Abstract
Nuclear factor-κB (NF-κB) ligand (RANKL) was shown to induce osteoclast differentiation by increasing the expression of c-Fos, NFATc1 and TRAP. Salubrinal treatment to bone marrow macrophage (BMM) cells, however, significantly blocked NFATc1 expression and osteoclast differentiation by RANKL. Overexpression of NFATc1 further confirmed that NFATc1 is a key factor affected by salubrinal in osteoclast differentiation by RANKL. Unexpectedly, NFATc1 and c-Fos mRNA expressions were not affected by salubrinal, implicating that NFATc1 expression is regulated at a translational stage. In support of this, salubrinal increased the phosphorylation of a translation factor eIF2α, decreasing the global protein synthesis including NFATc1. In contrast, a phosphorylation mutant plasmid pLenti-eIF2α-S51A restored RANKL-induced NFATc1 expression and osteoclast differentiation even in the presence of salubrinal. Furthermore, knockdown of ATF4 significantly reduced salubrinal-induced osteoblast differentiation as evidenced by decreased calcium accumulation and lowered expressions of the osteoblast differentiation markers, alkaline phosphatase and RANKL in MC3T3-E1 osteoblast cells. Salubrinal treatment to co-cultured BMM and MC3T3-E1 cells also showed reduction of osteoclast differentiation. Finally, salubrinal efficiently blocked osteoporosis in mice model treated with RANKL as evidenced by elevated bone mineral density (BMD) and other osteoporosis factors. Collectively, our data indicate that salubrinal could affect the differentiation of both osteoblast and osteoclast, and be developed as an excellent anti-osteoporosis drug. In addition, modulation of ATF4 and NFATc1 expressions through eIF2α phosphorylation could be a valuable target for the treatment of osteoporosis.
Keywords: Osteoclast, Osteoblast, Salubrinal, eIF2α, NFATc1
1. Introduction
Bone development in vertebrate animals is maintained by two co-ordinated actions of osteoblast (bone formation) and osteoclast (bone resorption). In fact, many bone disorders reflect imbalanced activities of osteoblast and osteoclast, leading to the increased (osteopetrosis) and decreased (osteoporosis) bone mass [1].
Osteoblast is derived from mesenchymal stem cells, whereas multinuclear osteoclast is formed by fusion of mononuclear macrophages derived from hematopoietic stem cells [2,3]. Osteoblast produces and secretes the structural components of bone matrix and releases minerals that contribute to bone formation. A few agents, such as 1, 25-dihydroxyvitamin D3, parathyroid hormone (PTH) and PTH-related peptide (PTHrP) are known to stimulate osteoblastic stromal cells [1]. In addition, osteoblast activation produces two factors, macrophage colony-stimulating factor-1 (MCSF-1) and receptor activator of nuclear factor-κB (NF-κB) ligand (RANKL), that in combination are sufficient and necessary for osteoclast differentiation [4–6]. MCSF-1 induces osteoclast proliferation while RANKL is required for the formation of giant osteoclast [7,8]. RANKL stimulation triggers the recruitment of TNF receptor-associated factor 6 (TRAF6), resulting in the activation of downstream signaling molecules, including c-Jun N-terminal protein kinase (JNK), p38, ERK and NF-κB. Transcription factors including c-Fos and NFATc1 are also up-regulated by RANKL, leading to the increased expression of osteoclast associated receptor (OSCAR), tartrate-resistant acid phosphatase (TRAP) and so on [9].
It is known that intra-cellular stresses are closely related with senescence and skeletal disorders such as osteoporosis [10,11]. Upon endoplasmic reticulum stress (ER-stress) induction, unfolded protein response (UPR) signaling through the activation of three separate pathways, inositol-requiring enzyme-1 (IRE1), double-stranded RNA-activated protein kinase-like ER kinase (PERK) and activating transcription factor-6 (ATF6), was reduced with aging [12,13]. In addition, ER-stress sensor PERK was associated with lowered bone mineral density (BMD) [14]. These observations indicate a close association of ER-stress with osteoporosis. However, the underlying mechanism still remains unclear.
Salubrinal is a compound selectively inhibiting the activity of a protein phosphatase complex containing growth arrest and DNA damage-inducible protein (GADD34) and thus prevents dephosphorylation of eIF2α downstream of PERK, resulting in increased phosphorylation and inactivation of eIF2α [15]. Neuronal cell death by ER-stress, associated with Parkinson’s disease and Huntington’s disease, was also reported to be protected by salubrinal treatment [16,17]. However, the effect of salubrinal on osteoclastogenesis has never been reported.
