); Nf68 (E,F); Trβ2 (G, H) mRNA in E13.5 Sfrs1 wild-type and KO retinal sections. A higher magnification image of the Trβ2 in situ that are shown in panels G and H are depicted in I and J. ISH with DIG-labeled anti-sense RNA probes detecting Brn3b (A′,B′); Nf68 (C′,D′); Rph3a (E′,F′) and Ap2Beta (G′H′) on retinal sections of P0 Sfrs1 wild-type and cKO retina. ISH with DIG-labeled anti-sense RNA probes on P7 Sfrs1wt/wt and Sfrs1-cKO retinal sections detecting Brn3b (A″,B″), Nf68 (C″,D″), S-opsin (E″,F″), Ndrg4 (G″,H″), Otx2 (J″,K″), Nr2E3 (L″,M″) mRNA. Pax6 Immunofluorescence (green) and DAPI which is false colored in magenta in a wild-type and KO from a P7 retina (O″,P″). The solid arrowhead shows the horizontal cells, while the open arrowhead points to the amacrine cells.
Neurons generated during early and mid embryonic development died in the Sfrs1-cKO retina
To investigate the persistence of the embryonically generated neurons, the above mentioned strategy was employed on P0 retinal sections. This analysis showed that at P0 the ISH signal for Brn3b+ and Nf68+ ganglion cells was reduced in the Sfrs1-cKO retina (Fig. 5B′, 5D′). At P0, Nf68 also labels horizontal cells (Fig. 5C′, open arrowhead), which were significantly reduced in the Sfrs1-cKO retina (Fig. 5D′ solid arrowhead). Similarly, the ISH signal for rabphilin-3a and Ap2Beta, which mark amacrine cells, was also reduced (Fig. 5F′, 5H′). It must be noted that the images of the P0 wild-type retina represent only a small portion of the entire retina. In contrast, one half of the mutant retina was captured at the same magnification, which underscores the drastic reduction in the size of the mutant retina. Next, we employed ISH at P7 with these and other probes to detect embryonically produced neurons, and very few of these neurons were detected. Specifically, there were very few ganglion cells in the Sfrs1-cKO retina, as detected by ISH for Brn3b (Fig. 5B″, arrowhead). ISH for Nf68, another marker for ganglion cells, showed a few more ganglion cells compared to the Brn3b-positive ganglion cells in the mutant retina (Fig. 5D″). In addition, cone cells (as determined by the S-opsin signal) were significantly reduced in the mutant retina (Fig. 5F″). Similarly, amacrine cells and horizontal cells were significantly reduced in the Sfrs1-cKO retina as determined by Ndrg4 ISH (Fig. 5H″, 5G″). While Ndrg4 ISH would suggest that there was a drastic decrease in the number of amacrine cells, immunofluorescence with Pax6 revealed the presence of relatively more amacrine cells, albeit a reduced number when compared to the wild-type retina (Fig. 5O″,5P″, open arrowhead). At P7, Pax6 antibody also marks horizontal cells, which were drastically decreased in the mutant retina compared to the wild-type littermate (Fig. 5O″, 5P″ solid arrowhead). In contrast, a similar reduction of cells produced postnatally was not observed. As seen by Nr2e3 and Otx2 ISH signal, the mutant retina did not exhibit a significant decrease in the thickness of the ONL, where primarily the Nr2E3+ rod photoreceptors reside, or in the scleral half of the INL, where the postnatally generated Otx2+ bipolar cells reside (Fig. 5J″–M″).
The significant decrease in the embryonically generated neurons was further quantified by dissociated cell in situ hybridizations (DISH) (Trimarchi et al., 2007). In this method, the total number of cells that were positive for ISH signal was divided by the total number of DAPI+ cells. As seen in Fig. 6C, by P7 there was a drastic decrease in the number of Nf68+ ganglion cells, Pax6+ amacrine cells and Blue-opsin+ cone photoreceptors. In contrast, there was no significant change in the numbers of glutamine-synthetase+ Müller glia, or Nrl+ rod photoreceptors. In the case of Chx10+ bipolar cells, there was a minor decrease in the Sfrs1-cKO compared to its wild-type littermate (Fig. 6D). In summary, the reduction in the number of embryonically generated neurons in the Sfrs1-cKO retina suggests that cell death during embryonic development was most likely that of the postmitotic cells.
Fig. 6. Quantification of the different cell types in the Sfrs1-cKO retina.
Quantification by dissociated in situ hybridization on P7 wild-type (n=3) and cKO (n=3) retinal cells with DIG-labeled RNA probes for specific cell types. A representative image with cells probed with NRL (red) in Sfrs1-WT and cKO and the nuclei are labeled with DAPI (Blue) (A,B); The relative percentage of ganglion cells (Nf68), cone photoreceptors (short wave opsin), and amacrine cells (Pax6) in wild-type and mutant retina are represented in C. The relative percentage of bipolar cells (Chx10), rod photoreceptors (rhodopsin), and Müller glia (GS) are shown in D.
