Abstract
Prebiotics are non-digestible substrates that stimulate the growth of beneficial microbial populations in the intestine, especially Bifidobacterium species. Among them, fructo- and galacto-oligosaccharides are commonly used in the food industry, especially as a supplement for infant formulas. Mechanistic details on the enrichment of bifidobacteria by these prebiotics are important to understand the effects of these dietary interventions. In this study the consumption of galactooligosaccharides was studied for 22 isolates of Bifidobacterium longum subsp. infantis, one of the most representative species in the infant gut microbiota. In general all isolates showed a vigorous growth on these oligosaccharides, but consumption of larger galactooligosaccharides was variable. Bifidobacterium infantis ATCC 15697 has five genes encoding β-galactosidases, and three of them were induced during bacterial growth on commercial galactooligosaccharides. Recombinant β-galactosidases from B. infantis ATCC 15697 displayed different preferences for β-galactosides such as 4′ and 6′-galactobiose, and four β-galactosidases in this strain released monosaccharides from galactooligosaccharides. Finally, we determined the amounts of short chain fatty acids produced by strain ATCC 15697 after growth on different prebiotics. We observed that biomass and product yields of substrate were higher for lactose and galactooligosaccharides, but the amount of acids produced per cell was larger after growth on human milk oligosaccharides. These results provide a molecular basis for galactooligosaccharide consumption in B. infantis, and also represent evidence for physiological differences in the metabolism of prebiotics that might have a differential impact on the host.
Keywords: Bifidobacterium longum subsp. infantis, Prebiotics, Galactooligosaccharides, β-Galactosidase
1. Introduction
The Bifidobacterium genus is composed of Gram-positive strictly anaerobic rods, which are common inhabitants of the intestinal tract of humans (Lee and O’Sullivan, 2010). They are dominant in the infant gut microbiota, especially in breast-fed infants (Yatsunenko et al., 2012), where they can represent up to the 90% of the total bacteria in this environment (Boesten et al., 2011). Bifidobacteria still represent a significant proportion of the adult gut microbiota (Eckburg et al., 2005), however different species can be found in both environments (Mangin et al., 2006; Roger et al., 2010).
Bifidobacteria show remarkable adaptations to use and metabolize complex oligosaccharides as a carbon and energy source (Lee and O’Sullivan, 2010). In breast-fed infants, the main carbon sources available for the developing intestinal microbiota are human milk oligosaccharides (HMO; (Kunz et al., 2000)) and certain bifidobacteria can gain access to N- and O-glycans in milk proteins or mucins (Garrido et al., 2012b; Ruas-Madiedo et al., 2008). Only a few bacterial species have been shown to use these substrates (Marcobal et al., 2010), and the molecular mechanisms involved in HMO consumption in bifidobacteria are beginning to be understood (Garrido et al., 2012a). In adults, diet delivers the intestinal microbiota a great variety of oligo- and polysaccharides, which are resistant to enzymatic degradation in the intestinal lumen and therefore reach distal portions of the intestine. Different Bifidobacterium species are capable of metabolizing complex oligosaccharides usually from plant origin such as cellodextrins and amyloses (Pokusaeva et al., 2011), raffinose (Dinoto et al., 2006), arabinooligosaccharides (Lagaert et al., 2010; Van Laere et al., 1997), xylooligosaccharides (Gilad et al., 2010), fructooligosaccharides and inulin (Omori et al., 2010; Perrin et al., 2001; Rossi et al., 2005), galactans and galactooligosaccharides (GOS; (Barboza et al., 2009; Goulas et al., 2009a; Hinz et al., 2005; O’Connell Motherway et al., 2011)) among several others.
Several infant formulas are supplemented with FOS and GOS with the aim of replicating some of the beneficial effects of human milk, in special its bifidogenic effect (Bakker-Zierikzee et al., 2005; Brunser et al., 2006). GOS are synthetically produced by microbial β-galactosidases (Gosling et al., 2010), which under specific conditions can perform transglycosylation reactions with lactose as the starting material. These enzymes are widespread in bifidobacteria, and some of them have remarkable yields in GOS synthesis (Hinz et al., 2004; Hung and Lee, 2002; Rabiu et al., 2001). GOS can have a degree of polymerization (DP) between 3 and 15 (Barboza et al., 2009) and are composed of galactose oligomers in β1–3/4/6 linkages with a terminal glucose residue (Coulier et al., 2009; Gosling et al., 2010). These substrates have been extensively studied for their prebiotic status, promoting the growth of beneficial microorganisms such as bifidobacteria and lactobacilli (Andersen et al., 2011; Davis et al., 2011), therefore providing putative health benefits (Gibson et al., 2004). GOS structures resemble galactan chains found in plant oligosaccharides abundant in adult diets, which might explain how these microorganisms consume GOS.
The mechanisms by which infant bifidobacteria are enriched by these prebiotics probably include oligosaccharide transporters and β-galactosidases specific for these substrates. In Bifidobacterium longum subsp. infantis ATCC 15697, two genes encoding solute-binding proteins (SBPs) from ABC transporters were specifically induced during growth on GOS (Garrido et al., 2011). β-galactosidases are widespread enzymes in bifidobacteria, and they display diverse substrate specificities (Goulas et al., 2009b). Two β-galactosidases in Bifidobacterium infantis HL96 have been previously studied (Hung and Lee, 2002; Hung et al., 2001) regarding their transglycosylation properties, and recently two β-galactosidases in the strain ATCC 15697 were shown to be active on different linkages found in HMO (Yoshida et al., 2012), and their activity likely complements β-hexosaminidases in this bacterium (Garrido et al., 2012c). Of increasing interest are also the metabolites produced after bifidobacterial fermentation of sugars, especially short chain fatty acids (SCFA) such as acetate and lactate as they represent one of the main protective mechanisms of bifidobacteria for its host (Fukuda et al., 2011). In this study we have characterized the consumption of GOS in a panel of B. infantis isolates from infant feces, and we further investigated the mechanisms involved in GOS degradation and metabolism in the type strain ATCC 15697.
2. Materials and methods
2.1. Microorganisms and media
Strains used in this study (Supplementary Table 1), were obtained from the American Type Culture Collection (Manassas, VA), and the University of California Davis Viticulture and Enology Culture Collection (Davis, CA). De Mann, Rogose and Sharp (MRS) broth supplemented with 0.05% w/v l-cysteine (Sigma–Aldrich, St. Louis, MO) was used for routine growth of B. infantis under anaerobic conditions (Coy Laboratory Products, Grass Lake, MI) at 37 °C in an atmosphere consisting of 5% carbon dioxide, 5% hydrogen, and 90% nitrogen. Chemically competent Escherichia coli BL21 Star and Top10 cells were obtained from Invitrogen (Carlsbad, CA), and transformants were cultured at 37 °C in Luria Broth with 50 µg/ml carbenicillin (Teknova, Hollister CA) when necessary.