In this study, it was revealed that increased phosphorylation of eIF2α by salubrinal significantly reduced osteoclast differentiation in mouse bone marrow cells. Ectopic expression of NFATc1 or phosphorylation mutant eIF2αS51A, however, increased osteoclast differentiation even in the presence of salubrinal. In addition, salubrinal enhanced osteoblast differentiation through eIF2α-ATF4 signaling pathway [18]. This is the first report demonstrating the anti-osteoporosis activity of salubrinal by inhibiting eIF2α dephosphorylation and ATF4 and NFATc1 expressions.
2. Materials and methods
2.1. Reagents and antibodies
α-MEM, fetal bovine serum, and penicillin were purchased from Invitrogen (Carlsbad, CA), and salubrinal purchased from Calbiochem (Darmstadt, GERMANY). Antibodies against β-actin, Flag and TRAP staining solution were obtained from Sigma Aldrich (St. Louis, MO). Recombinant human soluble MCSF and mouse RANKL were from PeproTech EC (London, United Kingdom). Antibodies against c-Fos, NFATc1, eIF2α, ATF4 and TRAP were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies against phospho-c-jun, phospho-ERK, ERK, phospho-JNK, JNK, phospho-p38, p38, phospho-IκB, and IκB were obtained from Cell Signaling Technology (Danvers, MA).
2.2. BMMs isolation and osteoclast differentiation
Mouse bone marrow cells were obtained from femurs and tibias of 6-week-old ICR mouse and were incubated in α-MEM complete media containing 10% fetal bovine serum, 100 U/ml penicillin in a 100 mm culture dish in the presence of MCSF (30 ng/ml) for 3 days. Adherent cells after removal of non-adherent cells were used as bone marrow macrophages (BMMs). To generate osteoclasts, BMMs (4×104 cells/well) were cultured for 4 days with MCSF (30 ng/ml) and RANKL (25 ng/ml) in 48-well (1 ml/well) tissue culture dishes with or without salubrinal pretreatment. On the 4th day, the cells were fixed with 10% formalin for 10 min, permeabilized with 0.1% Triton X-100, and then stained with tartrate-resistant acid phosphatase (TRAP), using the Leukocyte Acid Phosphatase Assay Kit (Sigma-Aldrich).
2.3. MC3T3-E1 cell differentiation and co-culture system
Clonal osteoblastic MC3T3-E1 cells were cultured in normal media (NM) in α-MEM (Gibco, Grand Island, NY) with 10% FBS, 20 mM HEPES and 1% penicillin-streptomycin. To induce differentiation, cells were seeded into a 12-well culture dish and further incubated until confluence. Cells were then transferred to α-MEM differentiation medium (DM) containing with 10% FBS, 1% penicillin-streptomycin, 10 mM β-glycerophosphate and 100 mg/ml ascorbic acid, and incubated for an additional 7-21 days. Mineralization of MC3T3-E1 cells was determined by Alizarin Red staining after fixation with 70% ethanol. To generate osteoclasts, BMMs (4×104 cells/well) and MC3T3-E1 (2×104 cells/well) were co-cultured for 5 to 6 days in differentiation media (DM).
2.4. Cell viability assay
The cell cytotoxicity assay was performed using a Cell counting kit-8 (Dojindo Molecular Technology, Japan) according to the manufacturer’s instructions. BMMs (1×103 cells/well) were cultured in the presence of M-CSF (30 ng/ml) and RANKL (25 ng/ml) in 96-well plates (200 μl/well) with or without salubrinal at varying concentrations (0–50 μM) for 48 hr. After 1 hr of CCK-8 treatment (10 μl), the plate was read at 450 nm (650 nm reference) using a 96-well plate recorder.
2.5. Western blot analysis
BMMs or osteoclasts were lysed in a buffer containing 50 mM Tris–HCl, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM sodium fluoride, 1 mM sodium vanadate, 1% deoxycholate, and protease inhibitors. The lysates were centrifuged at 15,000 ×g for 30 min and the supernatants were collected. After measurement of protein concentrations, equal amount of proteins (30 μg) were subjected to 8–10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred to polyvinylidene difluoride membranes (Milipore, Bedford, MA, USA). Membranes were blocked with 5% non-fat dried milk for 1 hr and were then incubated with the appropriate primary antibodies to be detected by enhanced chemiluminescence solution (ECL).