The previous conclusion that the loss of Sfrs1 function led to the cell death of postmitotic cells was further investigated by a birthdating experiment. A pregnant female at E16 was injected with BrdU, which is incorporated in cells undergoing DNA synthesis (S-phase). With each subsequent cell division, the amount of BrdU is reduced by half. Neurons produced right after the BrdU pulse would retain at least half the amount of BrdU incorporated initially with no further dilution. Analysis at P7 of the Sfrs1-cKO retinae stained for BrdU and Pax6 showed a drastic decrease in the number of heavily labeled BrdU positive cells in the ONL, INL and the GCL (Fig. 7). Here, the Pax6 anti-body was used to mark amacrine cells, which are one of the major cell types produced at the time of the BrdU pulse. Quantification revealed that in the Sfrs1-cKO retinae there was a two-fold decrease in the number of heavily labeled BrdU+ cells compared to its wild-type littermate (Fig. 7). This is in keeping with the death of postmitotic cells, although failure to produce postmitotic cells in the normal numbers by Sfr1-cKO progenitor cells might also contribute to this decrement.
Fig. 7. Survival and/or production of neurons during embryonic development.
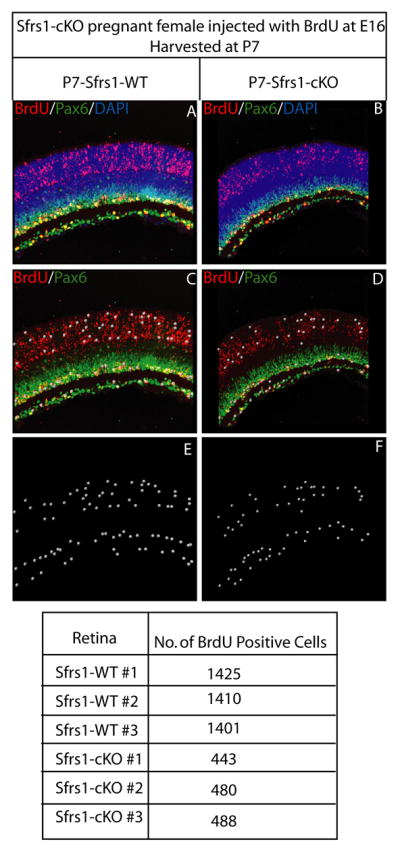
Immunofluorescence on P7 Sfrs1wt/wt and Sfrs1-cKO retinal sections from mice that were pulsed with BrdU at E16. P7 retinal sections from Sfrs1 wild-type and cKO mice stained for BrdU (red), Pax6 (green) and DAPI (blue) (A,B). Cells that were heavily labeled for BrdU were identified with an automated spot detection algorithm (IMARIS, Bitplane Inc.) utilizing a diameter threshold of 8μm and pixel intensity threshold of 20.641 and is rendered as white spots (C,D). The rendered spots without the fluorescence are shown in E and F. Table underneath shows the quantification of these spots in both the wild-type and the mutant retina.
Sfrs1-cKO retina underwent further degeneration during postnatal development
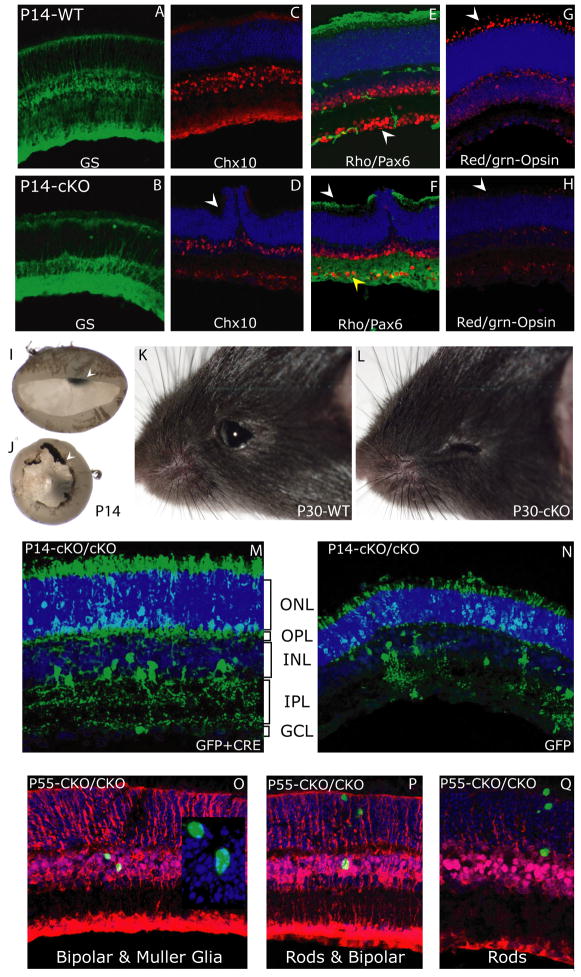
Next, we investigated the neuronal composition of the wild-type and cKO retina at P14, a time when retinal development is complete. Analysis at this stage revealed that the Sfrs1-cKO retina had undergone further degeneration as seen by the rosette formation (Fig. 8D, arrowhead). Furthermore, the mutant retina also had the ciliary body fused to the retina such that it could not be separated from the neural retina (Fig. 8J, arrowhead), which in the case of the wild-type littermate was easily removed (Fig. 8I, arrowhead). Despite the severe morphological deterioration of the mutant retina at P14, it still appeared to have a relatively unaltered percentage of Müller glia as seen by IF with glutamine synthetase antibody (Fig. 8A, 8B). Similarly, rod photoreceptors also appeared relatively unaltered in the mutant retina (Fig. 8F, solid arrowhead). The staining for rhodopsin showed an increased staining in the IPL, which was not observed in the wild-type retina. Since the rhodopsin antibody is a mouse monoclonal antibody, the IPL staining is most likely a cross-reactivity of the mouse secondary antibody to the immune cells. This was confirmed by the lack of IPL staining in the P14 mutant retina stained with a rabbit polyclonal antibody against rhodopsin (Supplemental Fig. 3). In contrast to the Müller glia and rhodopsin Ifs, Pax-6, Chx10 and red/green opsin positive cells were significantly reduced in the Sfrs1-cKO retina as compared to the wild-type (Fig. 8C–H). This dysmorphic retina