2.2. Consumption of GOS by B. infantis isolates
Bifidobacterium isolates in Supplementary Table 1 were grown overnight in MRS and inoculated at 5% in modified MRS (mMRS), containing no carbon source, and supplemented with 0.05% cysteine (Sigma) and 0.5% commercial GOS (Purimune, GTC Nutrition, Golden, CO) or 0.5% lactose as a growth control. Growth was monitored using a PowerWave microplate spectrophotometer (BioTek Instruments, Winoosky, VT) at 37 °C for 48 h, reading absorbance at 600 nm. Each experiment was done in triplicate, and controls with no carbon source and no bacteria were subtracted from growth values. Aliquots of the reactions (1 µl) were spotted in TLC Silica gel plates (Sigma). A mixture of n-propanol, acetic acid and water in a 2:1:1 ratio was used as solvent. Plates were dryed and sprayed with 0.5% α-naphthol and 5% H2SO4 in ethanol, and developed at 150 °C for 10 min.
2.3. Gene expression analysis
For RNA extraction, B. infantis ATCC 15697 was grown on the chemically defined media Zhang-Block-Mills 1 (ZMB-1; (Zhang et al., 2009)) to which 2% w/v of glucose (Sigma), lactose (Sigma), GOS (GTC nutrition) or purified HMO (Ward et al., 2006) were added. Growth was monitored in a plate reader as described above. Cells at exponential phase were pelleted at 12,000 × g for 2 min, resuspended in 1 ml of RNA later (Ambion, Austin, TX), stored at 4 °C overnight and then at −80 °C until use. RNA extraction was performed using the RNAqueous Ambion kit (Ambion) and cDNA was obtained from 10 µg of RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). For the relative quantification of β-galactosidase and related genes, the Fast Sybr Green Master Mix (Applied Biosystems, Foster City, CA) was used, using the gene Blon_0393, cysteinyl-tRNA synthetase, as the endogenous control (Parche et al., 2006). Reaction conditions were as recommended by manufacturer. Primer efficiency was normalized in each plate using standard curves. The Primer3 software was used for primer design (Supplementary Table 2), and the Q-gene software was used for relative quantification analysis.
2.4. β-galactosidase gene cloning
B. infantis ATCC 15697 genomic DNA was obtained from overnight cultures on MRS, using the MasterPure Gram Positive DNA Purification Kit (Epicentre Biotechnologies, Madison, WI), following the manufacturer instructions. Primers used for PCR cloning are shown in Supplementary Table 2. PCR reactions contained 0.2 mM dNTPs (Fermentas, Glen Burnie, MD), 1 ng DNA, 0.5 µM of each primer, and 2 U of Phusion DNA Polymerase (Finnzymes, Vantaa, Finland) in a 150 µl final volume. PCR was performed in a Verity 96 well thermal cycler (Applied Biosystems), using the following program: initial denaturation at 98 °C for 2 min; 35 cycles of denaturation at 98 °C 30 s, annealing at 58 °C for 30 s, and extension at 72 °C 3 min; and a final extension at 72 °C for 7 min. PCR products were gel purified (Qiaquick Gel Extraction Kit, Qiagen, Valencia, CA) and cloned into pET101 using the Champion pET101 Directional TOPO Expression Kit (Invitrogen), following manufacturer instructions. Plasmids were transformed into BL21 star E. coli cells as well as Top10 cells for plasmid storage, and transformants were confirmed for the correct insert sequence by plasmid sequencing using primers T7prom and T7term (Invitrogen).
2.5. Recombinant protein expression and purification
E. coli BL21 transformants were grown in 200 ml LB broth with 50 µg/ml carbenicillin in a shaker at 250 rpm (Innova-4000, New Brunswick Scientific, Edison, NJ) at 37 °C until cultures reached an O.D. of 0.6. Recombinant proteins were induced for 6 h with the following optimized conditions: 0.5 mM IPTG (USB) at 24 °C for Blon_0268 and Blon_2416; 0.5 mM IPTG at 28 °C for Blon_2016; 1 mM IPTG at 24 °C (Blon_2123) and 0.5 mM IPTG at 28 °C for Blon_2334. Cultures were centrifuged in 50 ml falcon tubes at 4000 rpm in an Eppendorf 5804 centrifuge (Eppendorf, Hauppauge, NY) for 20 min at 4 °C, and pellets were kept at −80 °C until use. Cells were resuspended in Bugbuster Protein Extraction Reagent (EMD Chemicals), using 5 ml of the buffer for every 100 ml of culture. Lysozyme (Sigma Aldrich, 50 µl of 50 mg/ml stock), and DNAse I (Roche Applied Sciences; 20 µl of 10,000 U stock) were added to help in bacterial lysis. The suspensions were vortexed and incubated for 10 min at room temperature, and centrifuged for 20 min at 13,200 rpm at 4 °C. Supernatants were recovered and applied to 1 ml Bio-Scale Mini Profinity IMAC cartridges, connected to an EP-1 Econo-pump (Bio-Rad, Hercules CA). Protein purification was performed as recommended by the manufacturer, but proteins were eluted using an imidazole gradient between 20 and 250 mM in the washing buffer. Recombinant β-galactosidases were checked for purity and correct molecular weight using 10% SDS-PAGE gels (Bio-Rad). Elution buffer was exchanged for PBS using Amicon Ultra-15 Centrifugal Filter Units, with a cut-off of 50 kDa (Millipore). Protein concentrations were determined using the Bio-Rad protein assay, with a standard curve using Bovine Serum Albumin (Sigma).
2.6. Determination of enzymatic kinetic parameters
Enzymatic assays were carried out using ortho-nitrophenyl-β-galactoside (ONPG; Sigma) at a concentration of 2 mg/ml and 1–10 µg of each recombinant enzyme. Optimum pH for each enzyme was determined using McIlvaine buffers, with values from 4.0 to 8.0. Reactions were performed in triplicate in 96 microwell plates, and contained 80 µl of each buffer, 15 µl of substrate, and 5 µl of enzyme. Reactions were incubated for 10 min at 37 °C, and stopped adding equal volumes of 1 M Na2CO3. Absorbance at 420 nm was determined using a Synergy 2 microplate reader (Biotek). For determination of optimum reaction temperatures, enzymatic assays were performed at optimum pH and at 4 °C, 30 °C, 37 °C, 45 °C, 55 °C and 65 °C. Relative activity was determined from OD420 values. Kinetic constants were obtained using substrate concentrations in the range of 0.1–4 mM of ONPG and 1–100 µg of each enzyme. Reactions were performed at optimum pH and temperature, and times were preestablished to fall within the initial rate of reaction. Amounts of o-nitrophenol produced in each reaction were calculated from a standard curve and OD420 values. Non-linear regression was used to determine Km and Vmax, fitting the experimental values to the Michaelis Menten equation, using the tool Solver on Microsoft Excel.
2.7. β-galactosidase substrate specificity determination
Recombinant enzymes were coincubated in phosphate buffer and 2 µg of the following substrates at their optimum pH and temperatures: d-lactose (Sigma), 3′ galactosyl lactose (Carbosynth, Berkshire UK), Galβ1-4Gal (4′ galactobiose; V-labs, Covington, LA), Galβ1-6Gal (6′ galactobiose; V-labs), and 10 µg of commercial GOS (GTC Nutrition). Reactions were carried out in 10 µl for the specified times at 37 °C, and inactivated at 95 °C for 5 min. 1 µl of each reaction was analyzed in TLC plates as described above.