2.6. [35S] labeling and specific antibody pull-down assay
BMMs were exposed to [35S]-methionine/cysteine for 1 hr, and after 24 hr incubation with or without salubrinal treatment, whole cell lysates were extracted and radio-labeled proteins were analyzed by autoradiography after SDS-PAGE. For antibody pull-down assay, the lysates were centrifuged at 15,000 rpm for 30 min at 4 °C, and 1 mg of soluble protein was incubated with 1 mg/ml NFATc1 antibody. The reaction was performed for 3 hr with gentle rotation until 30 μl of protein G plus-agarose beads (Santa Cruz) were added to the mixtures for further rotation at 4 °C, 30 min. Beads were pelleted by centrifugation at 3000 rpm for 2 min and washed 7 times with cold lysis buffer. After boiling in sample buffer, the eluted proteins were subjected to SDS-PAGE.
2.7. Quantitative PCR analysis
Total RNA was prepared using an RNeasy Mini kit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions, and cDNA was synthesized from 3 μg of total RNA (Superscript II Preamplification System; Invitrogen). Real-time PCR was performed using CFX96™ Real-time system with SYBR FAST KAPA iCycler qPCR kit following the PCR conditions: 40 cycles of denaturation at 95 °C (15 s) and amplification at 60 °C (1 min). All reactions were run in triplicates and the data were normalized to the housekeeping gene β-actin. Relative differences in PCR results were evaluated using the comparative cycle threshold method. Primer sets were: mouse c-Fos, 5′-ACTTCTTGTTTCCGGC-3′ (forward), 5′-AGCTTCAGGGTAGGTG-3′ (reverse); mouse NFATc1, 5′-C CGTTGCTTCCAGAAAATAACA-3′ (forward), 5′-TGTGGGATGTGAACTCG GAA-3′ (reverse); and mouse β-actin, 5′-TCTGCTGGAAGGTGGACAGT-3′ (forward), 5′- CCTCTATGCCAACACAGTGC-3′ (reverse); ALP, 5′-AGGA CATCGCCACTCAACTC-3′ (forward), 5′-GGTTCCAGACTGGTTACTGTCA-3′ (reverse); RANKL, 5′- AGCCGAGACTACGGCAAGTA-3′ (forward), reverse, 5′-AAAGTACAGGAACAGAGCGATG-3′ (reverse).
2.8. Lentivirus generation and infection
Transfection was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). For the production of sh-RNA lentivirus, pHR’-CMVΔR8.2Δvpr and pHR’-CMV-VSV-G (protein G of vesicular stomatitis virus) were cotransfected into 293 T cells with pLKO.1-puro-sh-Luciferase (sh-Luc), -sh-ATF4 or pLenti-puro-empty, -NFATc1, -eIF2α S51A as previously described [19]. After incubation in fresh medium for 24 hr, culture supernatants of the lentivirus-producing cells were collected. For lentiviral infection, BMMs were cultured in a medium containing MCSF (30 ng/ml) for 48 hr, and the medium was replaced with culture supernatants of lentivirus together with polybrene (6 μg/ml) and MCSF (30 ng/ml) for 12 hr. Infected cells were then cultured in the presence of MCSF for another 24 hr prior to the stimulation with RANKL. For MC3T3-E1 cells, viral-infected cells were selected with puromycin (2 μg/ml) for 24 hr.
2.9. In vivo experiments
Eight week old C57BL/6 mice were obtained from The Jackson Laboratory. Mice were treated according to the ethical guidelines for animal safety and experimentation. Recombinant human RANKL was from R&D system. Models of osteoporosis induced by RANKL have been described previously [20]. In these models, more than 6 mice were examined in each group. Salubrinal (1 mg/kg) or DMSO was injected intraperitoneally 24 hr before the first RANKL injection, and the mice (n=6) subsequently received simultaneous injections of salubrinal (1 mg/kg, IP) or DMSO and RANKL (0.5 mg/kg, IP) or PBS at 24 hr intervals for 3 days.
2.10. Statistical analysis
Values are presented as the mean±S.D. values from three or more experiments. Data were analyzed with the Student’s t test or ANOVA test for comparisons between two mean values. A value of P<0.05 was considered significant.
3. Results
3.1. Salubrinal inhibits RANKL-induced osteoclast differentiation from BMM cells
In an attempt to determine the effect of salubrinal on osteoclast differentiation, bone marrow macrophage (BMM) cells isolated from mice were treated with both MCSF-1 (30 ng/ml) and RANKL (25 ng/ml) in the presence or absence of salubrinal and the appearance of TRAP-positive, multinucleated cells was counted. Salubrinal significantly reduced osteoclast differentiation in a dose-dependent manner (Fig. 1A and B), with no cell toxicity even at the concentration of 50 μM (Fig. 1C). To see whether the differentiation inhibitory effect of salubrinal is related with RANKL-induced early signaling pathways, phosphorylation of JNK, p38, ERK, c-jun as well as IκB-α was examined with or without salubrinal. Although activation or phosphorylation of these kinases occurred within 5 min of RANKL stimulation, salubrinal had no effect on these signaling molecules (Fig. 1D).