ultimately underwent complete degeneration by P30 as seen by the absence of the entire eye in the Sfrs1-cKO mouse (Fig. 8L). We attempted to dissect out the eye from the P30 Sfrs1-cKO mice, but it revealed a small dysmorphic tissue of RPE cells without the lens or the retina (data not shown). In summary, the data gathered at P14 indicates that despite the severe morphological defects observed in the mutant retina, cell types including rod photoreceptors, Müller glia and bipolar cells were observed to be relatively unaffected, while others such as ganglion cells, amacrine cells, cone photoreceptors and horizontal cells seem to have undergone further loss.
Fig. 8. Postnatal retinal development in the Sfrs1-cKO mouse.
Immunofluorescence on P14 Sfrs1wt/wt and Sfrs1-cKO retinal sections with anti-glutamine synthetase (Müller glia) (A, B); anti-Chx10 (C, D) (bipolar cells and Müller glia); anti-rhodopsin (rod photoreceptors) and anti-Pax6 (amacrine and horizontal cells) (E, F); anti-red/green opsin (cone photoreceptors) (G, H). Image of the wild-type and mutant retina showing the ciliary margin attached to the mutant retina (I, J). Image of an adult (P30) mouse eye, Sfrs1wt/wt (K) and Sfrs1-cKO (L). P14 retinal sections stained with anti-GFP on P14 Sfrs1-cKO mice that were electroporated with Cre-GFP and LoxP-Stop-LoxP-GFP (M) and GFP alone (N) plasmids at P0. P55 retinal sections stained with anti-GFP (green) and anti-glutamine synthetase (red) antibody from mice that were infected with H2B-GFP-IRES-Cre virus at P0 (O–Q).
Sfrs1 was not required for the maintenance of late born neurons
The genetic ablation of Sfrs1 function in the retina revealed that Sfrs1 function was required for the survival of neurons produced embryonically, but not postnatally. Consequently, the Sfrs1-cKO retina had a disproportionate decrease in the ganglion cells, cone photoreceptors, horizontal cells and amacrine cells, compared to the rod photoreceptors, bipolar cells and Müller glia. To investigate whether rod photoreceptors, Müller glia, and bipolar cells were resistant to the loss of Sfrs1 function, P0 in vivo electroporation was employed. This strategy used two plasmids, one expressing Cre and the other expressed GFP in a conditional manner. The GFP reporter plasmid was such that the transcription of GFP was blocked by a stop cassette flanked by two loxP sites. Thus, only after Cre activity would the expression of GFP be initiated. Consequently, all the GFP+ neurons produced by these progenitor cells were presumed to be Sfrs1 deficient. Production of rod photoreceptors was normal as seen by GFP+ cells in the ONL with proper outer segments (Fig. 8M). In addition, amacrine cells appeared to be produced normally. While bipolar cells and Müller glia were also produced during this time, the promoter driving GFP failed to express reproducibly in these two cell types, making it difficult to asses their relative levels. To investigate the production and maintenance of bipolar cells and Müller glia, the same virus used for embryonic ablation of Sfrs1 was used to infect P0 retinal progenitor cells followed by analysis at P55. Indeed, several GFP+ clones in the Sfrs1-cKO retina were observed (Fig. 8O, 8P and 8Q). Based on the position and the shape of the nuclei, the GFP+ cells were determined to be rod photoreceptors, bipolar cells, and Müller glia (Figure 8O–Q and Supplemental Fig. 4). In summary, Sfrs1 function is required in a temporal manner, such that neurons born postnatally are resistant to the loss of its function.