2.8. Evaluation of relative affinities of β-galactosidases
Equimolar concentrations (0.2 mM) of ONPG (Sigma), lactose (Sigma), 4′ galactobiose (V-labs), 6′ galactobiose (V-labs), 3′ galactosyl lactose (Carbosynth, UK) and lacto-N-tetraose (V-labs) were coincubated with the same amount (1–20 µg) of each of the five recombinant µ-galactosidases for 10 min at their optimum temperatures and pH in McIlvaine buffer in a 10 µl volume. Reactions were inactivated by incubation at 95 °C for 5 min. The Galactose Assay Kit (Biovision, Mountain View CA) was used to quantify galactose concentrations present in each sample, following the manufacturer instructions. Fluorescence was quantified using a standard curve in a Synergy 2 microplate reader. Values were normalized considering the amount of galactose released from ONPG as 100%.
2.9. Production of SCFA by B. infantis ATCC 15697
The production of acetate, lactate and formate by B. infantis was tested on seven different substrates: glucose (Sigma), lactose, HMO, LNT (V-labs), GOS, FOS (raftilose Synergy 1, Orafti, Malvern, PA) and inulin (raftiline HP, Orafti, Malvern, PA). B. infantis ATCC 15697 was grown overnight in MRS and inoculated at 5% in modified MRS, replacing sodium acetate by sodium chloride (10 g/l peptone, 5 g/l yeast extract, 5 g/l sodium chloride, 2 g/l ammonium citrate, 0.2 g/l magnesium sulfate, 0.05 g/l manganese sulfate, 2 g/l dipotassium phosphate, 1 g/l tween 80, and 0.05% cysteine). Media was supplemented with equal amounts (2% w/v) of each carbon source mentioned above. The incubations were carried out at 37 °C in anaerobic chamber in triplicate (Coy Laboratory Products, Grass Lake, MI). After 48 h of incubation the final optical density was measured in a spectrophotometer at 600 nm ((Shimadzu Scientific Instruments, Columbia MD), and the media was recovered by centrifuging at 12,000 × g for 10 min). Supernatants were analyzed using the l-lactate assay kit (Bioassay Systems, Hayward, CA), acetate assay kit (Bioassay Systems), and formate assay kit (Biovision, Milpitas, CA) according to the manufacturer instructions. For calculation of fermentation kinetic parameters, OD values were converted to dry weight in a ratio of 0.39 (Rossi et al., 2005), and these values were used with the amounts of SCFA in grams and initial concentration of each substrate (2 g/100 ml) to estimate the product yield of substrate (YP/S), the biomass yield of substrate (Yx/S) and the product yield of biomass (YP/x). Substrate consumption was assumed to be 100%.
3. Results
3.1. Consumption of GOS by B. infantis isolates
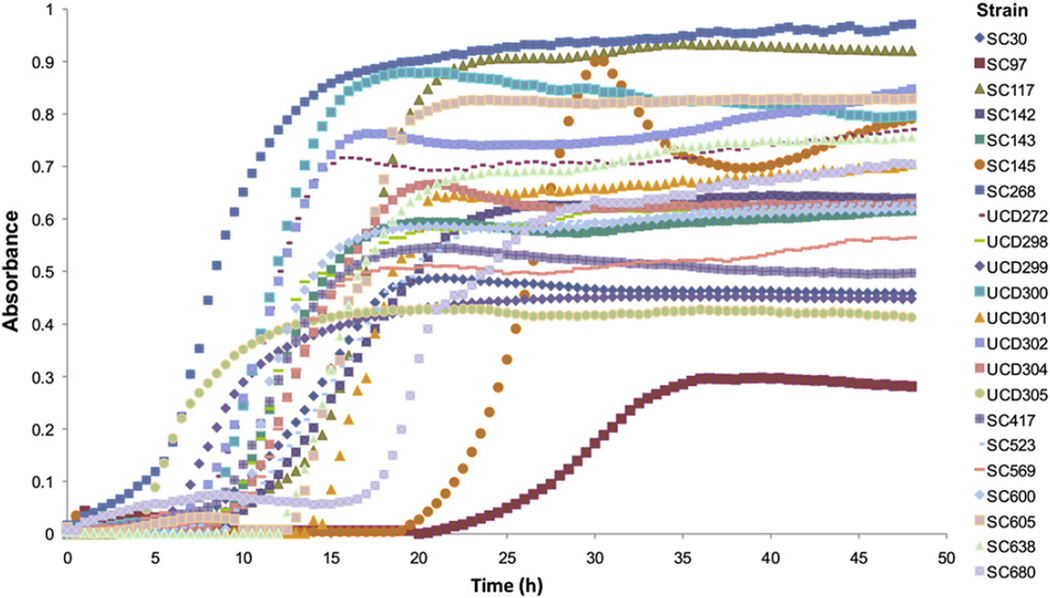
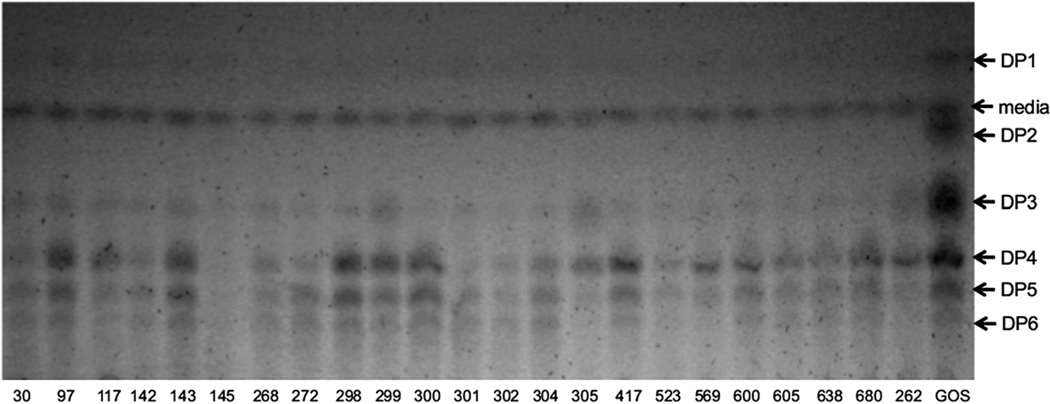
Twenty-two strains of B. infantis isolated from infant feces were studied in vitro for their ability to grow using 0.5% commercial GOS as the sole carbohydrate source. All isolates excepting SC97 showed a moderate to vigorous growth on GOS (Fig. 1). We also analyzed the supernatants of the fermentations in TLC plates (Fig. 2). All isolates were able to deplete the small amount of mono- and disaccharides found in commercial GOS, and DP3 was also consumed to a great extent. In general, disappearance of individual GOS with DP >3 was correlated with a higher final OD600, and conversely lower growth on GOS was related to a lack of consumption of larger GOS. For example, strains SC30, SC145 and UCD302 displayed the higher OD values on GOS and a noticeable reduction in all DP compared to the control with no bacteria added (Fig. 2). These results indicate that consumption of larger DP GOS is strain-dependent.