Fig. 1.
Salubrinal inhibits RANKL-induced osteoclast differentiation of BMM cells. (A and B) Mouse bone marrow cells were cultured with MCSF (30 ng/ml) and RANKL (25 ng/ml) at various concentrations of salubrinal. (A) After 4 days, cells were fixed and subjected to TRAP staining, (B) and the number of TRAP-positive, multinucleated osteoclasts was counted. All the bars show mean SE±from a representative triplicate experiment. The significance was determined by Student’s t-test (*P<0.5; **P<0.1). (C) BMM cells were cultured for 3 days with MCSF-1 (30 ng/ml) and RANKL (25 ng/ml) at the indicated concentrations of salubrinal. Cell viability was examined using CCK-8 solution kit as described in Materials & Methods. All the bars are mean SE±from a representative triplicate experiment. (D) BMM cells were pretreated with salubrinal (10 μM) or vehicle (DMSO) for 6 hr in the presence of MCSF (30 ng/ml) followed by stimulation with RANKL (50 ng/ml) for the indicated times. The whole cell lysates were prepared and subjected to western blot analysis with specific antibodies.
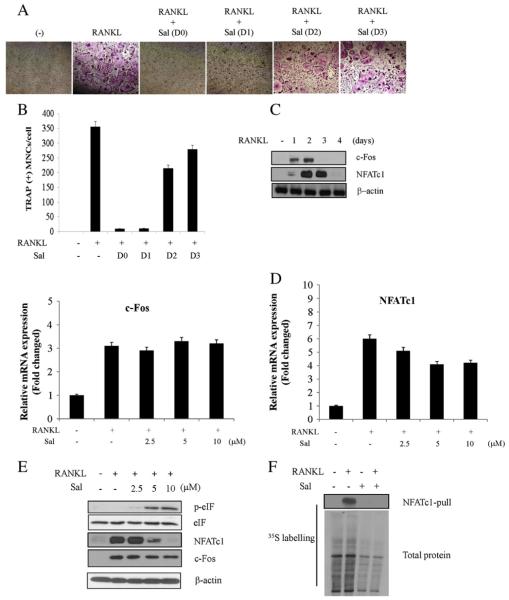
3.2. Time-dependent differential effect of salubrinal on RANKL-induced osteoclast differentiation
Osteoclastic differentiation of BMM cells could be observed within 4 days of RANKL treatment (data not shown). To determine the effective time the differentiation could be blocked by salubrinal, BMM cells were challenged with salubrinal at various times after RANKL treatment. It was found that the inhibitory effect of salubrinal on osteoclast differentiation could be obtained only when BMM cells had been treated with the compound within one day of RANKL stimulation (Fig. 2A and B). Salubrinal did not show differentiation inhibition when treated at later times. Thus, it was necessary to identify the proteins affected by salubrinal. In this regard, RANKL treatment induced orderly expression of c-Fos and NFATc1 which are known to be involved in osteoclast differentiation (Fig. 2C). Interestingly, however, RANKL-induced mRNA expression was not (c-Fos) or only modestly (NFATc1) affected by salubrinal (Fig. 2D), suggesting translational regulation of the proteins after salubrinal treatment. Given that NFATc1 is a positive feedback regulator for its transcription [8], it seems that NFATc1 protein degradation precedes mRNA reduction, partly explaining the slight reduction of NFATc1 mRNA by salubrinal as shown in Fig. 2D. Salubrinal was reported to inhibit dephosphorylation of eIF2α while maintaining the attenuation of protein synthesis after ER-stress induction [15]. Thus, eIF2α phosphorylation was increased while NFATc1 was dramatically reduced by salubrianl (Fig. 2E). Given that phosphorylation of eIF2α reduced global translation initiation and polypeptide biosynthesis [21], cells were treated with salubrinal to examine its effect on general protein synthesis. As shown in Fig. 2F, salubrinal significantly reduced global protein synthesis, with a dramatic effect on NFATc1. Unexpectedly, however, the expression of c-Fos was slightly reduced by salubrinal although the promoter region of NFATc1 has c-Fos binding sites.
Fig. 2.