Discussion
Sfrs1 is associated with G2/M phase of early retinal progenitor cell cycle and is itself temporally regulated via AS
During early embryonic development, Sfrs1 RNA was enriched in the apical side of the retina (Fig. 1A), which suggests that it is associated with the M phase of the cell cycle. This was confirmed by the overlapping expression pattern of Cdc20, and PH3, both of which mark the M phase of the cell cycle (Supplemental Fig. 1). Interestingly, similar apical expression pattern for Sfrs1 was observed in the olfactory epithelium (Supplemental Fig. 1). Together this suggests that Sfrs1 might regulate the G2/M phase transition in early progenitor cells. This is indeed the case in chicken DT-40 cells, where loss of Sfrs1 function results in G2 cell cycle block (Li et al., 2005).
In addition, Sfrs1, an ASF, is not only regulated at the transcriptional level during retinal development, but is also alternatively spliced in a temporal manner (Fig. 1). The newly identified isoform of Sfrs1 includes intron 3, which shifts the frame of the coding sequence which results in a premature stop codon (Fig. 1C). Consequently, this transcript would be subjected to non-sense mediated decay (NMD). However, Sfrs1 does not have a canonical stop codon, in that the position of its normal stop codon is not 50–55 nucleotides upstream of the 3′ exon-exon junction (EJC). Thus in the case of an intron-3 retention, which introduces a premature stop codon, the transcript would not be expected to undergo NMD. If this new isoform of Sfrs1 was not degraded, then it is possible that it produces a protein. Indeed, this is what we observed by immunoblot analysis (Fig. 1E). Furthermore, the novel isoform of Sfrs1, which has a frameshift, results in a truncated protein which lacks the RS domain. This alters the localization of this isoform of Sfrs1, since the RS domain serves as the nuclear localization signal (NLS) to which transportin-β binds and shuttles Sfrs1 protein into the nucleus (Lai et al., 2000). The importance of this NLS is confirmed by the cell culture experiment which shows that the RS-domain lacking isoform of Sfrs1 when fused to GFP is observed predominantly in the cytoplasm (Fig. 1F). This evidence is also corroborated by the immunoblot analysis in which the lower molecular weight immunoreactive band for Sfrs1 is observed predominantly in the postnatal cytoplasmic retinal fraction (Fig. 1E). By RT-PCR analysis, this new isoform was predominantly expressed postnatally, which suggests that Sfrs1 at this stage may have a novel function, such as regulating mRNA stability and/or translation. Since previous reports have shown that the lack of an RS domain does not alter its ability to bind mRNA and regulate translation (Sanford et al., 2005). Thus, the new isoform of Sfrs1 could have an additional function in translational regulation during postnatal retinal development. Indeed, Sfrs1 has been shown to act as an adaptor protein that recruits molecules responsible for regulation of cap-dependent translation of specific mRNAs (Michlewski et al., 2008). In all, this data underscores the role of AS in modulating activity of proteins by regulating subcellular localization.
The importance of Sfrs1 in retinal development first became apparent upon examination of newborn Sfrs1-cKO mice, which had small and dysmorphic retinas (Fig. 2A). This phenotype is primarily a result of cell death during embryonic retinal development (Fig. 3). Interestingly, this is in contrast to the role of Sfrs1 in cardiac development, which proceeds normally during the embryonic period in the absence of Sfrs1-mediated AS (Xu et al., 2005). However, the postnatal remodeling of the heart is abnormal in Sfrs1-cKO mice, resulting in cardiomyopathy and death (Xu et al., 2005). It is interesting to note that the presence of the phenotype in the two tissues is correlated with the absence of the Sfrs1b isoform. In other words, the times at which Sfrs1b isoform is expressed normally, are the times when loss of Sfrs1 function does not result in a phenotype. One possibility is that another ASF that regulates the AS of Sfrs1 normally might compensate for the loss of Sfrs1 function. Thus, the presence of Sfrs1b isoform serves as a proxy for the presence of the compensatory ASF, which in the heart is present embryonically, while in the retina it is present postnatally.