Fig. 1.
In vitro growth of strains of B. infantis using 0.5% commercial GOS as the sole media carbon source.
Fig. 2.
Analysis of the consumption of GOS by B. infantis isolates by TLC. Supernatants represent strains in Fig. 1, and a control prepared under the same conditions but with no bacteria added was included (GOS lane). Plates were run in n-butanol-acetic acid-water 2:1:1 and developed with α-naphthol. Numbers correspond to strains in Supplementary Table 1.
3.2. Distribution of β-galactosidases in B. infantis
To provide molecular details on the consumption of GOS in B. infantis, we focused on the genome of B. infantis ATCC 15697, which contains five genes predicted to encode β-galactosidases, EC 3.2.1.23 (Table 1). Some of these genes are in the proximity of carbohydrate transporters (Supplementary Fig. 1), suggesting a coregulated transcription. For example Blon_0268 and Blon_2334 are located next to sugar permeases, and Blon_2416 is in a gene cluster containing an ABC transporter with affinity for oligosaccharides and a Glycosyl Hydrolase (GH) family 43. Blon_2334 is part of the HMO cluster I (Sela et al., 2008), which contains several genes predicted to be important in metabolism of HMO in this bacterium. These enzymes belong to either GH2 or GH42, as defined by the Carbohydrate-Active Enzymes database (www.cazy.org; (Cantarel et al., 2009)). The presence of these genes in some of the B. infantis isolates used in this study has been previously determined by comparative genome hybridization ((LoCascio et al., 2010); Supplementary Table 3). While Blon_2016, Blon_2123 and Blon_2334 were present in all the strains, Blon_0268 was only found in strain UCD301 and Blon_2416 was lacking in strains UCD298, UCD299 and UCD300, altogether with Blon_2414, an upstream gene encoding a SBP induced by GOS in strain ATCC 15697.
Table 1.
Enzyme kinetic parameters and optimums for B. infantis β-galactosidases.
| Optimum pH |
Optimum temperature |
Km (mM) | kcat (s−1) |
kcat/Km (s−1 M−1) |
|
|---|---|---|---|---|---|
| Blon_0268 | 5.0 | 45 °C–55 °C | 0.60 | 5.11 | 8.50 × 103 |
| Blon_2016 | 5.0 | 45 °C | 0.70 | 1365.58 | 1.94 × 106 |
| Blon_2123 | 5.0–6.0 | 45 °C–55 °C | 1.06 | 269.92 | 2.5 × 105 |
| Blon_2334 | 7.5 | 37 °C | 0.29 | 420.04 | 1.47 × 106 |
| Blon_2416 | 5.0–6.5 | 45 °C | 1.09 | 7.21 | 6.58 × 103 |
3.3. Gene expression of β-galactosidases in B. infantis on GOS
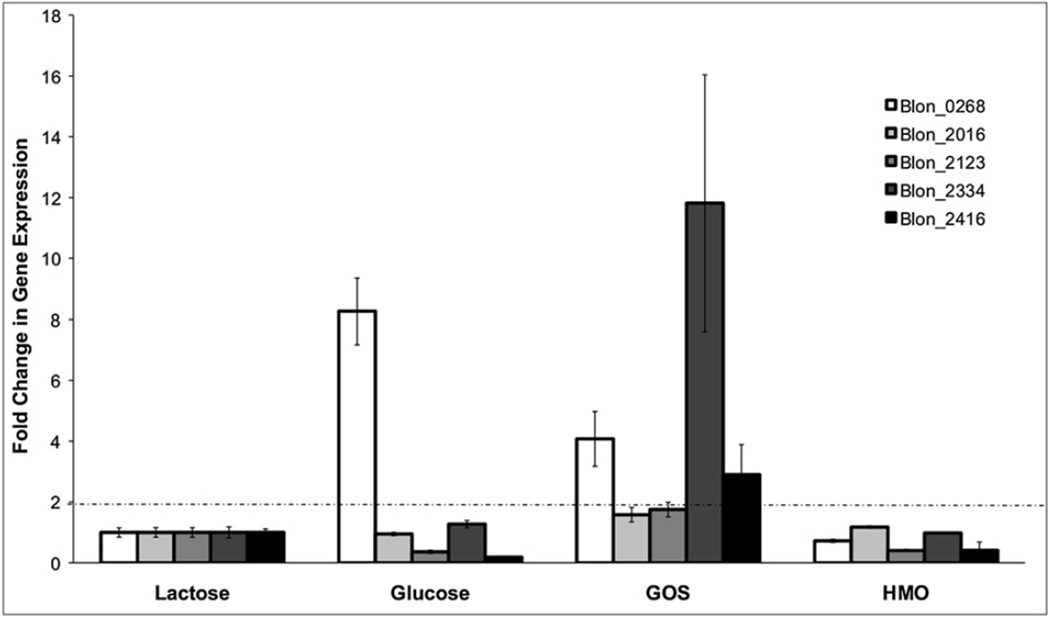
A relative quantification of the expression levels for each β-galactosidase gene was performed on B. infantis grown to exponential phase using lactose, glucose, GOS or HMO as the sole carbon source. Results were normalized to gene expression levels of cells growing on lactose (Fig. 3). We considered significant a level of induction more than two fold, as previously determined to be comparable to proteomic data (Garrido et al., 2011). Two genes, Blon_2123 and Blon_2416, were repressed at least four fold in cells grown on glucose. In contrast, Blon_0268 was induced eight fold during growth on glucose relative to lactose. When a pool of HMO was used as the sole carbon source, none of the enzymes was induced relative to lactose. However, Blon_2016 and Blon_2334 are constitutively expressed at high levels when B. infantis grows on HMO and lactose (Sela et al., 2008; Yoshida et al., 2012). Finally, growth on commercial GOS had the greatest impact on the transcription of B. infantis β-galactosidases, with Blon_2334 induced over tenfold and Blon_0268 and Blon_2416 also induced over two fold by these oligosaccharides.
Fig. 3.
Relative quantification of the gene expression of B. infantis β-galactosidases after logarithmic growth on the substrates indicated in the x-axis. Dashed lines indicate a two-fold change in gene expression.
We also analyzed the expression levels for genes adjacent to Blon_0268 and Blon_2334 (Supplementary Fig. 2), encoding for transporters of the major facilitator superfamily. Even though the carbohydrate affinity of these transporters (Blon_0267, Blon_2331 and Blon_2332) is unknown, their induction by GOS, as well as glucose (Blon_0268), is suggestive that their affinities are related to these substrates.