Time-dependent differential effect of salubrinal on RANKL-induced osteoclast differentiation. (A and B) After BMM cells were stimulated with RANKL (25 ng/ml), salubrinal (10 μM) was treated to BMM cells at the indicated times. (A) Cells were cultured for 4 days after RANKL treatment and subjected to TRAP staining, (B) and the number of multinucleated osteoclasts was counted. All the bars represent mean SE±from a representative triplicate experiment. (C) BMM cells were stimulated with RANKL (25 ng/ml) for indicated times and the total cell lysates were prepared for subjection to western blot analysis with specific antibodies. (D and E) BMM cells after stimulation with RANKL (25 ng/ml) for 24 hr were further exposed to the indicated concentrations of salubrinal for another 24 hr. (D) Total RNA was extracted and subjected to quantitative PCR analysis. All the bars are mean SE±from a representative triplicate experiment. (E) The whole cell lysates prepared were subjected to western blot analysis with specific antibodies. (F) BMM cells were incubated 24 hr with or without RANKL (25 ng/ml) followed by pulse labeling for 24 hr with 35S-labeled Pro-Mix. Salubrinal was added into the medium 1 hr after Pro-Mix treatment. Whole cell lysate were prepared and immunoprecipitation was performed with NFATc1 antibody for autoradiography after SDS-PAGE (top). Total protein synthesis was analyzed (bottom). All the bars represent mean SE±from a representative triplicate experiment. The significance was determined by Student’s t-test (*P<0.5; **P<0.1).
3.3. Essential role of NFATc1 or eIF2α in restoration of osteoclast differentiation
To investigate whether NFATc1 is a key molecule for the anti-osteoclastogenic function of salubrinal, NFAc1 was transiently transfected into BMM cells using lentiviral vectors (Fig. 3A). Salubrinal inhibited RANKL-induced osteoclast differentiation as expected but showed little inhibition when NFATc1 was overexpressed in cells (Fig. 3B). The number of differentiated cells was also increased by exogenous NFATc1 expression (Fig. 3C). Based upon the observations demonstrating that increased eIF2α phosphorylation and decreased NFATc1 expression could be achieved by salubrinal treatment to RANKL-induced BMM cells (Fig. 2E and F), and that mutations abolishing eIF2α phosphorylation by converting serine 51 to alanine broadly reinitiate protein translation and affect stress resistance [22], the phosphorylation resistant eIF2α mutant (pLenti-eIF2α-S51A) was expressed in BMM cells. As expected, BMM cells containing pLenti-eIF2α-S51A were resistant to salubrinal in reducing RANKL-induced osteoclast differentiation (Fig. 3B). The number of TRAP-positive osteoclast cells was also increased by pLenti-eIF2α-S51A (Fig. 3C). In accordance with its effect on osteoclast differentiation, pLenti-eIF2α-S51A rescued NFATc1 expression (Fig. 3D). RANKL-induced eIF2α phosphorylation was also significantly reduced by pLenti-eIF2α-S51A (Fig. 3D), accompanied by the restored protein synthesis (Fig. 3E).
Fig. 3.
Essential role of NFATc1 or eIF2α for restoration of osteoclast differentiation. (A–D) BMMs were infected with lenti-viral vectors, pLenti (empty vector), pLenti-NFAT1 or pLenti-eIF2α-S51A. (A and D) BMM cells were infected with pLenti-NFATc1 or pLenti-eIF2α-S51A followed by stimulation with RANKL (50 ng/ml) 48 hr with or without salubrinal (10 μM) pretreatment. Whole protein lysates were prepared and subjected to western blot analysis with specific antibodies. (B and C) Infected BMM cells as in (A) were stimulated with RANKL (25 ng/ml) in the presence or absence of salubrinal (10 μM). (B) After 4 days, TRAP assay performed, (C) and the number of multinucleated osteoclasts was counted. (E) BMM cells infected with pLenti-eIF2α-S51A were stimulated with RANKL (50 ng/ml) for 24 hr and pulsed with 35S-labeled Pro-Mix 1 hr before salubrinal (10 μM) treatment. After 24 hr, whole cell lysate was prepared for subjection to western blot analysis and autoradiography. All the bars mean show SE±from a representative triplicate experiment. The significance was determined by Student’s t-test (*P<0.5; **P<0.1).