Sfrs1 loss of function does not cause the death of retinal progenitor cells
As described earlier, cell death upon loss of Sfrs1 function was observed in chicken DT-40 cells, where death is due to a G2 cell cycle block (Li et al., 2005),. Thus, it is possible that a similar G2 block occurs in some retinal progenitor cells, which then might lead to cell death. However, the majority of the progenitor cells in the Sfrs1-cKO retina did not die, as there was no apparent difference in the number of Ki-67+ and PH3+ cells in the mutant retina compared to the wild-type littermates (Fig. 3A–H). In addition, infection of Sfrs1fl/fl progenitor cells with retrovirus expressing Cre and nuclear GFP produced GFP+ neurons, which would not have occurred if the initially infected progenitor cell would have died. These clones comprised many differentiated GFP+ neurons (Fig. 4C), which could have arisen only if the initially infected Sfrs1-cKO progenitor cell produced more progenitor cells, that then produced neurons. Similarly, several nLacZ+ neurons were detected in the P7 mutant retina (Fig. 3E). Again, the presence of the nLacZ+ cells reflect a history of Cre activity in the progenitor cells, which in turn implies excision of Sfrs1 gene in those progenitor cells. In all, loss of Sfrs1 function does not cause the death of the majority of retinal progenitor cells. However, death of a minority of progenitor cells cannot be excluded.
Sfrs1 loss of function causes the death of postmitotic cells generated in the early to mid-embryonic period
If the loss of Sfrs1 function did not lead to the death of progenitor cells, then it must be that of the postmitotic cells. This possibility is consistent with the pattern of TUNEL+ cells in the Sfrs1-cKO retinae. Specifically, during early development (E12.5) the TUNEL+ cells were located in the central retina where neurons are first generated. As development proceeded (E18.0) the TUNEL+ cells were located more peripherally (Fig. 3). This central to peripheral pattern that is observed for cell death is similar to the pattern of postmitotic cell production in the retina (Farah and Easter, 2005; Young, 1985a). Furthermore, immunofluorescence and ISH analysis to detect differentiated neurons at P0 and P7 revealed that ganglion cells, horizontal cells, cone cells and amacrine cells were significantly reduced (Fig. 5). Interestingly, these are the cell types that are produced during embryonic development, coincident with the period of cell death in the Sfrs1-cKO retina. In contrast, the later-born rod photoreceptors, bipolar cells, and Müller glia were not as reduced (Fig. 5, and Fig. 6). The disproportionate loss of early-born cell types was also seen in the retrovirally marked clones of Sfrs1-cKO cells (Fig. 4C). Taken together, these data indicate that the cells in the Sfrs1-cKO retina that undergo apoptosis are early born postmitotic cells. Previous reports have shown that loss of ganglion cells early in development can have an effect on proliferation (Mu et al., 2005). However, in the case of Sfrs1-cKO retina, we observed differentiating ganglion cells (Fig. 5D), suggesting that the critical signals that might be important for proliferation might be produced.
Examination of markers for each of the early generated cell types revealed some interesting similarities and differences. Ganglion cells are always the earliest born neurons in all vertebrate retinas. Some of these cells appear to die early, since cells expressing ganglion cell markers were reduced at E13.5 and P0 (Fig. 5A′–D′). These were most likely among the TUNEL+ cells seen from E12.5–E18. However, some ganglion cells persisted until P7. These surviving ganglion cells may have been born around P0 when there was very little cell death in the Sfrs1-cKO retina. Alternatively, these surviving ganglion cells may have been produced by a retinal progenitor cell that did not undergo a Cre-mediated Sfrs1 ablation. Finally, these ganglion cells might not be dependent upon Sfrs1 for survival during their early stages of development. As ganglion cells are a heterogeneous class of neurons, this latter possibility might be explained by differential dependence on Sfrs1 by different types of ganglion cells. Regardless of the reason for their survival at P7, many of these died later, since by P14, there was a further reduction in the number of cells expressing ganglion cell markers. Similarly, cone photoreceptors are born only during the embryonic period. These also appeared to have initiated their differentiation in the Sfrs1-cKO retina, as seen by expression of TRβ2. Unlike ganglion cells, which appeared to have migrated to the proper laminar location, the cone photoreceptors appeared scattered across the ONBL, rather than in their normal location near the RPE (Fig. 5I, 5J). This correlates with the presence of TUNEL+ cells across the ONBL (Fig. 3J, 3L) and the reduction in cells with cone markers later in development (Fig. 5P).
