Abstract
Inflammation associated reactive oxygen and nitrogen species (RONs), including peroxynitrite (ONOO−) and nitric oxide (NO· ), create base lesions that potentially play a role in the toxicity and large-scale genomic rearrangements associated with many malignancies. Nevertheless, little is known about the functional role of base excision repair (BER) deficiencies following exposure to RONs. Here, we explore the role of XRCC1 in modulating the levels of RONs-induced genotoxicity. XRCC1 is a scaffold protein critical for BER for which polymorphisms modulate the risk of cancer. We exploited CHO and human glioblastoma cell lines engineered to carry varied levels of BER components to study XRCC1. Cytotoxicity and SSB-intermediate levels were evaluated following exposure to the ONOO− donor, SIN-1, and to gaseous NO·. XRCC1 null cells are slightly more sensitive to SIN-1 toxicity. To explore the potential importance of XRCC1 in response to NO· -induced lesions, we used small-scale bioreactors to expose cells to NO· and found that XRCC1 does not impact genotoxicity in CHO cells, suggesting a minimal role for XRCC1 in response to RONs. However, using a molecular beacon assay to measure AAG-mediated lesion removal in vitro, we found that XRCC1 facilitates AAG-initiated BER of two key NO· -induced lesions: 1,N6-ethenoadenine and hypoxanthine. Furthermore, overexpression of AAG rendered XRCC1 cells sensitive to NO· -induced DNA damage and toxicity. These results show that AAG is a key glycosylase in response to NO· exposure and that the cellular and functional impact of XRCC1 depends upon the level of AAG. These studies are some of the first to assess the functional role of XRCC1 in response to NO·, and demonstrate the importance of BER balance when considering the impact of XRCC1 polymorphisms in response to RONs.
Keywords: XRCC1, base excision repair, inflammation, nitric oxide, SIN1, glycosylase
1.0 Introduction
The association between chronic inflammation and cancer is well established, with upwards of 15% of all malignancies associated with chronic inflammation [1–5]. Of particular interest is the role played by reactive oxygen and nitrogen species (RONs) in the initiation and promotion of malignancy. During inflammatory states, neutrophils and macrophages become activated and release genotoxic RONs, including nitric oxide (NO· ) and superoxide [6, 7]. These RONs are not only toxic to target cells, but they also induce collateral damage on neighboring tissue. We hypothesize that DNA damage caused by NO· and NO· -derived chemicals can cause genotoxicity and genomic instability that initiates and promotes malignancy.
NO· is an important signaling molecule at lower concentrations [8–10], but can be genotoxic at higher concentrations [11, 12]. Indeed, via its reactions with superoxide and oxygen to form ONOO− and nitrous anhydride (N2O3) respectively, NO· has been shown to induce numerous DNA lesions both in vitro [12, 13] and in vivo [14]. For example, ONOO− can oxidize guanine, making 8-nitroguanine [15], which can spontaneously depurinate leaving an abasic site [6, 15]. Furthermore, ONOO− can make direct DNA single strand breaks (SSBs) or damage DNA bases in the presence of CO2, via the action of nitrosoperoxycarbonate [16, 17]. In contrast, N2O3 primarily creates base deamination products; converting guanine to xanthine, adenine to hypoxanthine and cytosine to uracil, all of which induce transition mutations [13]. Additionally, RONs can cause lipid peroxidation, creating secondary metabolites that can subsequently alkylate DNA and create mutagenic alkylation lesions such as 1,N6-ethenoadenine (εA) [18, 19]. The alkylation lesion εA blocks replicative polymerases [20]; is mutagenic causing A:T→G:C transitions, A:T→T:A transversions [21, 22]; and is associated with the signature p53 and Kras mutations observed in rat and human liver cancers following exposure to vinyl chloride [23, 24].
The DNA lesions resulting from exposure to RONs can promote a number of deleterious phenotypes including toxicity [25, 26], elevated DNA double strand breaks (DSBs) [27], point mutations [14, 28] and large-scale genomic rearrangements [29]. Consequently, the repair of RON-mediated DNA damage may be an important mechanism for mitigating the toxic and tumorigenic effects of these lesions. Indeed, a recent study shows that mice deficient in alkyl adenine DNA glycosylase (AAG), the DNA glycosylase that repairs NO· -induced εA, have an increased susceptibility to inflammation-induced cancer [30]. Thus, an individual’s capacity to repair inflammation-mediated DNA lesions may represent a hitherto poorly defined risk factor for inflammation-mediated malignancy [30–34].
In mammalian cells, base excision repair (BER) is responsible for repairing many of the inflammation-mediated lesions described above. Briefly, this pathway consists of five, enzyme-catalyzed steps: lesion excision, strand scission, gap tailoring, DNA synthesis and ligation. For a detailed review of BER, the authors refer the reader elsewhere [31, 35–38]. Importantly all five enzymatic steps in the BER pathway are putatively facilitated by the x-ray repair cross-complementing group 1 (XRCC1) protein. Indeed, XRCC1 is a scaffold protein that, although lacking its own enzymatic activity, facilitates the enzymatic roles of numerous other BER pathway proteins. For example AAG, the DNA glycosylase that carries out excision of 3-methyl adenine (3me-A) [39], hypoxanthine (Hx) [39], εA [30] and other alkylpurines [40], has been shown to be physically and functionally associated with XRCC1 [39]. In addition, XRCC1 interacts with APE1, PARP1 and DNA polymerase β (Polβ) [35, 41, 42]. As a consequence of its central role in base excision repair, XRCC1 deficient cells are deficient in the repair of alkylation [43, 44] and ionizing radiation-induced [45–47] DNA damage. Furthermore, XRCC1 deficient cells are acutely sensitive to killing by alkylating agents, such as methyl methane sulfonate (MMS) [48, 49]. Additionally, XRCC1 deficient cells have higher baseline and post-MMS-exposure sister chromatid exchanges (SCEs) [50, 51], which are a measure of large-scale genomic rearrangements and thus represent a genomic instability mutator phenotype. Thus, despite having no enzymatic activity, XRCC1’s ability to act as a scaffold that accelerates BER clearly has many important biological effects.
Inflammatory RONs, such as NO·, are highly genotoxic. Nevertheless, very little is known about the susceptibility of XRCC1 deficient cells following exposure to inflammatory genotoxins. Previous studies have shown that XRCC1 deficient Chinese hamster ovary (CHO) cells have elevated levels of SCEs following exposure to NO· [51], suggesting an important role for XRCC1 during inflammation. Furthermore, XRCC1 deficient mouse [48] and CHO [52] cells are moderately sensitive to hydrogen peroxide [53]. Collectively, these findings suggest that XRCC1 may be important in the repair of RON-mediated DNA damage and may modulate toxicity and cancer risk. Indeed, various polymorphic variants in XRCC1 have been shown to be either positively [54] or negatively [55–57] associated with various inflammation-associated malignancies. These findings underscore the importance of defining the role of XRCC1 in response to inflammatory genotoxins, both for cancer risk assessment and for therapeutic intervention.
Here, we have analyzed the role of XRCC1 in the context of two cell types. In CHO cells, we found that XRCC1 prevents SIN1 and MMS-induced toxicity, but does not impact NO· -induced toxicity. However, using a molecular beacon assay with extracts from human glioblastoma cells to measure glycosylase-mediated base lesion removal, we found that XRCC1 facilitates AAG-initiated BER of two NO· -induced DNA base lesions, εA and hypoxanthine. Furthermore, in the human glioblastoma cell lines, BER imbalance mediated by overexpression of AAG sensitized cells to NO· -induced toxicity and DNA damage as well as the response of these cells to MMS. Together these data indicate that AAG is an important DNA glycosylase in the response to NO· -exposure, and that the role of XRCC1 depends upon the level of AAG. Therefore, the impact of an XRCC1 deficiency is context dependent, wherein under conditions of BER imbalance, there is increased susceptibility to NO· -induced genomic instability.
2.0. Materials and Methods
2.1 Cells and Cell Culture
AA8 and EM9 CHO cells were obtained from ATCC (Manassas, VA) and the human complemented EM9 cells were engineered, and kindly provided by the lab of Larry Thompson [46]. CHO cells were cultured in 10% FBS (Atlanta Biologics, Lawrenceville, GA) in DMEM (Cat#11965, Invitrogen, Carlsbad, CA) and penicillin/streptomycin (100U/ml; 100μg/ml) (Sigma, St. Louis, MO). CHO cells were cultured in 150mm dishes (Falcon, BD) and were kept growing exponentially, being passaged approximately once every three days. Cell passaging was conducted by aspirating media, rinsing in warm PBS and treating cells with warm trypsin (Invitrogen, Carlsbad, CA) for 5–10 min, quenching with 10mL media and then transferring 1mL of the resultant cell suspension to 30 mL media.