3.4. Induction of ATF4 dependent osteoblast differentiation and RANKL induction in MC3T3-E1 cells by salubrinal
Because bone homeostasis could be balanced by the degree of bone resorption by osteoclasts and bone formation by osteoblasts, the effect of salubrinal on MC3T3-E1 osteoblastic cells was examined. ATF4 was reported to up-regulate osteoblast differentiation [23] and salubrinal induced ATF4 expression via eIF2α phosphorylation [24,25]. We examined if salubrinal could induce osteoblastic mineralization through ATF4. Salubrinal increased ATF4 expression in MC3T3-E1 cells (Fig. 4A) and significantly promoted calcium accumulation (marker of bone nodule formation) in the cells as shown by alizarin red-S staining (Fig. 4B). ATF4 depletion, however, noticeably diminished calcium accumulation (Fig. 4B), suggesting the involvement of ATF4 in salubrinal-induced osteoblast differentiation. In addition, salubrinal-induced expressions of alkaline phosphatase (ALP) and RANKL, representative markers of osteoblast differentiation, were reduced by ATF4 knockdown (Fig. 4C).
Fig. 4.
Induction of ATF4 dependent osteoblast differentiation and RANKL induction by salubrinal in MC3T3-E1 cells. (A) MC3T3-E1 cells were infected with lentiviral shATF4 and treated with salubrinal (30 μM, 24 hr). Total cell lysates were prepared for immunoblot blot analysis. (B) Cells infected with shATF4 were treated with varying concentrations of salubrinal for 21 days and were subjected to alizalin staining for the measurement of intracellular calcium accumulation. (C) Cells infected with shATF4 and treated with salubrinal for 5 days were lysed and total RNA was extracted for quantitative PCR analysis. (D and E) BMM and MC3T3-E1 cells were co-cultured in normal medium (NM) or differentiation medium (DM) in the presence of varying concentration of salubrinal for 5 days. (D) TRAP staining was performed for the measurement of differentiated BMM cells, (E) and the number of TRAP-positive multinucleated osteoclasts was counted. All the bars represent mean SE±from a representative triplicate experiment. The significance was determined by Student’s t-test (*P<0.5; **P<0.1).
Osteoclast differentiation can often be regulated directly or indirectly by environmental cells, such as osteoblasts or stromal cells producing osteoclastogenic factors including RANKL [6]. Given the complicated results showing that salubrinal reduced RANKL-induced NFATc1 expression in osteoclast cells (Fig. 2E) while increasing RANKL level in osteoblast cells (Fig. 4C), it could be speculated that a factor, like RANKL, secreted from osteoblast cells in the presence of salubrinal could induce the differentiation of osteoclast cells. Thus, mouse BMMs cells were co-cultured with differentiating MC3T3-E1 osteoblast cells pretreated with salubrinal. Medium from the differentiating cells MC3T3-E1 (DM) was found to induce osteoclast differentiation of BMM cells (Fig. 4D). Salubrinal treatment, however, blocked osteoclast differentiation as shown by the reduction of TRAP-positive cells (Fig. 4D and E). These results suggested that osteoclast differentiation could be induced by RANKL produced from activated osteoblast cells but be blocked by salubrinal through the inhibition of NFATc1 expression.
3.5. Evaluation of salubrinal in the bone destruction model mice
Finally, to confirm the effect of salubrinal in vivo, bone loss model mice were prepared by RANKL injection [20]. Measurement of total femoral bone mineral density (BMD) using Dual-emission X-ray absorptiometry (DEXA) revealed that salubrinal significantly reduced RANKL-induced osteoporotic bone loss (Fig. 5A). Furthermore, 2D micro-CT image also showed that RANKL-induced bone destruction was notably reduced by salubrinal treatment (Fig. 5B and C).
Fig. 5.
Evaluation of salubrinal in bone destruction model mice. (A) Salubrinal (1 mg/kg) was injected intraperitoneally 24 hr before the first RANKL injection, and the mice (n=6) subsequently received simultaneous injections of salubrinal (1 mg/kg, IP) and RANKL (0.5 mg/kg, IP) at 24 hr intervals for 3 days. Total femoral bone mineral density (BMD) was measured using pDEXA X-ray bone densitometer. BMD was calculated using the bone mineral content (BMC) in the measured area. (B) Distal femurs and tibiae from NC (negative control), RANKL (positive control) and RANKL plus salubrinal mice were examined by micro-CT. Two-dimensional reconstruction of tibiae and femurs revealed increased bone mass in salubrinal treated mice compared with control littermates. (C) Histograms representing the 2D trabecular structural parameters in tibiae and femurs; bone volume per tissue volume (BV/TV), trabecular separation (Tb. Ts), trabecular thickness (Tb. Th) and trabecular number (Tb. N). All the bars are mean SE±from a representative triplicate experiment. The significance was determined by ANOVA test (*P<0.5; **P<0.1).