3.4. Kinetic parameters of β-galactosidases in B. infantis
In order to study some of the properties of these glycosyl hydrolases, they were cloned and expressed in E. coli, and purified with an N-terminal his-tag. ONPG was used for evaluating different kinetic parameters. Optimum pH values were relatively acidic for Blon_0268 and Blon_2016, and more neutral for the other three enzymes (Table 1). As observed with other β-galactosidases, the optimum temperature varied between 45 °C and 55 °C for all enzymes, except Blon_2334 which showed the highest activity at 37 °C. Using these conditions, kinetic parameters were determined using ONPG. Turnover rates were the highest for Blon_2016, and together with Blon_2334, these glycosidases showed the greatest kinetic efficiency given by a kcat/Km ratio over 1 × 106 s−1 M−1. These values are in agreement with those obtained byYoshida et al. (2012), and they were 1000 times higher than the kinetic efficiencies observed for Blon_0268 and Blon_2416 on ONPG.
3.5. Relative affinities and substrate specificity of B. infantis β-galactosidases
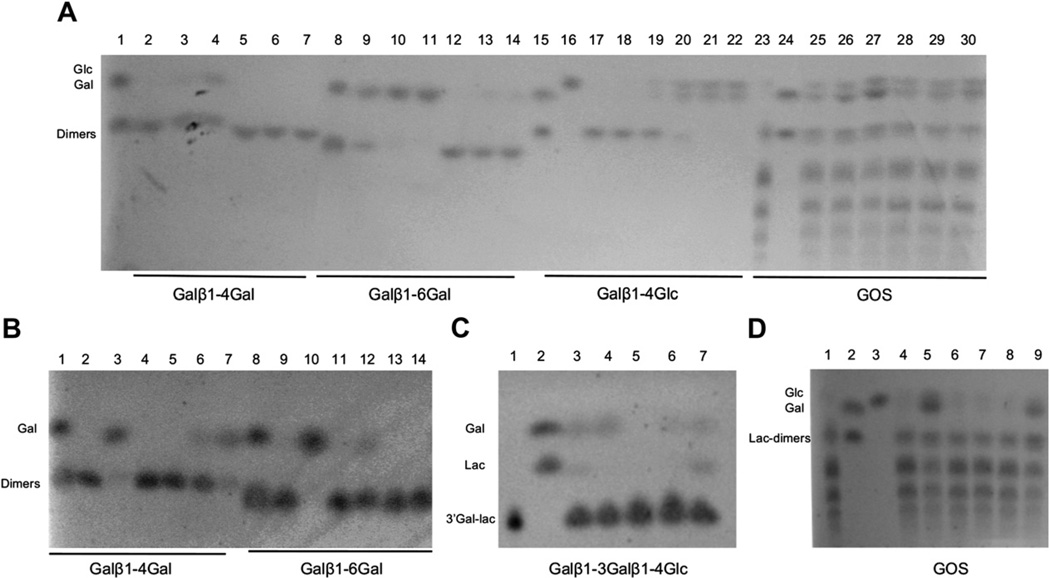
We also studied the activity of these enzymes on common β-galactosides. 4- and 6-galactobiose (Galβ1-4Gal and Galβ1-6Gal) and galactosyl-lactose (Galβ1-3Galβ1-4Glc) are common products in transglycosylation reactions (Gosling et al., 2010). Moreover, plant polysaccharides also contain these linkages as building blocks. As observed on TLC plates and considering the relative affinities for these substrates (Table 2), Blon_0268 displayed an overall preference for 6′ galactobiose (Fig. 4A, lanes 2–4; 9–11 and Fig. 4C, lane 3). Conversely, Blon_2416 showed a preference for 4′ galactobiose over galactosyl lactose (Fig. 4B lanes 6–7 and 13–14; Fig. 4C, lane 7). Blon_2123 was only partially active on 6′ galactobiose.
Table 2.
Relative affinities of B. infantis β-galactosidases for individual substrates. Results are expressed as a percentage normalized to the amount of galactose released from ONPG.
| Blon_0268 | Blon_2016 | Blon_2123 | Blon_2334 | Blon_2416 | |
|---|---|---|---|---|---|
| ONPG | 100 | 100 | 100 | 100 | 100 |
| Lactose | 78.9 | 19.9 | – | 128.1 | – |
| 4′ galactobiose | 9.6 | – | – | – | 267.6 |
| 6′ galactobiose | 153.2 | 60.3 | 137.5 | – | – |
| 3′Gal-lac | 18.7 | 11.6 | – | – | 6.1 |
| LNT | – | 123.4 | – | 14.5 | – |
Fig. 4.
Determination of the substrate specificities of B. infantis β-galactosidases for different galactosyl linkages by TLC indicated in the bottom of the figure. (A) Coincubations of Blon_0268 and Blon_2334 for 5, 20 and 60 min with different β-galactosides. Lanes 1 and 8: standards; lanes 2–4: Blon_0268 with 4-galactobiose; lanes 5–7: Blon_2334 with 4-galactobiose, 5-20–60 min; lanes 9–11: Blon_0268 with 6-galactobiose; lanes 12–14: Blon_2334 with 6-galactobiose. Lane 15: Lactose and galactose; lane 16: glucose; lanes 17–19: Blon_0268 with lactose; lanes 20–22: Blon_2334 with lactose. Lane 23: Commercial GOS; lane 24: lactose and galactose; lanes 25–27: Blon_0268 with GOS; lanes 28–30: Blon_2334 with GOS. (B) Coincubations of Blon_2016, Blon_2123 and Blon_2416 with 4′ or 6′ galactobiose for 20′ and 60′. Lanes 1 and 8: galactose and 4′ or 6′ galactobiose standards; lanes 2–3: Blon_2016 on 4-galactobiose; lanes 4–5: Blon_2123 on 4-galactobiose; lanes 6–7: Blon_2416 on 4-galactobiose; lanes 9–10: Blon_2016 on 6-galactobiose; lanes 11–12: Blon_2123 on 6-galactobiose; lanes 13–14: Blon_2416 on 6-galactobiose. (C) Glycolytic activity on 3′ galactosyl lactose. Enzymes were incubated with this substrate for 60′. Lane 1: 3′ galactosyl lactose; lane 2: galactose and lactose; lane 3: Blon_0268; lane 4: Blon_2016; lane 5: Blon_2123; lane 6: Blon_2334; lane 7: Blon_2416. (D) Coincubations of Blon_2016, Blon_2123 and Blon_2416 with commercial GOS for 20′ and 60′. Lane 1: GOS; lane 2: lactose and galactose; lane 3: glucose; lanes 4–5: Blon_2016; lanes 6–7: Blon_2123; lanes 8–9: Blon_2416. Gal: galactose, Glc: glucose, Lac: lactose.
Blon_2334 showed higher lactase activity compared to the other B. infantis β-galactosidases (Fig. 4A, lanes 20–22). Blon_0268 and Blon_2016 showed only a minor activity on this substrate (Fig. 4A, lanes 17–19 and Table 2). Blon_2016 releases galactose very efficiently from type 1 HMO such as lacto-N-tetraose (LNT; Galβ1-3GlcNAcβ1-3Galβ1-4Glc), and it had the highest enzymatic efficiency given by the kcat/Km ratio on ONPG (Table 1). The results presented in this study indicate that this enzyme can cleave other galactosyl linkages, such as those found in 4′ and 6′ galactobiose (Fig. 4C, lanes 2–3 and 8–9), 3′ galactosyl lactose (Fig. 4C, lane 4), and GOS (Fig. 4D, lanes 4–5), displaying however a preference for LNT (Table 2).