Engineering of glioblastoma LN428 cells has previously been published [58, 59]. The cell line LN428 (herein referred to as LN428WT) is an established glioblastoma derived cell line with mutations in p53 and deletions in p14ARF and p16 and is WT for PTEN. Briefly, human AAG expressing cells were generated by transfection of the AAG expression plasmid pRS1422 using FuGene 6 transfection reagent (Roche, Indianapolis, IN) according to the manufacturer’s protocol. Transfected cell lines were cultured in G418 for two weeks, and individual clones stably expressing human AAG were selected. The lentiviral shuttle vectors (control: pLKO.1-puro-turbo green fluorescent protein [GFP]) were purchased from Sigma, as was the XRCC1 shRNA shuttle vector (Gene ID: NM_006297, Clone ID: NM_006297.1–1489s1c1, Target sequence: CCAGTGCTCCAGGAAGATATA), via the UPCI Lentiviral facility. Lentiviral particles were generated by co-transfection of four plasmids [pLKO.1-puro-TurboGFP or pLKO.1-puro-XRCC1shRNA together with pMD2.g(VSVG), pVSV-REV and pMDLg/pRRE] into 293-FT cells using FuGene 6 Transfection Reagent (Roche, Indianapolis, IN), as described previously [58]. Forty-eight hours after transfection, lentivirus-containing supernatant was collected and passed through 0.45 μM filters to isolate the viral particles. Lentiviral transduction was performed as described earlier [58]. The LN428WT cells were cultured in alpha MEM (Invitrogen, Carlsbad, CA), 10% fetal bovine serum (Atlanta Biologics, Lawrenceville, GA), antibiotic/antimycotic (Sigma), gentamicin, L-glutamine (Sigma). The XRCC1-KD and the LN428/GFP glioblastoma cell lines were cultured in the same media as the LN428WT cells described above supplemented with 1 μg/mL puromycin (Sigma). The AAGOE glioblastoma cell line (LN428 cells modified for elevated expression of AAG) was cultured in the same media as the LN428WT cells and supplemented with 600 μg/mL Geneticin (InVitrogen) and the AAGOE/XRCC1-KD glioblastoma cell line (LN428 cells modified for elevated expression of AAG and loss of XRCC1 expression) was cultured in the same media as the LN428WT cells and supplemented with 600μg/mL Geneticin (InVitrogen) plus 1 μg/mL puromycin (Sigma).
2.2 Cell extract preparation and Immunoblot
CHO whole cell extracts were prepared using RIPA buffer according to the manufacturer’s instructions (Pierce, Rockford, IL). Protein concentration assessment in cell extracts was done by BCA quantification (Pierce, Rockford, IL) according to the manufacturer’s instructions. Volumes containing 20–100 μg of protein were loaded into a precast 4–20% gradient Tris-HCl gel (BioRad, Hercules, CA). Protein was transferred onto a nitrocellulose membrane and XRCC1 protein was probed using a rabbit polyclonal primary antibody against XRCC1 (Abcam, Cambridge, MA) and an HRP-conjugated secondary goat antibody against rabbit IgG. For the loading control a rabbit monoclonal antibody (Cell Signaling, Beverly, MA) against β-actin was used. LN428 nuclear extracts were prepared and protein concentrations were determined as described previously [58]. 20 μg of protein was loaded on a precast 4–20% Tris-glycine gel (Invitrogen) or 25 mg of protein was loaded onto a precast 4–20% Mini-PROTEAN TGX gel (Bio-Rad). For the analysis of the human glioblastoma cell lines, the following primary antibodies were used in the immunoblot assays: anti-human MPG (Mab; clone 506-3D) [58]; anti PCNA (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-XRCC1 (Bethyl Labs; Montgomery, TX).
2.3. CHO MMS and SIN-1 Exposures for Comet Assay
Log phase CHO cells were rinsed with warm PBS and passaged as described above. Approximately 24 hours later, cells were trypsinized, counted and re-suspended in 1% low melting point (LMP) agarose (Invitrogen, Carlsbad, CA) at 37°C at a density of 2.5×105 cells/mL. A volume of 500μL (1.25×105 cells) of gel containing re-suspended cells was then pipeted onto 37°C pre-warmed, agarose-precoated glass slides and cover-slipped. The cells were allowed to settle onto the slides for 10 minutes and were then moved to 4°C to allow the gel to solidify. Coverslips were removed and the cells were exposed to either MMS or SIN-1 at the specified concentrations in PBS (Invitrogen, Carlsbad, CA). For the repair experiments, following exposure, the slides were rinsed with PBS and were transferred to CHO media for the specified period of time before being lysed for the alkaline comet assay, as described below (Section 2.6).
To investigate the DNA repair capacity of CHO cell lines exposed to MMS, the CometChip, a high throughput version of the comet assay, was used as described by us previously [60]. This approach enabled four cell lines (AA8, EM9, H9T3-6-3, H9T3-7-1) to be evaluated simultaneously in a standard 96-well format. Briefly, at least 10,000 cells were added to each well and allowed to settle into arrayed microwells by gravity. Excess cells were aspirated after 15 minutes and the arrayed cells were encapsulated in a layer of 1% low melting point agarose. After exposure to the specified dose of MMS, the wells were rinsed with PBS and replenished with fresh growth medium for the specified period of time before lysis was conducted for the alkaline comet assay.
2.4 CHO and Glioblastoma NO· Exposures
For NO· exposures, 1×106 CHO or glioblastoma cells were seeded in media into 60mm tissue culture dishes (BD Falcon, Franklin Lakes, NJ). Approximately 24 hours later, dishes were secured to the bottom of a 100mL bioreactor and exposed to gases for specified periods of time, as previously described [29, 61, 62]. Following exposure, a sample of media was taken for nitrite quantification and the cells were trypsinized, counted, and analyzed for clonogenic survival and DNA damage using the alkaline comet assay.
2.5 Clonogenic Survival Assay
Clonogenic survival assay was conducted using standard protocols. Briefly, CHO and glioblastoma cells were seeded and approximately 24 hours later, cells were exposed to SIN-1 or MMS for 30min at 37°C. The cells were then rinsed with PBS and incubated in media for 10–14 days to form colonies. The colonies were then fixed in 1:1 Methanol:PBS and stained with 0.5% crystal violet in 1:1 methanol:PBS for 10min. Colonies were rinsed with de-ionized water, allowed to dry and were then counted. P values were calculated by student’s t-test on the mean of at least three replicate experiments.
2.6 Alkaline Comet Assay
To measure the accumulation of base excision repair intermediates following exposure to a given genotoxin, a modified version of the alkali comet assay (single cell gel electrophoresis) was employed [63]. Briefly, cells immobilized in 1% agarose were lysed for at least one hour in lysis buffer containing 2.5 M NaCl, 100 mM Na2EDTA, and 10 mM tris and 1% triton X-100 at pH 10. Following lysis, cells in the gel are held in electrophoresis buffer (0.3 M NaOH, 1 mM Na2EDTA) for 40 minutes for alkali unwinding. Following the alkali unwinding incubation, electrophoresis was performed at 1V/cm, 300mA for 30 min. Slides were neutralized in 0.4 M tris, before staining with 500 μg/mL ethidium bromide. Analysis was carried out by Metafer v.3.6.7 (Metasystems, Waltham, MA). At least 100 nucleoids per condition were analyzed and the median calculated. Each experiment was replicated at least 3 times and the mean of all replicates plotted. P values were calculated by student’s t-test. To measure the accumulation of base excision repair intermediates following MMS exposure, a single high throughput comet gel was used to simultaneously assess all cell types and repair time points. After conducting the standard comet assay protocol described above, analysis was carried out using automated software described previously [60].
2.7 AAG-specific Molecular Beacon Assay
Oligodeoxyribonucleotides were designed, as we have described [59] to form a stem-loop structure with 13 nucleotides in the loop and 15 base pairs in the stem. Carboxyfluorescein (6-Fam) is a fluorescent molecule that is quenched by Dabcyl in a non-fluorescent manner via Förster Resonance Energy Transfer (FRET) [64, 65]. Oligodeoxyribonucleotides were purchased from Integrated DNA Technologies (Coralville, USA):
FD-Con, 6-FAM-dGCACTATTGAATTGACACGCCATGTCGATCAATTCAATAGTGC-Dabcyl, where 6-FAM is carboxyfluorescein and Dabcyl is 4-(4′-dimethylaminophenylazo) benzoic acid;
FD-MPG1, 6-FAM-dGCACTXTTGAATTGACACGCCATGTCGATCAATTCAATAGTGC-Dabcyl, where X is 1,N6-ethenoadenine (εA);
FD-MPG2, 6-FAM-dGCACTXTTGAATTGACACGCCATGTCGATCAATTCAATAGTGC-Dabcyl, where X is hypoxanthine (Hx);
FD-THF, 6-FAM-dGCACTXTTGAATTGACACGCCATGTCGATCAATTCAATAGTGC-Dabcyl, where X is terahydrofuran (THF), an abasic site analog [66].