Amacrine cells were also significantly reduced in the Sfrs1-cKO retina, as revealed by the reduction in ISH signal for Ndrg4. Again, not all amacrines were gone by P7, since anti-Pax6 labeling showed some amacrine cells in the Sfrs1-cKO retina at this age (Fig. 5Q, 5R, 5W-X). Ndrg4 marks a subset of amacrine cells, while Pax6 is a pan-amacrine cell marker (de Melo et al., 2003; Hitchcock et al., 1996). Thus, the more severe reduction in Ndrg4 ISH signal relative to the reduction in Pax6 signal could reflect some difference in dependence upon Sfrs1 among amacrine cell types. Birthdating experiments with [3H]-thymidine have shown that the peak of amacrine cell production is around E16.5–E17.5 and ends around P4 (Farah and Easter, 2005; Young, 1985a; Young, 1985b). In addition, the loss of Sfrs1 in P0 progenitors following electroporation of Cre showed that the majority of amacrine cells produced during postnatal development did not die (Fig. 6K). Thus, the significant loss of Ndrg4+ amacrine cells in Sfrs1-cKO mouse that was observed by P7 is likely due to the death of the embryonically produced amacrine cells. As amacrine cells are a heterogeneous group of neurons, it is possible that certain subtypes of amacrine cells might be born during embryonic development, and that these types die in the Sfrs1-cKO retina, while the subtypes born postnatally are different types of amacrine cells that are not dependent on Sfrs1 for survival. Based on this interpretation, we posit that there are at least two classes of amacrine cells as defined by their dependence on Sfrs1 for survival.
Unlike the aforementioned cell types, rod photoreceptors are born in significant numbers throughout retinal development. As discussed above, many of the early born cells died prior to P0. To determine if this pattern of death was also true for rod photoreceptors, BrdU birthdating was used to label rod photoreceptors born embryonically. A BrdU pulse was administered at E16 to the wt and Sfrs1-cKO animals to label S phase cells. Cells that undergo their terminal cell division retain much of the BrdU label, and at E16 this should include rod photoreceptors (Carter-Dawson and LaVail, 1979). As seen in Fig. 7, the number of BrdU+ cells in the ONL of the P7 Sfrs1-cKO retina was significantly reduced compared to the wild-type littermate retina. Since we have shown that the majority of progenitor cells survive in the absence of Sfrs1, and that postmitotic cells are produced, it is likely that rod photoreceptors were produced at E16 and they subsequently died. The other possibility is that cone photoreceptors are also produced and the reduction in the heavily labeled BrdU+ cells in the ONL is due to the reduction in the cone photoreceptors and not the rod photoreceptors. This is unlikely, since 95% of the cone photoreceptors are produced by E14 and very few (<5%) are produced at E16 (Rapaport et al., 2004). In contrast, rod photoreceptor production is initiated around E15 and by E16 approximately 10% of the rod photoreceptors are being produced. Thus, the reduction in the BrdU+ cells in the ONL is most likely due to rod photoreceptor death. However, in the mutant retina as determined by their position, there are rod photoreceptors that are heavily labeled with BrdU. This might be due to the mosaicism of the Chx10::Cre. Interestingly, rod photoreceptors born from later stage progenitor cells did not die. This was obvious following introduction of Cre at P0 via either electroporation or viral infection. These alternative strategies confirm the ability of the postnatally generated neurons to survive the loss of Sfrs1 function. This was necessary because the genetic ablation of Sfrs1 resulted in degeneration of the eye during postnatal development.
Later degeneration of the retina might be due to non-cell autonomous effects
The death of early born neurons in the Sfrs1-cKO retina was followed by a profound loss of cells such that no recognizable ocular structures remained by P30 (Fig. 8J). This degeneration may be secondary to the loss of the early born neurons. For example, the disproportionate growth of the lens, which abuts the retina at P0, may lead to aberrant metabolism, or other physiologically relevant alterations, in the retina. In addition, the Chx-10::Cre is also expressed in the ciliary margin, which in the case of Sfrs1-cKOmice may also contribute to the degeneration of the overall architecture of the eye (Rowan and Cepko, 2004; Rowan et al., 2004). As seen in Fig. 8J, in the mutant retina, the ciliary body was fused to the retina such that upon dissection, it was difficult to separate the neural retina from the ciliary body. This may suggest that the differentiation of the ora serrata, which marks the boundary of the peripheral neural retina and the ciliary body, was compromised (Wang et al., 2003). This defect may contribute to the degeneration of the Sfrs1-cKO eye via loss of proteins that make up the vitreous body (VB) and the ILM (Coca-Prados and Escribano, 2007). The majority of the proteins that make up the ILM and the VB, with the exception of agrin, are produced by the lens and the ciliary body (Halfter et al., 2008). The absence of these protein might result in a compromised ILM of the mutant retina (Fukai et al., 2002). In addition, there is no VB in the P0 Sfrs1-cKO eye (data not shown). Consequently, there is no intraocular pressure in the mutant retina. These secondary effects could contribute to the postnatal degeneration of the eye in the Sfrs1-cKO animals.