4. Discussion
The endoplasmic reticulum (ER) is an organelle that has essential roles in multiple cellular processes and occupies a unique position responsible for the control of protein quality [26]. A number of intracellular stressors induce ER-stress directly or indirectly, triggering an evolutionarily conserved response termed the unfolded protein response (UPR). Among the many factors in UPR signaling, eIF2α and ATF4 are the focus in this study since eIF2α is the target of salubrinal and regulates the expression of ATF4 in ER-stress. Although the involvement of eIF2α in osteoclast differentiation and of ATF4 in osteoblast differentiation was evaluated in this study, their roles in other cells remain to be determined. However, regardless of their important contributions to osteoporosis, NFATc1 is expected to be critical for proper regulation of osteoporosis based on the data obtained in our study.
A simplified summary of the intracellular mechanism of salubrinal is described in Fig. 6. Upon ER-stress, activated PERK induces transient phosphorylation of eIF2α for attenuation of new protein synthesis. Re-initiation of protein synthesis can be achieved by dephosphorylation of eIF2α by protein phosphatase activity of GADD34. Salubrinal blocks eIF2α dephosphorylation, maintaining the phosphorylated status of eIF2α. Although global protein synthesis could be suppressed by eIF2α phosphorylation, ATF4 was reported to be increased by eIF2α phosphorylation [24]. Thus, salubrinal is expected to increase ATF4 level, consequently leading to the induction of osteoblast differentiation and RANKL expression. In contrast, eIF2α phosphorylation in salubrinal-treated cells could also suppress NFATc1 expression in response to RANKL and down-regulate osteoclast differentiation.
Fig. 6.

Simplified model for anti-osteoporosis activity of salubrinal. Salubrinal inhibits dephosphorylation of eIF2α and maintains phosphorylated status of eIF2α, up-regulating ATF4 expression and osteoblast differentiation. On the other hand, osteoclast differentiation by RANKL is suppressed by salubrinal. Reduction of NFATc1 expression seems to be critical for the inhibitory effect of salubrinal on osteoclast differentiation.
A prototypical bone disease osteoporosis that is characterized by reduced bone strength is observed most frequently in postmenopausal women and in elderly men [27], but the mechanism has not yet been elucidated in detail. Recent studies have demonstrated that ER-stress proteins are associated with age-related disorders; eIF2α mutant mice survived 18 hr after birth [22], PERK is essential for skeletal development [28], and sensitivity of UPR system including eIF2α phosphorylation and ATF4 expression declines with age [29]. In this regard, ER-stress regulation through eIF2α and ATF4 could be a good system for anti-osteoporosis. So far, there have been two classes of osteoporosis therapeutic agents; anti-resorption drugs likely bisphosphonates targeting osteoclasts, and anabolic drugs against bone formation likely parathyroid hormone (PTH) targeting osteoblasts. Although these drugs are effective, most of them have drawbacks showing side-effects and incomplete recovery, part of which due to single target application, either bone resorption or formation [30]. In this respect, our study showed that salubrinal is a small molecule having dual functions for both cell types, and hence it is strongly suggested that salubrinal could be developed as a new type of anti-osteoporosis therapeutics affecting both osteoclast and osteoblast differentiation. In addition, further exploitation of this salubrinal-eIF2α-ATF4 connection would be valuable for identifying new generation of osteoporosis regulatory proteins and small molecules.
Acknowledgements
This work was supported by the World Class Institute (WCI) Program (WCI 2009-002), Global R&D Center (GRDC) Program, World Class University (R31-2008-000-10103) and in part by Basic Research Program (2012-0003778) of the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST), Technology Development Program for Agriculture and Forestry, Ministry for Agriculture, Forestry and Fisheries, and also supported by KRIBB Research Initiative Program.
Footnotes
Conflict of interest and other disclosures All authors state that they have no conflicts of interest.