When the DNA is in a stem-loop structure, the 6-FAM at the 5′ end and the Dabcyl at the 3′ end are brought into close proximity, enabling efficient quenching of 6-FAM by Dabcyl. If the lesion (εA or Hx) is removed by AAG and the DNA backbone is hydrolyzed by APE1 (THF), the 6-FAM-containing oligonucleotide (4 bases in length) will dissociate from the hairpin at 37°C and the 6-FAM dissociation from the DNA hairpin prevents the quenching by Dabcyl. The increase in 6-FAM-mediated fluorescence is proportional to the amount of the lesion removed. Any increase in fluorescence in control beacon with a normal adenine would be the result of non-specific cleavage of the DNA backbone.
Nuclear protein extracts were prepared as previously described [59]. Approximately 500 μL of nuclear protein extracts were dialyzed twice using the Slide-A-Lyzer Dialysis Cassette with a 7,000 molecular weight cut-off (Pierce; Rockford, IL). The samples were dialyzed 2x for 90 min each at 4°C in the following buffer: 50 mM Hepes pH7.5, 100 mM KCl, 0.5 mM EDTA, 20% Glycerol and 1 mM DTT. Reactions were performed using 10 μg of dialyzed protein extract and beacon substrate (final conc = 40 nM) in the following buffer: 25 mM HEPES-KOH pH7.8, 150 mM KCl, 0.5 mM EDTA, 1% Glycerol, 0.5 mM DTT. Fluorescence was measured, every 20 seconds for 60 minutes, using a StepOnePlus real-time PCR system and expressed as arbitrary units (AU). The fluorescence data was then analyzed as we have described [59]. Briefly, background fluorescence (from incubation of the beacon alone) was eliminated by subtracting the fluorescence values of a control well containing no protein extract from all wells. The fluorescence value of the 5-minute time point was selected as the zero value for each well. We subtracted this value from all other time points in that well so all graphs begin from zero AU and 5 minutes after initiating the reaction. Five minutes was selected as the point from which to begin comparisons because time points earlier than four minutes contained variations in absolute fluorescence measurements independent of molecular beacon and cell line (not shown). The mean of three separate trials was plotted with error bars representing the standard error of the mean. The mean fluorescence values and the values for the standard error of the mean for each are listed in Dataset S1 in the Supplement.
2.8 MMS-induced Cytotoxicity Assay in Glioblastoma Cells
MMS-induced cytotoxicity was determined by an MTS assay, a modified MTT assay as described previously [58]. Briefly, cells (LN428WT, XRCC1-KD, AAGOE and AAGOE/XRCC1-KD) were seeded in 96-well dishes at 2000 cells/well. After 24 hours, cells were exposed to serial dilutions of MMS (6 wells per dose) in growth media for 4 hours at 37°C. MMS-containing medium was replaced with fresh medium and the plates were incubated at 37°C for an additional 48 hours at which point the total cell number was determined by a modified MTT assay (MTS; Promega) [67]. Metabolically active cells were quantified by the bioreduction of the MTS tetrazolium compound by recording absorbance at 490 nm using a microplate reader. Results were calculated from the average of three separate experiments and are reported as the % of treated cells relative to the cells in control wells (% Control).
2.9 Quantitative RT-PCR Analysis
Validation of shRNA-mediated reduction of XRCC1 expression was initially determined by quantitative RT-PCR (qRT-PCR) using an Applied Biosystems StepOnePlus system. Briefly, 80,000 cells were lysed and reverse transcribed using the Applied Biosystems Taqman® Gene Expression Cells-to-CT™ Kit. Each sample was analyzed in triplicate and the results are an average of all three analyses. Analysis of mRNA expression was conducted as per the manufacturer (ΔΔCT method) using Applied Biosystems TaqMan® Gene Expression Assays (XRCC1: Hs00959834_m1) and normalized to the expression of human β-actin (part #4333762T).
3.0. Results and Discussion
3.1. Role of XRCC1 in response to NO· and ONOO− exposure in CHO cells
Reactive oxygen and nitrogen species released by activated immune cells can cause myriad DNA lesions. Among these potentially deleterious reactive species are NO· and superoxide, which can combine to form ONOO−, a dominant reactive nitrogen species formed under physiological conditions [6]. Our first objective was therefore to investigate whether cells deficient in XRCC1 are susceptible to ONOO− and NO·.
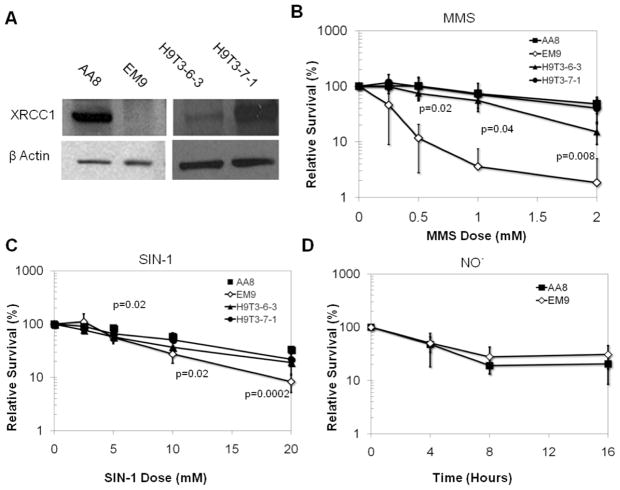
To learn about the role of XRCC1 in response to NO· and ONOO−, we exploited EM9 CHO cells, which are effectively null for XRCC1 as they carry a frameshift mutation that results in expression of only a third of the XRCC1 protein [68] (Fig. 1A). The EM9 mutant cells were initially isolated from their parental AA8 CHO cell line by a generalized mutant screen for sensitivity to DNA damaging agents and are sensitive to killing by ethyl methane sulfonate (EMS), MMS and ionizing radiation [45, 47, 49]. Additionally, EM9 cells display defective base excision repair [49, 69] and have elevated frequency of baseline [49, 70] and post-genotoxin exposure sister chromatid exchanges (SCEs). As controls, we used the parental AA8 CHO cell line and XRCC1-deficient EM9 cells complemented with the human XRCC1 cDNA (H9T3-6-3 and H9T3-7-1 cells, gift of L. Thompson) (Fig. 1A)[46].
Figure 1. Role of XRCC1 in modulating toxicity of ONOO− and NO· in CHO cells.
A. XRCC1 WT (AA8) and null (EM9) CHO cells, as well as human XRCC1 complemented EM9 (H9T3-6-3 and H9T3-7-1) cells were used to assess the toxicity of (B) MMS, (C) the ONOO− donor SIN-1 and (D) gaseous NO·, using a clonogenic survival assay. P values were produced by a two-tailed t-test and only values below 0.05 are displayed.
Following exposure to SIN-1, gaseous NO· or MMS, CHO cells were assessed for survival via a clonogenic survival assay. XRCC1 deficient cells have previously been demonstrated to be deficient in the repair methylation damage, and are therefore highly sensitive to MMS [43, 44, 46, 49, 69, 71]. We therefore exposed cells to MMS as a reference against which to benchmark the effects of exposure to inflammatory RONs. As previously established [44, 46, 49], XRCC1 deficient CHO EM9 cells were significantly more susceptible to MMS exposure than XRCC1 WT AA8 cells and human XRCC1-complemented H9T3-6-3 and H9T3-7-1 cells (Fig. 1B). In contrast, XRCC1 deficient cells were only slightly more sensitive to SIN-1 exposure (p< 0.05 for doses over 5mM) when compared to WT CHO cells or human XRCC1-complemented CHO cells (Fig. 1C). These results are consistent with studies showing that XRCC1 deficient cells are mildly sensitive to oxidizing agents [48]. In contrast to SIN-1, XRCC1 null EM9 cells were no more sensitive than WT cells to NO· exposure (Fig. 1D), even at doses that are toxic to WT cells.