Temporal requirement of Sfrs1 function for the survival of retinal neurons
In summary, the current investigation suggests that the key determinant of whether cells survive in the Sfrs1-cKO retina is when they are born, as opposed to the subtype of neuron. Several lines of evidence point to this conclusion. First, the number of TUNEL+ cells decreased late in embryonic development, such that the only TUNEL+ cells at E18 were those in the periphery. This is consistent with resistance to death beginning in the center, and advancing to the periphery, which overlaps with the patterns of genesis of each cell type. Second, the genetic ablation of Sfrs1 early in development led to the loss of many early born neurons. This includes amacrine cells and rod photoreceptors, and both survive the loss of Sfrs1 when they are generated postnatally. Survival of postnatal neurons is demonstrated by P0 electroporation of Cre and P0 viral infection. This is coincident with the presence of the Sfrs1b isoform. Thus, as described before, the presence of the Sfrs1b isoform may serve as a proxy for the presence of a compensatory ASF. Finally, we propose a model in which AS mediated by Sfrs1 is required for the terminal differentiation and/or maintenance of neurons produced during early to mid embryonic development, but not for neurons produced during postnatal period (Fig. 9). Consequently, there is a significant depletion of early born cell types, while the later born cell types remain unaltered. The data presented here underscores the dynamic role of ASFs during vertebrate neuronal development and warrants further investigations that focus on either the changes in splice patterns of groups of genes or the gain and loss of function analysis of other ASFs. In addition, identifying other ASFs that are coexpressed with Sfrs1 may shed light onto the compensatory ASF that may render postnatally generated neurons resistant to the loss of Sfrs1 function.
Fig. 9. Role of Sfrs1 during retinal development.
A. Schematic representation (green & yellow) of the expression pattern of the two isoforms of Sfrs1 during development. A schematic representation showing the requirement of Sfrs1 in dark gray, and the lack of requirement light gray to white. The resistance to loss of Sfrs1 function during postnatal development overlaps with the occurrence of the Sfrs1b isoform (yellow), which may serve as a proxy for the expression of a compensatory ASF. B. A single progenitor in the Sfrs1wt/wt retina is shown undergoing cell divisions and producing retinal neurons and glia from E10.5 to P14. Mitotic cells are shown as (
 ), while postmitotic cells are shown as (
), while postmitotic cells are shown as (
 ). The black line in the middle depicts embryonic time with P0 indicating birth. Below the black line there are boxes with labels such as ganglion cells, horizontal cells, cone photoreceptors, amacrine cells, rod photoreceptors, bipolar cells and Müller glia, which show the birth order of all cell types. While the upper panel depicts a wild-type progenitor undergoing cell division and postmitotic cell production, the lower panel shows a single progenitor in the Sfrs1-cKO retina undergoing cell division and producing retinal neurons with (
). The black line in the middle depicts embryonic time with P0 indicating birth. Below the black line there are boxes with labels such as ganglion cells, horizontal cells, cone photoreceptors, amacrine cells, rod photoreceptors, bipolar cells and Müller glia, which show the birth order of all cell types. While the upper panel depicts a wild-type progenitor undergoing cell division and postmitotic cell production, the lower panel shows a single progenitor in the Sfrs1-cKO retina undergoing cell division and producing retinal neurons with (
 ) denoting cell death (B,C).
) denoting cell death (B,C).
Supplementary Material
Acknowledgments
We would like to thank Dr. Xiang-Dong Fu for generously sharing the Sfrs1 conditional knock out mouse. We would also like to acknowledge Jerome Gros and Jessica Whited for advice and helpful suggestions during the preparation of this manuscript. This project was funded by Howard Hughes Medical Institute.