Finally, after incubation of B. infantis β-galactosidases with commercial GOS for 1 h, we observed that four of them displayed significant hydrolytic activity on these prebiotics, as observed by an increase in time in the amount of galactose and glucose released from commercial GOS (Fig. 4A, lanes 23–30 and Fig. 4D).
3.6. Production of SCFA by B. infantis
Sugar metabolism in bifidobacteria differs from other bacterial metabolic pathways (Fushinobu, 2010) and is characterized by the presence of fructose-6-P phosphoketolase, an enzyme that generates acetyl-P and erythrose-4-P from fructose-6-P. This pathway, termed the “bifid shunt”, produces 2.5 mol of ATP per mole of glucose, as well as 3 mol of acetate and 2 mol of lactate that are either released to the media or used in de novo fatty acid synthesis (Sela et al., 2010). These yields are variable among different strains and growth conditions. Here, we determined the amounts and yields of SCFA produced by B. infantis ATCC 15697 after growth on different prebiotics including GOS as the sole carbon source. Supernatants reached exponential phase at similar times and were recovered at 48 h, where all cultures reached stationary phase. In order to compare production of SCFA across the different substrates, three kinetic yield coefficients were calculated (Table 3), estimating a 100% substrate consumption, which is supported by the efficient oligosaccharide consumption of GOS (Barboza et al., 2009), FOS and inulin (Perrin et al., 2001), and lactose, glucose, HMO and LNT (Asakuma et al., 2011; LoCascio et al., 2007; Sela et al., 2012) by B. infantis.
Table 3.
Kinetic parameters of B. infantis fermentations of simple sugars and prebiotics. YP/S: product yield of substrate (g/g−1), Yx/S: biomass yield of substrate (g/g−1), YP/x: product yield of biomass (g/g−1). LNT: Lacto-N-tetraose. Standard deviations were below 0.001 and not shown.
| YP/S acetate | YP/S lactate | YP/S formate | YP/S total | Yx/S | YP/x acetate | YP/x lactate | YP/x formate | YP/x total | Acetate to lactate ratio |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | 0.170 | 0.136 | 0.004 | 0.310 | 0.034 | 4.998 | 3.973 | 0.120 | 9.092 | 1.887 |
| Lactose | 0.170 | 0.145 | 0.000 | 0.315 | 0.044 | 3.888 | 3.318 | 0.004 | 7.210 | 1.758 |
| LNT | 0.137 | 0.106 | 0.001 | 0.244 | 0.023 | 5.885 | 4.542 | 0.030 | 10.457 | 1.943 |
| HMO | 0.129 | 0.094 | 0.000 | 0.223 | 0.014 | 9.180 | 6.713 | 0.000 | 15.893 | 2.051 |
| GOS | 0.167 | 0.158 | 0.000 | 0.326 | 0.044 | 3.838 | 3.635 | 0.004 | 7.476 | 1.584 |
| FOS | 0.126 | 0.082 | 0.004 | 0.211 | 0.019 | 6.482 | 4.234 | 0.182 | 10.898 | 2.297 |
| Inulin | 0.111 | 0.047 | 0.010 | 0.168 | 0.020 | 5.618 | 2.375 | 0.499 | 8.491 | 3.548 |
We observed that the product and biomass yields of substrate were higher during growth on lactose, GOS and glucose (Table 3), indicating that under these conditions B. infantis produces biomass and acids more efficiently. Although less biomass and SCFA were produced, growth on HMO, under these conditions we observed the highest yield of product over biomass, which indicates that B. infantis produces more acetate and lactate per cell. Formate is another end product of bifidobacterial fermentation of carbohydrates, and growth on inulin, FOS and glucose led to a significant production of this organic acid. Finally, the acetate:lactate ratio was determined for each substrate. Considering that the theoretical value in bifidobacteria is 1.5, we observed that growth on FOS and inulin, as well as HMO and LNT to a lesser extent, led to a considerable increase in this value.
4. Discussion
GOS are important food supplements that, among other health benefits, stimulate the growth of beneficial microorganisms in the gastrointestinal tract. In this work we studied several aspects of the utilization of commercial prebiotic GOS by B. infantis, a representative species of the developing infant gut microbiota. As a dietary substrate, GOS is not degraded by host intestinal enzymes. However, its relatively simple composition, consisting of repeats of β1–3/4/6 galactosyl linkages, allows several bacteria, including Bacteroides sp. and Clostridium sp. to degrade and consume GOS in vitro (Gibson et al., 2004). Surprisingly, GOS consumption in human volunteers resulted in significant and consistent enrichment of mainly one genus, Bifidobacterium, as shown recently (Davis et al., 2011). Several species of this genus have been shown to grow on these prebiotics as the sole media carbon source (Goulas et al., 2007; Hopkins et al., 1998; Van Laere et al., 2000), and for example it has been shown by MALDI-FT-ICR mass spectrometry that B. infantis ATCC 15697 is an efficient GOS consumer with an overall preference for GOS with DP 4 but consuming a range of molecules ranging DP 3 to 8 (Barboza et al., 2009). Unfortunately, these detection methods are expensive and not suitable for studies of GOS consumption at the larger scale. Here we studied GOS consumption in B. infantis at the strain level. We observed that all B. infantis isolates studied can grow on GOS with DP up to 3, and only some isolates showed a considerable consumption of larger GOS, which correlated with higher OD values during bacterial growth. These results indicate phenotypic differences in GOS consumption among different strains of the subspecies infantis. This variability is intriguing when considering the in vivo impact of these prebiotics for example in formula-fed infants.
Bacterial consumption of GOS requires transport molecules and specific glycosyl hydrolases that can release galactose to be used in central metabolic pathways. Specific transport systems have been described in Lactobacillus acidophilus NCFM (Andersen et al., 2011), and an operon devoted to consumption of galactans has been described in Bifidobacterium breve (O’Connell Motherway et al., 2011). The specificity of certain bifidobacterial β-galactosidases for galactosides has been defined (Goulas et al., 2009a,b; Hinz et al., 2004; Hung and Lee, 2002). We previously found two solute-binding proteins, part of ABC transporters, induced during growth on commercial GOS (Garrido et al., 2011), potentially participating in the import of GOS. In this study we determined that the gene expression of β-galactosidases Blon_0268, Blon_2334 and Blon_2416 is significantly increased during bacterial growth on GOS, and Blon_2016 is expressed constitutively at considerable levels (Yoshida et al., 2012). Interestingly, the encoded β-galactosidases in B. infantis show affinity for distinct galactosyl linkages. For example while Blon_0268 preferred β1–6 linkages, Blon_2416 was more specific for Galβ1-4Gal linkages and Blon_2334 had high lactase activity. The results presented in this work suggest that these enzymes, including Blon_2016 which has significant activity against several galactosyl linkages, act cooperatively releasing galactose from the commercial GOS preparation used in this study, which contains β1–3, β1–4 and β1–6 galactosyl linkages (Gosling et al., 2010). In aggregate, these observations provide a molecular framework for GOS consumption in the subspecies infantis. A recent comparative genomic hybridization analysis of some of the isolates included in this study (Table S3, (LoCascio et al., 2010)) shows that β-galactosidases Blon_0268 and Blon_2416 as well as SBP Blon_2414 are absent in isolates UCD298, UCD299 and UCD300. This might correlate with the little consumption of larger GOS and the poor growth on 0.5% GOS of these three isolates.