The observation that XRCC1 cells have a slight, yet significant increase in sensitivity to SIN-1 and not to NO· is surprising and may be due to the difference in the spectrum of lesions made by SIN-1 versus NO·. While SIN-1 and NO· may make many of the same lesions via ONOO−, including direct DNA SSBs [16, 72] and oxidized base lesions including 8-oxo-7, 8-dihydro-2′-deoxyguanosine [73], 8-nitro-2′-deoxyguanosine [15, 17] and 5-guanidino-4-nitroimidazole [74], NO· exposure can result in other chemistries. For example, NO· can cause base deamination via the action of nitrous anhydride (N2O3). Deamination of DNA by nitrous anhydride can form xanthine and hypoxanthine from guanine and adenine, respectively. While the lesions formed by oxidative damage are generally removed by bifunctional glycosylases, the deamination products are generally removed by monofunctional glycosylases. Therefore, the uncleaved abasic sites formed after removal of NO· -induced deamination products may be more readily tolerated by XRCC1 deficient cells, compared to the cleaved abasic sites formed at sites of ONOO−- induced oxidative lesions (or the directly induced SSBs) [75–77]
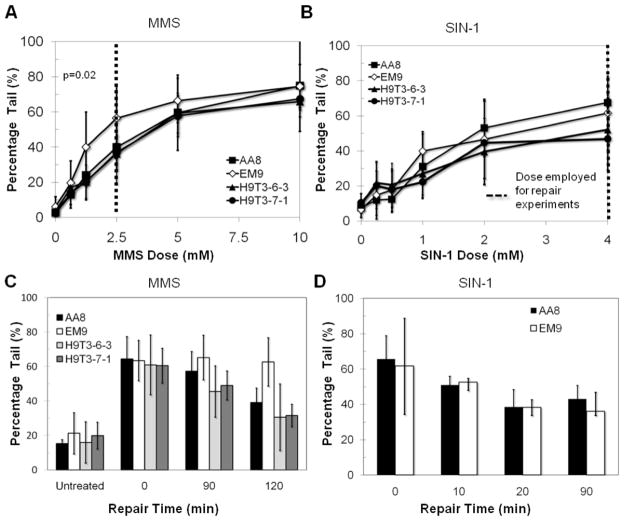
As an alternative approach for evaluating the biological role of XRCC1 in response to RONs, we investigated XRCC1’s impact on BER capacity following RON-induced DNA damage. To assess BER, we used the alkaline single cell gel electrophoresis (comet assay) using both the standard comet assay protocol and the high throughput format described by us previously [60]. The alkaline comet assay is a well established, gel-electrophoresis based assay that has been used to measure DNA SSBs, apurinic/apyrimidinic (AP) sites and alkali-labile base lesions in single cells [60, 63, 78, 79]. The assay can thus be used to measure the levels of DNA base excision repair intermediates, following exposure to inflammatory RONs. We found that XRCC1 deficient EM9 cells trend towards a greater accumulation of base excision repair intermediates compared to WT cells, as the cells are exposed to increasing concentrations of MMS (Fig. 2A), which is consistent with previous studies [41, 43, 44, 48, 71]. XRCC1 deficient EM9 cells also clearly show slower repair kinetics, as expected (Fig. 2C). In contrast to MMS, XRCC1 deficient cells displayed equivalent levels of BER intermediates as WT cells following exposure to increasing concentrations of SIN-1 (Fig. 2B) and the XRCC1 null EM9 cells repair SIN-1-induced BER-intermediates with the same efficiency as the AA8 WT cells (Fig. 2D). These data indicate that, at least in CHO cells, XRCC1 does not play a significant role in the repair of SIN-1-induced DNA damage, as assessed by the alkali comet assay.
Figure 2. Role of XRCC1 in the repair of MMS and SIN-1 mediated DNA damage.
Base excision repair intermediates measured in CHO cells exposed to (A) MMS and (B) SIN-1. Repair kinetics subsequently generated by treating CHO cells at the dose specified (---) for (C) MMS and (D) SIN-1. P values were produced by a student’s t-test and only values below 0.05 are displayed.
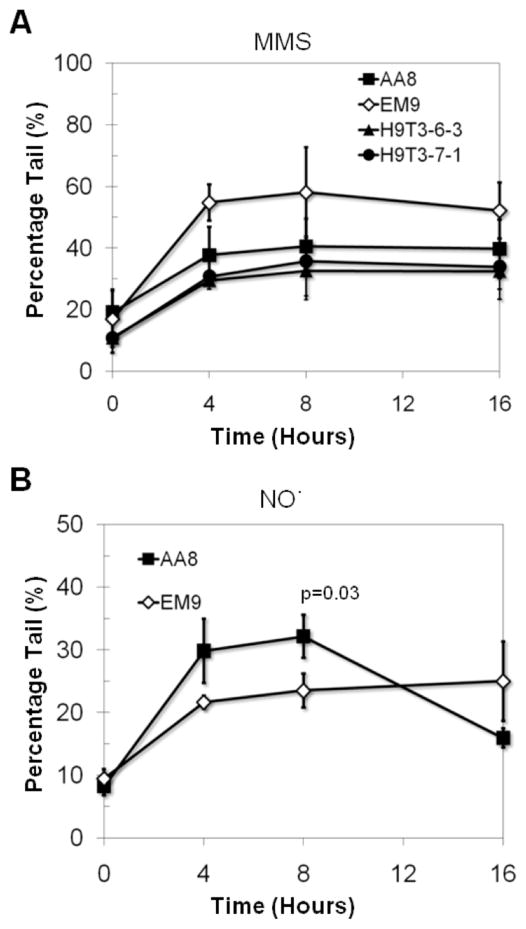
To explore the possibility that XRCC1 modulates repair of NO· -induced DNA damage, we used the comet assay to assess the BER intermediate levels in EM9 and AA8 CHO cells following exposure to increasing durations of exposure to NO·. A previously described bioreactor system was used to expose cells to NO· [61, 62]. Briefly, cells were exposed to gaseous NO· at a steady state concentration (~2μM for 10% NO· and ~11μM for 100% NO· ) for varying lengths of time to deliver increasing cumulative doses, typically expressed as μM/min, but expressed here as duration of exposure for simplicity. For comparison, and to control for any results due to the chronicity of exposure to NO·, conditions were developed for chronic MMS exposure that yield a comparable impact on the levels of DNA damage (as revealed by the comet assay). We found that the XRCC1 deficient EM9 cells displayed elevated levels of BER-intermediates following increasing duration of exposure to MMS (Fig. 3A), which is consistent with their slower repair kinetics (Fig. 2C). In contrast to MMS exposure, rather than seeing increased levels of BER intermediates in the XRCC1 deficient cells, following increasing duration of exposure to NO· we observed that the WT AA8 cells showed higher levels of NO· -induced BER intermediates (Fig. 3B).
Figure 3. Role of XRCC1 in the repair of MMS and NO· mediated DNA damage.
Base excision repair intermediates measured in CHO cells exposed to (A) MMS and (B) NO·. P values were produced by student’s t-test and only values below 0.05 are displayed.
The observation that the XRCC1 WT cells have higher levels of BER intermediates than the XRCC1 null EM9 cells may at first glance seem anomalous. However, since the comet assay detects AP sites and frank DNA SSBs, the comet assay effectively detects DNA species in-between the glycosylase step and the ligation step of the BER pathway. Therefore, the increase in BER intermediates in WT cells relative to the XRCC1 deficient cells following exposure to NO· may reflect XRCC1-facilitated removal of NO· -induced base damage. Since SIN-1 exposure did not result in the same BER-intermediate dynamics as those observed following exposure to NO·, one could speculate that XRCC1 may modulate the function of a glycosylase (e.g., AAG) that acts specifically on NO· -induced DNA damage and not SIN-1-induced damage, i.e. deamination products like xanthine and hypoxanthine. Indeed, other groups have shown that AAG can excise hypoxanthine [80–82], and that XRCC1 can enhance this activity [39]. We therefore set out to more directly explore the impact of XRCC1 on AAG-mediated excision of NO· -mediated DNA damage.
3.2 Role of XRCC1/ AAG interaction in human (glioblastoma LN428) cell extracts
Two key mutagenic lesions created by NO· and excised by AAG are hypoxanthine (Hx) and 1,N6-ethenoadenine (εA). Hx is the deamination product of adenine that is formed by NO· via the nitrosative action of nitrous anhydride (N2O3)—an auto-oxidation product of the reaction between NO· and molecular oxygen [6]. Hx mispairs with cytosine, causing A:T →G:C transition mutations. In contrast, εA can mispair with adenine, guanine or cytosine [83] and is a DNA damage-mediated product of RON-derived lipid peroxidation [18, 84, 85]. Furthermore, εA inhibits DNA replication [86] and is thus a toxic base lesion [87]. We therefore investigated whether, and to what extent, XRCC1 facilitates AAG-initiated BER of Hx and εA DNA lesions. To do this, we used LN428 glioblastoma cells that were WT (LN428WT) or that were engineered to be deficient in XRCC1 via lentiviral-mediated shRNA knockdown (XRCC1-KD). In addition, we used these same cells engineered to over-express AAG (AAGOE and AAGOE/XRCC1-KD), and cells engineered to express GFP (GFP) as a control for the exogenous constructs in the other cells. As shown, the LN428WT cells express a very low, undetectable level of AAG but express measurable levels of XRCC1, as determined by immunoblot analysis (Fig. 4A). To compare the expression levels of XRCC1 and AAG among the four cell lines, the immunoblot was scanned and quantified using NIH Image J and the associated analysis software package, normalizing the expression across the four cell lines to the AAGOE cells and to the expression of PCNA within each sample. As indicated in the plot of relative protein expression (Fig. 4B), the AAG expression levels are very similar when comparing the AAGOE and AAGOE/XRCC1-KD cells. Similarly, the expression of XRCC1 is similar when comparing the LN428WT and AAGOE cells. Importantly, no expression of XRCC1 was detected in the XRCC1-KD and AAGOE/XRCC1-KD cells.
Figure 4. XRCC1 facilitates AAG-mediated NO· -induced base lesion removal.