References
- Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–72. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- Cepko CL. Immortalization of neural cells via retrovirus-mediated oncogene transduction. Annu Rev Neurosci. 1989;12:47–65. doi: 10.1146/annurev.ne.12.030189.000403. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The patterning and onset of opsin expression in vertebrate retinae. Curr Opin Neurobiol. 1996;6:542–6. doi: 10.1016/s0959-4388(96)80062-6. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–95. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Coca-Prados M, Escribano J. New perspectives in aqueous humor secretion and in glaucoma: the ciliary body as a multifunctional neuroendocrine gland. Prog Retin Eye Res. 2007;26:239–62. doi: 10.1016/j.preteyeres.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Davis-Silberman N, Ashery-Padan R. Iris development in vertebrates; genetic and molecular considerations. Brain Res. 2008;1192:17–28. doi: 10.1016/j.brainres.2007.03.043. [DOI] [PubMed] [Google Scholar]
- de Melo J, Qiu X, Du G, Cristante L, Eisenstat DD. Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J Comp Neurol. 2003;461:187–204. doi: 10.1002/cne.10674. [DOI] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–18. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Farah MH, Easter SS., Jr Cell birth and death in the mouse retinal ganglion cell layer. J Comp Neurol. 2005;489:120–34. doi: 10.1002/cne.20615. [DOI] [PubMed] [Google Scholar]
- Fukai N, Eklund L, Marneros AG, Oh SP, Keene DR, Tamarkin L, Niemela M, Ilves M, Li E, Pihlajaniemi T, et al. Lack of collagen XVIII/endostatin results in eye abnormalities. Embo J. 2002;21:1535–44. doi: 10.1093/emboj/21.7.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Li L, Schlueter C, Duchrow M, Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E, Flad HD. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol. 1991;138:867–73. [PMC free article] [PubMed] [Google Scholar]
- Halfter W, Dong S, Dong A, Eller AW, Nischt R. Origin and turnover of ECM proteins from the inner limiting membrane and vitreous body. Eye. 2008 doi: 10.1038/eye.2008.19. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Macdonald RE, VanDeRyt JT, Wilson SW. Antibodies against Pax6 immunostain amacrine and ganglion cells and neuronal progenitors, but not rod precursors, in the normal and regenerating retina of the goldfish. J Neurobiol. 1996;29:399–413. doi: 10.1002/(SICI)1097-4695(199603)29:3<399::AID-NEU10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–42. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2006;103:11748–53. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–52. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Fujita M, Sakamoto H. Unique and redundant functions of SR proteins, a conserved family of splicing factors, in Caenorhabditis elegans development. Mech Dev. 2000;95:67–76. doi: 10.1016/s0925-4773(00)00339-7. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–23. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Kim DS, Ross SE, Trimarchi JM, Aach J, Greenberg ME, Cepko CL. Identification of molecular markers of bipolar cells in the murine retina. J Comp Neurol. 2008;507:1795–810. doi: 10.1002/cne.21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci U S A. 2001;98:10154–9. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–9. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–14. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–18. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Longman D, McGarvey T, McCracken S, Johnstone IL, Blencowe BJ, Caceres JF. Multiple interactions between SRm160 and SR family proteins in enhancer-dependent splicing and development of C. elegans. Curr Biol. 2001;11:1923–33. doi: 10.1016/s0960-9822(01)00589-9. [DOI] [PubMed] [Google Scholar]
- Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu XD, Adams JA. Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J Mol Biol. 2008;376:55–68. doi: 10.1016/j.jmb.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough RM, Cantor CR, Ding C. High-throughput alternative splicing quantification by primer extension and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Nucleic Acids Res. 2005;33:e99. doi: 10.1093/nar/gni098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–89. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–60. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Liang S, Maeda H, Frishman LJ, Klein WH. Ganglion cells are required for normal progenitor- cell proliferation but not cell-fate determination or patterning in the developing mouse retina. Curr Biol. 2005;15:525–30. doi: 10.1016/j.cub.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C, Cepko CL. Ultrasound-guided in utero injections allow studies of the development and function of the eye. Dev Dyn. 2008;237:1034–42. doi: 10.1002/dvdy.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–24. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- Rivera-Feliciano J, Tabin CJ. Bmp2 instructs cardiac progenitors to form the heart-valve-inducing field. Dev Biol. 2006;295:580–8. doi: 10.1016/j.ydbio.2006.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Rowan S, Chen CM, Young TL, Fisher DE, Cepko CL. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–52. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci U S A. 2005;102:15042–7. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–5. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- Sidman RL. Histogenesis of Mouse Retina Studied with Thymidine-H-3. 1961. [Google Scholar]
- Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol. 2007;502:1047–65. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Wang J, McLeod D, Henson DB, Bishop PN. Age-dependent changes in the basal retinovitreous adhesion. Invest Ophthalmol Vis Sci. 2003;44:1793–800. doi: 10.1167/iovs.02-0802. [DOI] [PubMed] [Google Scholar]
- Weinstein J. Cell cycle-regulated expression, phosphorylation, and degradation of p55Cdc. A mammalian homolog of CDC20/Fizzy/slp1. J Biol Chem. 1997;272:28501–11. doi: 10.1074/jbc.272.45.28501. [DOI] [PubMed] [Google Scholar]
- Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr, Ye Z, Liu F, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985a;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell proliferation during postnatal development of the retina in the mouse. Brain Res. 1985b;353:229–39. doi: 10.1016/0165-3806(85)90211-1. [DOI] [PubMed] [Google Scholar]
- Zahler AM. Purification of SR protein splicing factors. Methods Mol Biol. 1999;118:419–32. doi: 10.1385/1-59259-676-2:419. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–47. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.