Some of the health benefits attributed to bifidobacteria can be explained by their unique sugar metabolism (Russell et al., 2011). The bifid shunt is a very efficient pathway that generates large amounts of SCFA, mainly acetate and lactate, and proteomics has shown that all the enzymes in this pathway are active during growth on HMO in B. infantis (Sela et al., 2008). These acids are mainly secreted, preventing the growth of intestinal pathogens, and they shape the intestinal epithelium being used as energy sources as well as signaling molecules (Scholtens et al., 2012). As expected, SCFA production in bifidobacteria depends on the species as well as the carbon source utilized (Fukuda et al., 2011; Palframan et al., 2003; Perrin et al., 2001; Rossi et al., 2005). Here we determined that growth on different prebiotics or simple sugars leads to important differences in the amount of SCFA produced. GOS are structurally similar to lactose, and both carbohydrates were similarly fermented very efficiently leading to higher amounts of biomass and product compared to the other carbohydrates in this study (Table 3). This is probably related to bacterial adaptations to consumption of galactose-containing oligosaccharides and might be indicative of the efficiency of the Leloir pathway in B. infantis. When comparing the amount of SCFA produced per cell however (proportional to YP/x), B. infantis cells growing on HMO produced significantly more acetate and lactate per cell compared to other prebiotics. This trend was less pronounced in LNT, a much simpler individual HMO. These observations might be related to the simultaneous fermentation of different monosaccharides that are found in HMO (galactose, N-acetylglucosamine, fucose, sialic acid and glucose). In contrast, growth on simpler substrates such as lactose, glucose and GOS led to lower YP/x values. Interestingly, formate production was majorly appreciated only during growth on FOS and inulin, similar to Bifidobacterium animalis (Van der Meulen et al., 2004). Formate is produced at the expense of lactate by pyruvate-formate lyase, which increases the acetate/lactate ratio and provides an additional ATP (Perrin et al., 2001).
In conclusion, this study provides evidence for the diversity in GOS consumption among fecal isolates of B. infantis, a prominent member of the infant gut microbiota. We also provided information on the cognate molecular mechanisms involved in this process, in particular relative to gene expression and specificity of β-galactosidases in strain ATCC 15697, as well as differences in metabolic profiles among different prebiotics used for growth in vitro.
Supplementary Material
Acknowledgments
Daniel Garrido was funded in part through a Fulbright-Conicyt Chile scholarship and a National Milk Producer Federation scholarship. Santiago Ruiz-Moyano was supported in part by the Ministry of Education and Science of Spain and University of Extremadura, Spain. Rogelio Jimenez-Espinoza was sponsored by a UC MEXUS-CONACyT. This work was supported by grants from the University of California Discovery Grant Program (05GEB01NHB), the California Dairy Research Foundation, DSM Food Specialities and National Institutes of Health NICHD Awards R01AT007079, and R01HD061923.
Abbreviations
- GOS
galactooligosaccharides
- HMO
human milk oligosaccharides
- FOS
fructooligosaccharides
- LNT
lacto-N-tetraose
- SCFA
short chain fatty acids
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.fm.2012.10.003.
References
- Andersen JM, Barrangou R, Abou Hachem M, Lahtinen S, Goh YJ, Svensson B, Klaenhammer TR. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc. Natl. Acad. Sci. U S A. 2011;108:17785–17790. doi: 10.1073/pnas.1114152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 2011;286:34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker-Zierikzee AM, Alles MS, Knol J, Kok FJ, Tolboom JJ, Bindels JG. Effects of infant formula containing a mixture of galacto- and fructooligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br. J. Nutr. 2005;94:783–790. doi: 10.1079/bjn20051451. [DOI] [PubMed] [Google Scholar]
- Barboza M, Sela DA, Pirim C, Locascio RG, Freeman SL, German JB, Mills DA, Lebrilla CB. Glycoprofiling bifidobacterial consumption of galacto-oligosaccharides by mass spectrometry reveals strain-specific, preferential consumption of glycans. Appl. Environ. Microbiol. 2009;75:7319–7325. doi: 10.1128/AEM.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesten R, Schuren F, Ben Amor K, Haarman M, Knol J, de Vos WM. Bifidobacterium population analysis in the infant gut by direct mapping of genomic hybridization patterns: potential for monitoring temporal development and effects of dietary regimens. Microb. Biotechnol. 2011;4:417–427. doi: 10.1111/j.1751-7915.2010.00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunser O, Gotteland M, Cruchet S, Figueroa G, Garrido D, Steenhout P. Effect of a milk formula with prebiotics on the intestinal microbiota of infants after an antibiotic treatment. Pediatr. Res. 2006;59:451–456. doi: 10.1203/01.pdr.0000198773.40937.61. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulier L, Timmermans J, Bas R, Van Den Dool R, Haaksman I, Klarenbeek B, Slaghek T, Van Dongen W. In-depth characterization of prebiotic galacto-oligosaccharides by a combination of analytical techniques. J. Agric. Food Chem. 2009;57:8488–8495. doi: 10.1021/jf902549e. [DOI] [PubMed] [Google Scholar]
- Davis LM, Martinez I, Walter J, Goin C, Hutkins RW. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One. 2011;6:e25200. doi: 10.1371/journal.pone.0025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoto A, Marques TM, Sakamoto K, Fukiya S, Watanabe J, Ito S, Yokota A. Population dynamics of Bifidobacterium species in human feces during raffinose administration monitored by fluorescence in situ hybridization-flow cytometry. Appl. Environ. Microbiol. 2006;72:7739–7747. doi: 10.1128/AEM.01777-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- Fushinobu S. Unique sugar metabolic pathways of bifidobacteria. Biosci. Biotechnol. Biochem. 2010;74:2374–2384. doi: 10.1271/bbb.100494. [DOI] [PubMed] [Google Scholar]
- Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One. 2011;57:e17315. doi: 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 2012a;3:415S–421S. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Nwosu C, Ruiz-Moyano S, Aldredge D, German JB, Lebrilla CB, Mills DA. Endo-beta-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex n-glycans from human milk glycoproteins. Mol. Cell. Proteomics. 2012b;11:775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Mills DA. Release and utilization of N-acetyl-d- glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe. 2012c;18:430–435. doi: 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 2004;17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- Gilad O, Jacobsen S, Stuer-Lauridsen B, Pedersen MB, Garrigues C, Svensson B. Combined transcriptome and proteome analysis of Bifidobacterium animalis subsp. lactis BB-12 grown on xylo-oligosaccharides and a model of their utilization. Appl. Environ. Microbiol. 2010;76:7285–7291. doi: 10.1128/AEM.00738-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosling A, Stevens GW, Barber AR, Kentish SE, Gras SL. Recent advances refining galactooligosaccharide production from lactose. Food Chem. 2010;121:307–318. [Google Scholar]