(A) Immunoblot showing expression of AAG (MPG) and XRCC1 protein in wild type (LN428WT), AAG over-expressing cells (AAGOE), XRCC1 knockdown cells (XRCC1-KD), and AAG over-expressing cells with XRCC1 knocked down (AAGOE/XRCC1-KD). (B) Bar graph showing the relative protein expression of AAG and XRCC1 in LN428WT, AAGOE, XRCC1-KD and AAGOE/XRCC1-KD cells. The immunoblot in (A) was scanned and quantified using NIH Image J and the associated analysis software package, normalizing the expression across the four cell lines to the AAGOE cells and to the expression of PCNA within each sample. (C–F) Fluorescent molecular beacon assay indicating XRCC1 facilitated excision of NO· -induced base lesions by AAG from DNA double-stranded oligonucleotides containing (C) no lesion, (D) an abasic analog THF, (E) hypoxanthine (Hx) or (F) 1,N6-ethenoadenine (εA).
To measure AAG-dependent DNA glycosylase activity or APE1-dependent abasic site hydrolysis, we developed a fluorescence-based molecular beacon excision assay to measure the efficiency with which AAG can excise specific base lesions (Hx or εA) or that APE1 can hydrolyze an abasic-site analog (Tetrahydrofuran, THF) in real-time, in vitro, essentially as we have described [59]. We assessed the ability of cell extracts from LN428WT, XRCC1-KD, AAGOE and AAGOE/XRCC1-KD cells to excise εA, Hx or THF [59]. We observed minimal fluorescence when extracts were incubated with a control molecular beacon that contained no lesion (Fig. 4C). Since the signal (fluorescence) is generated following base lesion removal and abasic site hydrolysis by APE1, we also tested each lysate using a THF beacon to ensure that all four lysates could similarly cleave an abasic site. As expected, a similar increase in fluorescence was observed by all four of the cell extracts when evaluated for APE1 activity using the THF beacon (Fig. 4D). It should be noted that we did not observe any difference in activity between the XRCC1 expressing cells (LN428WT and AAGOE) and the XRCC1-KD cells (XRCC1-KD and AAGOE/XRCC1-KD), indicating that cleavage of the AP site by APE1 is not influenced by XRCC1. An earlier report using purified enzymes suggested that XRCC1 stimulates APE1-mediated 3′-5′ exonuclease activity but it is not likely that this activity of APE1 is measured in this assay [88]. However, our data is in contrast to an earlier report suggesting that XRCC1 and APE1 interact and that XRCC1 stimulates APE1 activity [42]. Clearly, such an XRCC1-dependent stimulation of APE1 activity is not observed here, at least for the hydrolysis of the THF abasic site analog.
Conversely, a molecular beacon containing hypoxanthine (Hx) resulted in a time-dependent increase in fluorescence when incubated with extracts from LN428WT, XRCC1-KD, AAGOE and AAGOE/XRCC1-KD cells, which is indicative of base release by the AAG glycosylase and cleavage of the resulting AP site (Fig. 4E). As expected, cells over-expressing AAG (AAGOE) yielded the highest increase in fluorescence, which is consistent with these cells being the most efficient at excising the Hx lesion. However, when XRCC1 expression is knocked down in the AAGOE cells (AAGOE/XRCC1-KD), there is a decrease in the observed fluorescence (Fig. 4E). The observed 25% reduction in glycosylase activity in the XRCC1 deficient cells (when measured at 40 min) indicates that XRCC1 facilitates AAG excision of Hx, which is consistent with a previous report using purified recombinant proteins [39]. In contrast to the cells overexpressing AAG, the molecular beacon assay yields very little signal for the LN428WT or the XRCC1-KD cell extracts. The nominal increase in fluorescence is probably due to the exceptionally low amounts of AAG in the LN428WT parental cell line. Indeed, the levels of AAG in the LN428WT parental cell line and the XRCC1-KD cell line are so low as to preclude noticeable detection by western blot (Fig. 4A). Thus, additional expression of AAG leads to a significant increase in the excision of εA and Hx.
To learn if these observations are generalizable to other AAG substrates, we queried whether 1,N6-ethenoadenine (εA) excision by AAG could be facilitated by XRCC1. We incubated cell extracts from the glioblastoma cells with a molecular beacon containing εA. Similar to the Hx molecular beacon, a time-dependent increase in fluorescence was observed following incubation with lysates from LN428WT, XRCC1-KD, AAGOE and AAGOE/XRCC1-KD cells. Of the cell lines examined, the AAGOE cells yielded the greatest increase in fluorescence and the AAGOE/XRCC1-KD cells yielded a significant (45% at 40 min) reduction in the kinetics of BER initiation (Fig. 4F). The difference in the observed fluorescence between the AAGOE cells and the AAGOE/XRCC1-KD cells indicates that XRCC1 does indeed facilitate AAG excision of εA. Collectively, these data indicate that XRCC1 is capable of stimulating the AAG-mediated excision of two inflammation-associated base lesions, Hx and εA, in vitro.
3.3 Role of XRCC1/AAG in response to NO· exposure in human (glioblastoma LN428) cells
Results from the molecular beacon assay show that XRCC1 can facilitate AAG-mediated BER of inflammation-induced base lesions in vitro. Given that CHO cells and human cells can differ significantly, we set out to measure the impact of XRCC1 on BER intermediates in glioma cells, following exposure to MMS and NO·. Following exposure to MMS, we observed a time-dependent increase in BER intermediates for all genotypes examined, as measured by comet assay and the observed increase in the tail moment following 50 μM MMS exposure (Fig. 5A). Importantly, we found that the LN428WT, XRCC1-KD, GFP and the AAGOE cells, accumulated similar levels of BER intermediates. This indicates that the increase in base-excision initiation that is the result of over expression of AAG is tolerated in these cells. Additionally, this result indicates that the deficiency in XRCC1 is similarly tolerated at these low doses of MMS (50 μM), with respect to the accumulation of BER-intermediates. However, when XRCC1 is suppressed in cells that over-express AAG (AAGOE/XRCC1-KD cells), there is a significant accumulation of BER intermediates (Fig. 5A). The accumulation of BER intermediates in cells that over-express AAG and are deficient in XRCC1 (AAGOE/XRCC1-KD) is consistent with a model in which XRCC1 is enhancing AAG activity but primarily facilitating the activity of a BER step downstream of the glycosylase. Thus AAG initiates BER and excises damaged bases from DNA to form BER intermediates that can then be detected by the comet assay, and the lack of XRCC1 prevents the efficient resolution of these BER intermediates. We speculate that the lack of XRCC1 results in the decreased efficiency of the 5′-dRP lyase activity of Polβ, previously shown to be the rate-limiting step in the monofunctional glycosylase initiated BER [89–92]. Indeed, it has been demonstrated that the XRCC1/Polβ interaction is important for the repair of both alkylation [44] and oxidative [52] DNA damage. Thus, the accumulation of BER intermediates in AAGOE/XRCC1-KD cells may primarily be due to an accumulation of BER intermediates occurring in-between the glycosylase step and the 5′-dRP lyase gap-tailoring step of Polβ. This result demonstrates the importance of XRCC1 in tolerating an imbalance in BER, caused by excess glycosylase activity. Therefore, when considering the potential impact of an observed increase in glycosylase activity, the efficiency of downstream components, including XRCC1, must be considered. For example, we have demonstrated recently that the sensitivity of human cancer cells to chemotherapeutic alkylating agents depends on the expression profile of both AAG (MPG) and Polβ [59, 93].
Figure 5. Role of XRCC1 and AAG in human glioblastoma cells following exposure to MMS.
(A) Base excision repair intermediates, as determined by the Comet Assay, measured in human glioblastoma cells following exposure to MMS (0.05mM in media). The % Tail (Y-axis) for each cell line is potted as a function of MMS exposure time (hours). P values were produced by a student’s t-test and only values below 0.05 are displayed. (B) MMS-induced cytotoxicity in LN428WT, AAGOE, XRCC1-KD and AAGOE/XRCC1-KD cells. After treatment (4 hrs MMS followed by 48 hrs in normal media), viable cells were determined using a modified MTT assay. Plots show the % viable cells as compared to untreated (control) cells. Means are calculated from six values in each experiment. Results indicate the mean ± S.E. of three independent experiments.
In order to determine the susceptibility of the human glioblastoma cell lines to killing by MMS, we exposed cells to increasing concentrations of MMS, ranging from 0 to 500 μM, for 4 hours (the time found to yield a significant difference among these 4 cell lines in the comet assay, Fig. 5A) and measured cell survival after 48 hours, as we have described previously [58, 59, 93]. We found that both cell lines with elevated expression of AAG (AAGOE and AAGOE/XRCC1-KD cells) were significantly more sensitive to MMS than those with low (undetectable) expression of AAG (LN428WT and XRCC1-KD cells) (Fig. 5B). The XRCC1-KD cells were not more sensitive to killing by MMS than LN428WT cells, likely due to the low level of AAG expression in these cells (see Fig. 4A). Consistent with the observation that the AAGOE/XRCC1-KD cells displayed highly elevated BER intermediates following exposure to MMS (Fig. 5A), the AAGOE/XRCC1-KD cells were more susceptible to killing by MMS than even the AAGOE cells (Fig. 5B). This suggests that the elevated BER intermediates observed in AAGOE/XRCC1-KD cells are highly toxic and that the loss of XRCC1 prevents downstream processing of these BER intermediates.