References
- [1].Goltzman D. Nature Reviews. Drug Discovery. 2002;1(10):784–796. doi: 10.1038/nrd916. [DOI] [PubMed] [Google Scholar]
- [2].Boyle WJ, Simonet WS, Lacey DL. Nature. 2003;423(6937):337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- [3].Aubin JE. Reviews in Endocrine & Metabolic Disorders. 2001;2(1):81–94. doi: 10.1023/a:1010011209064. [DOI] [PubMed] [Google Scholar]
- [4].Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveira-dos-Santos AJ, Van G, Itie A, Khoo W, Wakeham A, Dunstan CR, Lacey DL, Mak TW, Boyle WJ, Penninger JM. Nature. 1999;397(6717):315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- [5].Tanaka S, Takahashi N, Udagawa N, Tamura T, Akatsu T, Stanley ER, Kurokawa T, Suda T. The Journal of Clinical Investigation. 1993;91(1):257–263. doi: 10.1172/JCI116179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Takayanagi H. Nature Reviews. Immunology. 2007;7(4):292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- [7].Biskobing DM, Fan X, Rubin J. Journal of Bone and Mineral Research. 1995;10(7):1025–1032. doi: 10.1002/jbmr.5650100706. [DOI] [PubMed] [Google Scholar]
- [8].Novack DV, Teitelbaum SL. Annual Review of Pathology. 2008;3:457–484. doi: 10.1146/annurev.pathmechdis.3.121806.151431. [DOI] [PubMed] [Google Scholar]
- [9].Teitelbaum SL, Ross FP. Nature Reviews. Genetics. 2003;4(8):638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- [10].Manolagas SC. Endocrine Reviews. 31(3):266–300. doi: 10.1210/er.2009-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Terzioglu M, Larsson NG. Novartis Foundation Symposium. 2007;287:197–208. doi: 10.1002/9780470725207.ch14. (discussion 208–113) [DOI] [PubMed] [Google Scholar]
- [12].Naidoo N, Ferber M, Master M, Zhu Y, Pack AI. The Journal of Neuroscience. 2008;28(26):6539–6548. doi: 10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Naidoo N. Ageing Research Reviews. 2009;8(3):150–159. doi: 10.1016/j.arr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- [14].Liu J, Hoppman N, O’Connell JR, Wang H, Streeten EA, McLenithan JC, Mitchell BD, Shuldiner AR. Journal of Bone and Mineral Research. 2012;27(2):331–341. doi: 10.1002/jbmr.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. Science. 2005;307(5711):935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- [16].Smith WW, Jiang H, Pei Z, Tanaka Y, Morita H, Sawa A, Dawson VL, Dawson TM, Ross CA. Human Molecular Genetics. 2005;14(24):3801–3811. doi: 10.1093/hmg/ddi396. [DOI] [PubMed] [Google Scholar]
- [17].Reijonen S, Putkonen N, Norremolle A, Lindholm D, Korhonen L. Experimental Cell Research. 2008;314(5):950–960. doi: 10.1016/j.yexcr.2007.12.025. [DOI] [PubMed] [Google Scholar]
- [18].Ron D, Walter P. Nature Reviews. Molecular Cell Biology. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- [19].Soung NK, Kang YH, Kim K, Kamijo K, Yoon H, Seong YS, Kuo YL, Miki T, Kim SR, Kuriyama R, Giam CZ, Ahn CH, Lee KS. Molecular and Cellular Biology. 2006;26(22):8316–8335. doi: 10.1128/MCB.00671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tomimori Y, Mori K, Koide M, Nakamichi Y, Ninomiya T, Udagawa N, Yasuda H. Journal of Bone and Mineral Research. 2009;24(7):1194–1205. doi: 10.1359/jbmr.090217. [DOI] [PubMed] [Google Scholar]
- [21].Harding HP, Zhang Y, Ron D. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- [22].Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Molecular Cell. 2001;7(6):1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- [23].Elefteriou F, Ahn JD, Takeda S, Starbuck M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, Clement K, Vaisse C, Karsenty G. Nature. 2005;434(7032):514–520. doi: 10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- [24].Dever TE. Cell. 2002;108(4):545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- [25].Lewerenz J, Maher P. The Journal of Biological Chemistry. 2009;284(2):1106–1115. doi: 10.1074/jbc.M807325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim I, Xu W, Reed JC. Nature Reviews. Drug Discovery. 2008;7(12):1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- [27].Cummings SR, Melton LJ. Lancet. 2002;359(9319):1761–1767. doi: 10.1016/S0140-6736(02)08657-9. [DOI] [PubMed] [Google Scholar]
- [28].Wei J, Sheng X, Feng D, McGrath B, Cavener DR. Journal of Cellular Physiology. 2008;217(3):693–707. doi: 10.1002/jcp.21543. [DOI] [PubMed] [Google Scholar]
- [29].Paz Gavilan M, Vela J, Castano A, Ramos B, del Rio JC, Vitorica J, Ruano D. Neurobiology of Aging. 2006;27(7):973–982. doi: 10.1016/j.neurobiolaging.2005.05.010. [DOI] [PubMed] [Google Scholar]
- [30].Rachner TD, Khosla S, Hofbauer LC. Lancet. 377(9773):1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]