- Goulas TK, Goulas AK, Tzortzis G, Gibson GR. Molecular cloning and comparative analysis of four beta-galactosidase genes from Bifidobacterium bifidum NCIMB41171. Appl. Microbiol. Biotechnol. 2007;76:1365–1372. doi: 10.1007/s00253-007-1099-1. [DOI] [PubMed] [Google Scholar]
- Goulas T, Goulas A, Tzortzis G, Gibson GR. Comparative analysis of four beta-galactosidases from Bifidobacterium bifidum NCIMB41171: purification and biochemical characterisation. Appl. Microbiol. Biotechnol. 2009a;82:1079–1088. doi: 10.1007/s00253-008-1795-5. [DOI] [PubMed] [Google Scholar]
- Goulas T, Goulas A, Tzortzis G, Gibson GR. Expression of four beta-galactosidases from Bifidobacterium bifidum NCIMB41171 and their contribution on the hydrolysis and synthesis of galactooligosaccharides. Appl. Microbiol. Biotechnol. 2009b;84:899–907. doi: 10.1007/s00253-009-2009-5. [DOI] [PubMed] [Google Scholar]
- Hinz SW, van den Brock LA, Beldman G, Vincken JP, Voragen AG. Beta-galactosidase from Bifidobacterium adolescentis DSM20083 prefers beta(1,4)-galactosides over lactose. Appl. Microbiol. Biotechnol. 2004;66:276–284. doi: 10.1007/s00253-004-1745-9. [DOI] [PubMed] [Google Scholar]
- Hinz SW, Pastink MI, van den Broek LA, Vincken JP, Voragen AG. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl. Environ. Microbiol. 2005;71:5501–5510. doi: 10.1128/AEM.71.9.5501-5510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins MJ, Cummings JH, Macfarlane GT. Inter-species differences in maximum specific growth rates and cell yields of bifidobacteria cultured on oligosaccharides and other simple carbohydrate sources. J. Appl. Microbiol. 1998;85:381–386. [Google Scholar]
- Hung MN, Lee BH. Purification and characterization of a recombinant beta-galactosidase with transgalactosylation activity from Bifidobacterium infantis HL96. Appl. Microbiol. Biotechnol. 2002;58:439–445. doi: 10.1007/s00253-001-0911-6. [DOI] [PubMed] [Google Scholar]
- Hung MN, Xia Z, Hu NT, Lee BH. Molecular and biochemical analysis of two beta-galactosidases from Bifidobacterium infantis HL96. Appl. Environ. Microbiol. 2001;67:4256–4263. doi: 10.1128/AEM.67.9.4256-4263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G. Substrate specificity of three recombinant alpha-L-arabinofuranosidases from Bifidobacterium adolescentis and their divergent action on arabinoxylan and arabinoxylan oligosaccharides. Biochem. Biophys. Res. Commun. 2010;402:644–650. doi: 10.1016/j.bbrc.2010.10.075. [DOI] [PubMed] [Google Scholar]
- Lee JH, O’Sullivan DJ. Genomic insights into bifidobacteria. Microbiol. Mol. Biol. Rev. 2010;74:378–416. doi: 10.1128/MMBR.00004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 2010;76:7373–7381. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin I, Suau A, Magne F, Garrido D, Gotteland M, Neut C, Pochart P. Characterization of human intestinal bifidobacteria using competitive PCR and PCR-TTGE. FEMS Microbiol. Ecol. 2006;55:28–37. doi: 10.1111/j.1574-6941.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell Motherway M, Fitzgerald GF, van Sinderen D. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb. Biotechnol. 2011;4:403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori T, Ueno K, Muramatsu K, Kikuchi M, Onodera S, Shiomi N. Characterization of recombinant beta-fructofuranosidase from Bifidobacterium adolescentis G1. Chem. Cent. J. 2010;4:9. doi: 10.1186/1752-153X-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palframan RJ, Gibson GR, Rastall RA. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr. Issues Intest Microbiol. 2003;4:71–75. [PubMed] [Google Scholar]
- Parche S, Beleut M, Rezzonico E, Jacobs D, Arigoni F, Titgemeyer F, Jankovic I. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 2006;188:1260–1265. doi: 10.1128/JB.188.4.1260-1265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin S, Warchol M, Grill JP, Schneider F. Fermentations of fructooligosaccharides and their components by Bifidobacterium infantis ATCC 15697 on batch culture in semi-synthetic medium. J. Appl. Microbiol. 2001;90:859–865. doi: 10.1046/j.1365-2672.2001.01317.x. [DOI] [PubMed] [Google Scholar]
- Pokusaeva K, O’Connell-Motherway M, Zomer A, Macsharry J, Fitzgerald GF, van Sinderen D. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2011;77:1681–1690. doi: 10.1128/AEM.01786-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiu BA, Jay AJ, Gibson GR, Rastall RA. Synthesis and fermentation properties of novel galacto-oligosaccharides by beta-galactosidases from Bifidobacterium species. Appl. Environ. Microbiol. 2001;67:2526–2530. doi: 10.1128/AEM.67.6.2526-2530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology. 2010;156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 2005;71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes- Gavilan CG, Margolles A. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 2008;74:1936–1940. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DA, Ross RP, Fitzgerald GF, Stanton C. Metabolic activities and probiotic potential of bifidobacteria. Int. J. Food Microbiol. 2011;149:88–105. doi: 10.1016/j.ijfoodmicro.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annu. Rev. Food Sci. Technol. 2012;3:425–447. doi: 10.1146/annurev-food-022811-101120. [DOI] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Price NP, Mills DA. Metabolism of Bifidobacteria. In: Mayo B, van Sinderen D, editors. Bifidobacteria, Genomics and Molecular Aspects. 2010. pp. 45–70. [Google Scholar]
- Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 2012;78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meulen R, Avonts L, De Vuyst L. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl. Environ. Microbiol. 2004;70:1923–1930. doi: 10.1128/AEM.70.4.1923-1930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laere KM, Beldman G, Voragen AG. A new arabinofuranohydrolase from Bifidobacterium adolescentis able to remove arabinosyl residues from double-substituted xylose units in arabinoxylan. Appl. Microbiol. Biotechnol. 1997;47:231–235. doi: 10.1007/s002530050918. [DOI] [PubMed] [Google Scholar]
- Van Laere KM, Abee T, Schols HA, Beldman G, Voragen AG. Characterization of a novel beta-galactosidase from Bifidobacterium adolescentis DSM 20083 active towards transgalactooligosaccharides. Appl. Environ. Microbiol. 2000;66:1379–1384. doi: 10.1128/aem.66.4.1379-1384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. Bifidobacterium longum subsp. infantis uses two different beta-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–368. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- Zhang G, Mills DA, Block DE. Development of chemically defined media supporting high-cell-density growth of lactococci, enterococci, and streptococci. Appl. Environ. Microbiol. 2009;75:1080–1087. doi: 10.1128/AEM.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.