To explore the extent to which XRCC1 can facilitate AAG-initiated base excision of NO· -mediated DNA damage in live cells, we exposed glioblastoma cells to two steady state concentrations of ~2μM and ~11μM of NO·, achieved by exposure to 10% and 100% NO·, respectively. Interestingly, the response of the human glioblastoma cells to NO· exposure was different from the CHO cell response. Indeed, the glioblastoma cells tolerated the 10% NO· better than the CHO cells, resulting in only nominal increases in BER-intermediates (Fig. 6A). This result is consistent with previous findings that different cell types have different thresholds for, and thus respond differently to, NO· exposures [11, 61, 62]. Similar to the MMS exposures, BER intermediates in the XRCC1-KD cells, AAGOE cells and GFP control cells exposed to 10% and 100% NO· were similar to the levels in the LN428WT cells (Fig. 6A and B). In contrast, the AAGOE/XRCC1-KD cells showed higher levels of BER intermediates compared to XRCC1-KD cells (Fig. 6C). These data suggest that AAG removes RONS-induced base damage, and furthermore reveal that the impact of XRCC1 is dependent on AAG.
Figure 6. Role of XRCC1 and AAG in human glioblastoma cells following exposure to NO.
·.
Base excision repair intermediates in human glioblastoma cells exposed to (A) 10% NO·, (B) 100% NO· for LN428WT and AAG over-expressing cells (AAGOE) and (C) 100% NO· for XRCC1 deficient (XRCC1-KD) cells and XRCC1 deficient, AAG over-expressing cells (AAGOE/XRCC1-KD). Asterisk indicates that the average difference for AAGOE/XRCC1-KD versus XRCC1-KD is significantly greater than zero (95% CI). Toxicity in human glioblastoma cells following exposure to (D) 10% NO·, (E) 100% NO· for LN428WT and AAG over-expressing cells (AAGOE) and (F) 100% NO· for XRCC1 deficient (XRCC1-KD) cells and XRCC1 deficient, AAG over-expressing cells (AAGOE/XRCC1-KD).
To learn about the role of XRCC1 in modulating the toxic effects of NO·, all glioblastoma genotypes were challenged with both 10% (Fig. 6D) and 100% (Fig. 6E and F) NO·. Greater toxicity was observed for the 100% NO· exposure, and all genotypes were found to be sensitive to NO· induced toxicity. Furthermore, consistent with the levels of BER-intermediates measured in these cells, none of the glioblastoma cells assessed were relatively more sensitive to killing by NO·, when compared to the LN428WT cells (Fig. 6E). However, cells that over-express AAG and lack XRCC1 (AAGOE/XRCC1-KD) were significantly more sensitive to NO· toxicity than XRCC1 deficient cells (p = 0.04; Fig. 6F). Taken together, these results demonstrate that the role of XRCC1 in protecting cells from NO· -induced toxicity is dependent on the levels of AAG.
4.0 Conclusions
Understanding the role that reactive nitrogen and oxygen play in initiating and promoting malignancy is important for risk assessment and therapeutic intervention in inflammation-associated malignancies. Here, we examined a putative risk factor in inflammation-associated malignancy: the role of XRCC1 in the repair of DNA damage by the reactive nitrogen species ONOO- and NO·. We found that in CHO cells, XRCC1 is protective against ONOO− mediated toxicity, but this protection was at least partially independent of XRCC1’s role in base excision repair. These results suggest that XRCC1 may be modulating toxicity in a BER-independent fashion. Indeed, there is evidence that XRCC1 may play a role in Ku-independent, non-homologous end joining [94–96], a mechanism for the repair of DNA double strand breaks which can result in toxicity and other deleterious phenotypes [97]. Additionally there are several lines of evidence that suggest XRCC1 may play a role in homologous recombination, another pathway by which DNA DSBs can be repaired. For example, Taylor et al. showed that XRCC1 partially co-localizes with an important homologous recombination protein, Rad51, implicating XRCC1 in homologous recombination [98]. Further, the high levels of XRCC1 observed in testes relative to levels in other cell types [99] and the occurrence of these higher levels specifically in pachytene spermatocytes [100], implicate XRCC1 in meiotic recombination. These examples, and indeed our findings here, are consistent with role(s) for XRCC1, in addition to its role in base excision repair.
While we observed that XRCC1 may stimulate AAG excision of the inflammation mediated base lesions Hx and εA in vitro, as measured by the Molecular Beacon assay, in human glioblastoma cells XRCC1 appears to primarily act downstream of the glycosylase when cells are exposed to either NO· damage and for alkylation damage. This is particularly striking following exposure of the cells to MMS, where the loss of XRCC1 in the cells with elevated expression of AAG (AAGOE/XRCC1-KD) show extreme hypersensitivity to MMS. Furthermore, although the XRCC1-KD cells do not show an increase in the levels of NO· -induced BER intermediates, the human glioblastoma LN428WT and XRCC1-KD cells express almost no AAG, raising the possibility that XRCC1’s role in response to NO· is AAG-dependent. Overexpression of AAG indeed demonstrated a significant rise in BER intermediates in the XRCC1-KD cells (AAGOE/XRCC1-KD), indicating that AAG actively removes RONS-induced lesions in human cells, and that AAG acts upstream of XRCC1 in response to NO· and MMS exposure. Likewise, cells expressing high levels of AAG do not show a significant increase in the levels of BER intermediates unless XRCC1 is knocked down, especially at low doses of MMS. Taken together, the impact of deficiencies in XRCC1, such as those caused by polymorphic variants, appears to be dependent on the levels of AAG. Given the reports on over-expression of AAG in several cancers [101–104], it is suggested that the expression of AAG be considered when evaluating potential functional impact of XRCC1 polymorphic variants.
Supplementary Material
Dataset S1 contains the mean fluorescence values for each of the Molecular Beacon plots shown in Figure 4, as well as the values for the standard error of the mean for each.
Highlight.
XRCCl facilitates AAG-initiated BER of ethenoadenine and hypoxanthine, key lesions that are induced by reactive nitrogen species (RNS)
XRCCl has a modest impact on sensitivity to RNS-induced genotoxicity.
Overexpression of AAG renders XRCCl deficient cells sensitive to RNS-induced DNA damage and toxicity.
These data indicate that XRCCl and AAG modulate RNS genotoxicity, and suggest that the impact ofXRCCl polymorphisms is dependent upon the level of AAG.
Acknowledgments
We are grateful to Larry Thompson for the human complemented EM9 cells. We thank Debby Pheasant, Jen Calvo and Harshawardhan Banda for microscopy support and we thank the MIT Center for Environmental Health Sciences for its support (NIEHS P30-ES002109). We thank Gerald Wogan and Laura Trudel (MIT) for their input and technical support and also thank Sandy Schamus (Univ. of Pitt.) for help with the MTS assays. Technical support for use of the high throughput comet assay platform was provided by David Weingeist and David Wood. Work was supported by NIH Grants P01-CA026731 and 2-R01-CA079827-05A1 with partial support from U01-ES016045 (BE). This work was also supported by grants from the Pittsburgh Foundation and the National Institutes of Health (NIH) [GM087798; CA148629] to RWS and from the NIH [ES019498] to RWS and BE. Support for the UPCI Lentiviral Facility was provided to RWS by the Cancer Center Support Grant from the National Institutes of Health [P30 CA047904]. Support was also provided by the University of Pittsburgh Department of Pharmacology & Chemical Biology to DS.
Footnotes
RWS is a Scientific Consultant to Trevigen, Inc. The rest of the authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6.0 References
- 1.Pisani P, Parkin DM, Munoz N, Ferlay J. Cancer and infection: estimates of the attributable fraction in 1990. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1997;6:387–400. [PubMed] [Google Scholar]
- 2.Kuper H. Infections as a major preventable cause of human cancer. Journal of Internal Medicine. 2000;248:171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 3.Ohshima H, Tatemichi M, Sawa T. Chemical basis of inflammation-induced carcinogenesis. Archives of biochemistry and biophysics. 2003;417:3–11. doi: 10.1016/s0003-9861(03)00283-2. [DOI] [PubMed] [Google Scholar]
- 4.Balkwill F, Mantovani A. Inflammation and Cancer: back to Virchow? The Lancet. 2001;357:539–544. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 5.Coussens L. Inflammation and Cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. International journal of cancer. Journal international du cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annual Review of Biochemistry. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- 9.Cary SP, Winger JA, Derbyshire ER, Marletta MA. Nitric oxide signaling: no longer simply on or off. Trends in biochemical sciences. 2006;31:231–239. doi: 10.1016/j.tibs.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: implications in cellular signaling. Free radical biology & medicine. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burney S, Tamir S, Gal A, Tannenbaum SR. A mechanistic analysis of nitric oxide-induced cellular toxicity. Nitric Oxide. 1997;1:130–144. doi: 10.1006/niox.1996.0114. [DOI] [PubMed] [Google Scholar]
- 12.Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, Dunams TM, Cebula TA, Koch WH, Andrews AW, Allen JS, et al. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 13.Burney S, Caulfield JL, Niles JC, Wishnok JS, Tannenbaum SR. The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutation Research. 1999;424:37–49. doi: 10.1016/s0027-5107(99)00006-8. [DOI] [PubMed] [Google Scholar]
- 14.Gal A, Wogan GN. Mutagenesis associated with nitric oxide production in transgenic SJL mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:15102–15107. doi: 10.1073/pnas.93.26.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yermilov V, Rubio J, Ohshima H. Formation of 8-nitroguanine in DNA treated with peroxynitrite in vitro and its rapid removal from DNA by depurination. FEBS Lett. 1995;376:207–210. doi: 10.1016/0014-5793(95)01281-6. [DOI] [PubMed] [Google Scholar]
- 16.Tretyakova NY, Burney S, Pamir B, Wishnok JS, Dedon PC, Wogan GN, Tannenbaum SR. Peroxynitrite-induced DNA damage in the supF gene: correlation with the mutational spectrum. Mutat Res. 2000;447:287–303. doi: 10.1016/s0027-5107(99)00221-3. [DOI] [PubMed] [Google Scholar]
- 17.Yermilov V. Effects of carbon dioxide/bicarbonate on induction of DNA single strand breaks and formation of 8-nitroguanine, 8-oxoguanine, and base propenal mediated by peroxynitrite. FEBS Lett. 1996;399:67–70. doi: 10.1016/s0014-5793(96)01288-4. [DOI] [PubMed] [Google Scholar]
- 18.el Ghissassi F, Barbin A, Nair J, Bartsch H. Formation of 1,N6-ethenoadenine and 3,N4-ethenocytosine by lipid peroxidation products and nucleic acid bases. Chemical research in toxicology. 1995;8:278–283. doi: 10.1021/tx00044a013. [DOI] [PubMed] [Google Scholar]
- 19.Nair J, Gal A, Tamir S, Tannenbaum SR, Wogan GN, Bartsch H. Etheno adducts in spleen DNA of SJL mice stimulated to overproduce nitric oxide. Carcinogenesis. 1998;19:2081–2084. doi: 10.1093/carcin/19.12.2081. [DOI] [PubMed] [Google Scholar]
- 20.Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu AK, Wood ML, Niedernhofer LJ, Ramos LA, Essigmann JM. Mutagenic and genotoxic effects of three vinyl chloride-induced DNA lesions: 1,N6-ethenoadenine, 3,N4-ethenocytosine, and 4-amino-5-(imidazol-2-yl)imidazole. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]
- 22.Pandya GA, Moriya M. 1,N6-ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 23.Barbin A. Formation of DNA etheno adducts in rodents and humans and their role in carcinogenesis. Acta Biochim Pol. 1998;45:145–161. [PubMed] [Google Scholar]
- 24.Barbin A. Etheno-adduct-forming chemicals: from mutagenicity testing to tumor mutation spectra. Mutation Research. 2000;462:55–69. doi: 10.1016/s1383-5742(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 25.Szabo C, Zingarelli B, O’Connor M, Salzman AL. DNA strand breakage, activation of poly (ADP-ribose) synthetase, and cellular energy depletion are involved in the cytotoxicity of macrophages and smooth muscle cells exposed to peroxynitrite. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1753–1758. doi: 10.1073/pnas.93.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 27.Clemons NJ, McColl KE, Fitzgerald RC. Nitric oxide and acid induce double-strand DNA breaks in Barrett’s esophagus carcinogenesis via distinct mechanisms. Gastroenterology. 2007;133:1198–1209. doi: 10.1053/j.gastro.2007.06.061. [DOI] [PubMed] [Google Scholar]
- 28.deRojas-Walker T, Tamir S, Ji H, Wishnok JS, Tannenbaum SR. Nitric oxide induces oxidative damage in addition to deamination in macrophage DNA. Chemical research in toxicology. 1995;8:473–477. doi: 10.1021/tx00045a020. [DOI] [PubMed] [Google Scholar]
- 29.Kiziltepe T, Yan A, Dong M, Jonnalagadda VS, Dedon PC, Engelward BP. Delineation of the chemical pathways underlying nitric oxide-induced homologous recombination in mammalian cells. Chem Biol. 2005;12:357–369. doi: 10.1016/j.chembiol.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, Rickman BH, Rogers AB, Moroski-Erkul CA, McFaline JL, Schauer DB, Dedon PC, Fox JG, Samson LD. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. The Journal of clinical investigation. 2008;118:2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Frosina G. Tumor suppression by DNA base excision repair. Mini Rev Med Chem. 2007;7:727–743. doi: 10.2174/138955707781024544. [DOI] [PubMed] [Google Scholar]
- 33.Sweasy JB, Lang T, DiMaio D. Is base excision repair a tumor suppressor mechanism? Cell Cycle. 2006;5:250–259. doi: 10.4161/cc.5.3.2414. [DOI] [PubMed] [Google Scholar]
- 34.Tudek B. Base excision repair modulation as a risk factor for human cancers. Molecular aspects of medicine. 2007;28:258–275. doi: 10.1016/j.mam.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell research. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base Excision Repair and lesion-dependent sub-pathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zharkov DO. Base excision DNA repair. Cellular and molecular life sciences: CMLS. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campalans A, Marsin S, Nakabeppu Y, O’Connor RT, Boiteux S, Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst) 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; Washington, D.C: 2006. [Google Scholar]
- 41.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO Journal. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO Journal. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor RM, Thistlethwaite A, Caldecott KW. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Molecular and Cellular Biology. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong HK, Wilson DM., 3rd XRCC1 and DNA polymerase beta interaction contributes to cellular alkylating-agent resistance and single-strand break repair. J Cell Biochem. 2005;95:794–804. doi: 10.1002/jcb.20448. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz JL, Giovanazzi S, Weichselbaum RR. Recovery from sublethal and potentially lethal damage in an X-ray-sensitive CHO cell. Radiation Research. 1987;111:58–67. [PubMed] [Google Scholar]
- 46.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Molecular and Cellular Biology. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.vanAnkeren SC, Murray D, Meyn RE. Induction and rejoining of gamma-ray-induced DNA single- and double-strand breaks in Chinese hamster AA8 cells and in two radiosensitive clones. Radiation Research. 1988;116:511–525. [PubMed] [Google Scholar]
- 48.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson LH, Brookman KW, Dillehay LE, Carrano AV, Mazrimas JA, Mooney CL, Minkler JL. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutation Research. 1982;95:427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- 50.Thompson LH, Brookman KW, Minkler JL, Fuscoe JC, Henning KA, Carrano AV. DNA-mediated transfer of a human DNA repair gene that controls sister chromatid exchange. Molecular and Cellular Biology. 1985;5:881–884. doi: 10.1128/mcb.5.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zdzienicka MZ, van der Schans GP, Natarajan AT, Thompson LH, Neuteboom I, Simons JW. A Chinese hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis. 1992;7:265–269. doi: 10.1093/mutage/7.4.265. [DOI] [PubMed] [Google Scholar]
- 52.Dianova, Sleeth KM, Allinson SL, Parsons JL, Breslin C, Caldecott KW, Dianov GL. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res. 2004;32:2550–2555. doi: 10.1093/nar/gkh567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulkarni A, McNeill DR, Gleichmann M, Mattson MP, Wilson DM., 3rd XRCC1 protects against the lethality of induced oxidative DNA damage in nondividing neural cells. Nucleic Acids Research. 2008;36:5111–5121. doi: 10.1093/nar/gkn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Tse CK, Eaton A, Mohrenweiser HW, Newman B, Bell DA. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiology Biomarkers & Prevention. 2001;10:217–222. [PubMed] [Google Scholar]
- 55.Lee JM, Lee YC, Yang SY, Yang PW, Luh SP, Lee CJ, Chen CJ, Wu MT. Genetic polymorphisms of XRCC1 and risk of the esophageal cancer. International journal of cancer. Journal international du cancer. 2001;95:240–246. doi: 10.1002/1097-0215(20010720)95:4<240::aid-ijc1041>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 56.Ratnasinghe D, Yao SX, Tangrea JA, Qiao YL, Andersen MR, Barrett MJ, Giffen CA, Erozan Y, Tockman MS, Taylor PR. Polymorphisms of the DNA repair gene XRCC1 and lung cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2001;10:119–123. [PubMed] [Google Scholar]
- 57.Stern MC, Umbach DM, van Gils CH, Lunn RM, Taylor JA. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2001;10:125–131. [PubMed] [Google Scholar]
- 58.Tang J, Goellner EM, Wang XW, Trivedi RN, St Croix CM, Jelezcova E, Svilar D, Brown AR, Sobol RW. Bioenergetic Metabolites Regulate Base Excision Repair-Dependent Cell Death in Response to DNA Damage. Molecular Cancer Research. 2010;8:67–79. doi: 10.1158/1541-7786.MCR-09-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang JB, Svilar D, Trivedi RN, Wang XH, Goellner EM, Moore B, Hamilton RL, Banze LA, Brown AR, Sobol RW. N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro-oncology. 2011;13:471–486. doi: 10.1093/neuonc/nor011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li CQ, Pang B, Kiziltepe T, Trudel LJ, Engelward BP, Dedon PC, Wogan GN. Threshold effects of nitric oxide-induced toxicity and cellular responses in wild-type and p53-null human lymphoblastoid cells. Chemical research in toxicology. 2006;19:399–406. doi: 10.1021/tx050283e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang C, Trudel LJ, Wogan GN, Deen WM. Thresholds of nitric oxide-mediated toxicity in human lymphoblastoid cells. Chemical research in toxicology. 2003;16:1004–1013. doi: 10.1021/tx0340448. [DOI] [PubMed] [Google Scholar]
- 63.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nature protocols. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 64.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 65.Yaron A, Carmel A, Katchalski-Katzir E. Intramolecularly quenched fluorogenic substrates for hydrolytic enzymes. Anal Biochem. 1979;95:228–235. doi: 10.1016/0003-2697(79)90210-0. [DOI] [PubMed] [Google Scholar]
- 66.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. The Journal of biological chemistry. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 67.Berridge MV, Tan AS. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Archives of Biochemistry and Biophysics. 1993;303:474–482. doi: 10.1006/abbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 68.Shen MR, Zdzienicka MZ, Mohrenweiser H, Thompson LH, Thelen MP. Mutations in hamster single-strand break repair gene XRCC1 causing defective DNA repair. Nucleic Acids Research. 1998;26:1032–1037. doi: 10.1093/nar/26.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutation Research. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 70.Dillehay LE, Thompson LH, Carrano AV. DNA-strand breaks associated with halogenated pyrimidine incorporation. Mutation Research. 1984;131:129–136. doi: 10.1016/0167-8817(84)90052-x. [DOI] [PubMed] [Google Scholar]
- 71.Brem R, Hall J. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Research. 2005;33:2512–2520. doi: 10.1093/nar/gki543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doulias PT, Barbouti A, Galaris D, Ischiropoulos H. SIN-1-induced DNA damage in isolated human peripheral blood lymphocytes as assessed by single cell gel electrophoresis (comet assay) Free radical biology & medicine. 2001;30:679–685. doi: 10.1016/s0891-5849(00)00511-6. [DOI] [PubMed] [Google Scholar]
- 73.Inoue S, Kawanishi S. Oxidative DNA damage induced by simultaneous generation of nitric oxide and superoxide. FEBS Lett. 1995;371:86–88. doi: 10.1016/0014-5793(95)00873-8. [DOI] [PubMed] [Google Scholar]
- 74.Gu F, Stillwell WG, Wishnok JS, Shallop AJ, Jones RA, Tannenbaum SR. Peroxynitrite-induced reactions of synthetic oligo 2′-deoxynucleotides and DNA containing guanine: formation and stability of a 5-guanidino-4-nitroimidazole lesion. Biochemistry. 2002;41:7508–7518. doi: 10.1021/bi020148q. [DOI] [PubMed] [Google Scholar]
- 75.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 76.Spek EJ, Vuong LN, Matsuguchi T, Marinus MG, Engelward BP. Nitric oxide-induced homologous recombination in Escherichia coli is promoted by DNA glycosylases. Journal of bacteriology. 2002;184:3501–3507. doi: 10.1128/JB.184.13.3501-3507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spek EJ, Wright TL, Stitt MS, Taghizadeh NR, Tannenbaum SR, Marinus MG, Engelward BP. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. Journal of bacteriology. 2001;183:131–138. doi: 10.1128/JB.183.1.131-138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collins AR, Dobson VL, Dusinska M, Kennedy G, Stetina R. The comet assay: what can it really tell us? Mutation Research. 1997;375:183–193. doi: 10.1016/s0027-5107(97)00013-4. [DOI] [PubMed] [Google Scholar]
- 79.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 80.Asaeda A, Ide H, Asagoshi K, Matsuyama S, Tano K, Murakami A, Takamori Y, Kubo K. Substrate specificity of human methylpurine DNA N-glycosylase. Biochemistry. 2000;39:1959–1965. doi: 10.1021/bi9917075. [DOI] [PubMed] [Google Scholar]
- 81.Miao F, Bouziane M, O’Connor TR. Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Research. 1998;26:4034–4041. doi: 10.1093/nar/26.17.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci U S A. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Speina E, Kierzek AM, Tudek B. Chemical rearrangement and repair pathways of 1,N6-ethenoadenine. Mutation Research. 2003;531:205–217. doi: 10.1016/j.mrfmmm.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 84.Chung FL, Chen HJ, Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 85.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 86.Tolentino JH, Burke TJ, Mukhopadhyay S, McGregor WG, Basu AK. Inhibition of DNA replication fork progression and mutagenic potential of 1, N6-ethenoadenine and 8-oxoguanine in human cell extracts. Nucleic Acids Research. 2008;36:1300–1308. doi: 10.1093/nar/gkm1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bartsch H, Nair J. Oxidative stress and lipid peroxidation-derived DNA-lesions in inflammation driven carcinogenesis. Cancer Detection & Prevention. 2004;28:385–391. doi: 10.1016/j.cdp.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 88.Dyrkheeva NS, Khodyreva SN, Lavrik OI. Interaction of APE1 and other repair proteins with DNA duplexes imitating intermediates of DNA repair and replication. Biochemistry Biokhimiia. 2008;73:261–272. doi: 10.1134/s0006297908030048. [DOI] [PubMed] [Google Scholar]
- 89.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 90.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 91.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base Excision Repair Intermediates Induce p53-independent Cytotoxic and Genotoxic Responses. J Biol Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 92.Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 93.Trivedi RN, Wang XH, Jelezcova E, Goellner EM, Tang J, Sobol RW. Human methyl purine DNA glycosylase and DNA polymerase β expression collectively predict sensitivity to temozolomide. Molecular Pharmacology. 2008;74:505–516. doi: 10.1124/mol.108.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ahmed EA, de Boer P, Philippens ME, Kal HB, de Rooij DG. Parp1-XRCC1 and the repair of DNA double strand breaks in mouse round spermatids. Mutation Research. 2010;683:84–90. doi: 10.1016/j.mrfmmm.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 95.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 96.Charbonnel C, Gallego ME, White CI. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010;64:280–290. doi: 10.1111/j.1365-313X.2010.04331.x. [DOI] [PubMed] [Google Scholar]
- 97.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair. 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 98.Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Molecular and Cellular Biology. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoo H, Li L, Sacks PG, Thompson LH, Becker FF, Chan JY. Alterations in expression and structure of the DNA repair gene XRCC1. Biochemical and biophysical research communications. 1992;186:900–910. doi: 10.1016/0006-291x(92)90831-5. [DOI] [PubMed] [Google Scholar]
- 100.Zhou ZQ, Walter CA. Cloning and characterization of the promoter of baboon XRCC1, a gene involved in DNA strand-break repair. Somat Cell Mol Genet. 1998;24:23–39. doi: 10.1007/BF02677493. [DOI] [PubMed] [Google Scholar]
- 101.Cerda SR, Turk PW, Thor AD, Weitzman SA. Altered expression of the DNA repair protein, N-methylpurine-DNA glycosylase (MPG) in breast cancer. FEBS Lett. 1998;431:12–18. doi: 10.1016/s0014-5793(98)00697-8. [DOI] [PubMed] [Google Scholar]
- 102.Kim NK, Ahn JY, Song J, Kim JK, Han JH, An HJ, Chung HM, Joo JY, Choi JU, Lee KS, Roy R, Oh D. Expression of the DNA repair enzyme, N-methylpurine-DNA glycosylase (MPG) in astrocytic tumors. Anticancer Res. 2003;23:1417–1423. [PubMed] [Google Scholar]
- 103.Kim NK, An HJ, Kim HJ, Sohn TJ, Roy R, Oh D, Ahn JY, Hwang TS, Cha KY. Altered expression of the DNA repair protein, N-methylpurine-DNA glycosylase (MPG) in human gonads. Anticancer Res. 2002;22:793–798. [PubMed] [Google Scholar]
- 104.Lage H, Christmann M, Kern MA, Dietel M, Pick M, Kaina B, Schadendorf D. Expression of DNA repair proteins hMSH2, hMSH6, hMLH1, O6-methylguanine-DNA methyltransferase and N-methylpurine-DNA glycosylase in melanoma cells with acquired drug resistance. Int J Cancer. 1999;80:744–750. doi: 10.1002/(sici)1097-0215(19990301)80:5<744::aid-ijc19>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 contains the mean fluorescence values for each of the Molecular Beacon plots shown in Figure 4, as well as the values for the standard error of the mean for each.