Abstract
A unique cryo-electron microscopy facility has been designed and constructed at the University of Texas Medical Branch (UTMB) to study the three-dimensional organization of viruses and bacteria classified as select agents at biological safety level (BSL)-3, and their interactions with host cells. A 200 keV high-end cryo-electron microscope was installed inside a BSL-3 containment laboratory and standard operating procedures were developed and implemented to ensure its safe and efficient operation. We also developed a new microscope decontamination protocol based on chlorine dioxide gas with a continuous flow system, which allowed us to expand the facility capabilities to study bacterial agents including spore-forming species. The new unified protocol does not require agent-specific treatment in contrast to the previously used heat decontamination. To optimize the use of the cryo-electron microscope and to improve safety conditions, it can be remotely controlled from a room outside of containment, or through a computer network world-wide. Automated data collection is provided by using JADAS (single particle imaging) and SerialEM (tomography). The facility has successfully operated for more than a year without an incident and was certified as a select agent facility by the Centers for Disease Control.
Keywords: cryo-electron microscopy, single particle imaging, electron tomography, biological safety containment
Introduction
Studying “live” infectious agents, e.g. viruses and/or bacteria, presents a challenge both in design of the facility and in logistics of operation of such a laboratory. Studying the three-dimensional (3D) structure of viruses and bacteria is important for understanding key pathogenic components of the agents and their disease mechanisms. Determining how viruses and bacteria function and interact with each other, as well as studying their interactions with host cells, may reveal their “Achilles’ heels” and allow researchers to develop new and improved vaccines and therapeutics. Cryo-electron microscopy (Cryo-EM) allows one to preserve biological samples in nearly native state by flash freezing and immobilizing them in a solid matrix of vitreous water, which is essential for revealing their native or modified organization (Chiu, Downing, Dubochet et al., 1986; Dubochet, Adrian, Chang et al., 1988; Stewart, 1989; Unwin, 1986). However, in the case of a potential incident, aerosol infectious samples could present a substantial threat to the health of research and maintenance personnel. Therefore cryo-EM studies of aerosol-infectious pathogens should be performed in a high biological containment facility. This containment requirement presents a major challenge for the use of cryo-EM to study infectious agents. Such a laboratory would need Standard Operating Procedures to safely operate the microscope and other equipment in the containment as well as protocols for specimen preparation, handling and storage of cryo-EM grids, their disposal after experiments, emergency procedures, procedures for entry and exit containment, etc. In addition, a method of microscope decontamination would have to be developed to provide safe access for researchers and maintenance personnel in the event of an incident leading to contamination of the microscope column or vacuum system. High environmental standards required for cryo-EM performance are not particularly compatible with requirements for biological containment, including negative air pressure, number of air exchanges per hour, and hard wall surfaces preventing acoustic noise attenuation. Wearing personal protective equipment (PPE) during experiments further complicates the operation because respirators can impair researcher’s vision during specimen preparation and transfer to the microscope; multiple layers of gloves can reduce manual dexterity, crucial for specimen and grid handling; specimen treatment including flash-freezing must be performed in an enclosure (e.g. inside of a biological safety cabinet) and all the tools, bottles, waste, etc. should be disinfected after each use leading to increase of the wear and tear of cryo-plungers, tweezers, micro-pipettes, cryo grid boxes and other accessories and supplies.
A unique cryo-EM laboratory for studying infectious agents inside BSL-3 containment (Sherman, Freiberg, Razmus et al., 2010) became operational at UTMB in 2006. Unfortunately, the laboratory was flooded by hurricane Ike in 2008 but was recently rebuilt and is fully operational again. The entire laboratory is currently flood-proofed to an elevation of 14 feet above sea level to mitigate the risks from future storms. The laboratory has all the necessary equipment for sample preparation including cryo-plungers, a high-pressure freezer, cryo-ultramicrotome and a freeze substitution system. The containment has been designed following the Biosafety in Microbiological and Biomedical Laboratories (U.S. Department of Health and Human Services, 2007) recommendations for BSL-3 facilities and was optimized for cryo-EM operation. The Facility was the first of its kind in the world to provide researches with BSL-3 capabilities for structural studies of infectious agents. The facility has a high-end cryo-EM that is used to perform both single particle and tomography experiments. Standard Operating Procedures and specimen preparation and handling protocols were originally developed for the Facility in 2006 and were updated before the Laboratory became operational again in 2011, including procedures and provisions for many catastrophic events including fire, loss of power, air supply/exhaust failures, agent losses or spill incidents, bomb threats, etc. Weather-related issues were addressed by shutting the lab down early enough to allow for its decontamination before a required evacuation. There is a general disaster preparation plan for all BSL-3 labs on campus; the Cryo-EM lab has its own specific procedures to follow.
Before hurricane Ike, we developed a heat decontamination protocol to disinfect both the inside of the microscope column and its vacuum system (Sherman, Freiberg, Razmus et al., 2010). Although that protocol was successfully used for one year, its use was limited to heat-labile agents that cannot survive exposure to 60°C within a time frame of hours, and required heat lability testing of each specific agent. This requirement severely limited the range of agents that could be studied in the Laboratory. As an example, neither bacterial samples nor heat-stable viruses could be studied in BSL-3 containment because of the limitations of the decontamination protocol.
Restoration of the Facility after the hurricane Ike prompted us to consider options for more effective and versatile decontamination methods applicable to a broader range of infectious agents, including bacteria. Ideally, such methods would be safe for the microscope, and could be automated or at least semi-automated. We have tested several different disinfecting approaches and found that using chlorine dioxide gas as a sterilant is the best choice currently available. Here we report on our experience and the specific design of an automated system for decontamination of an electron microscope and automated data collection system in BSL-3 containment that has permitted the operation of a high-end cryo-EM to perform both single particle and tomography experiments.
Materials and methods
Virus production and purification
All virus manipulations were carried out in BSL-3 laboratories using recommended procedures (Sherman, Freiberg, Razmus et al., 2010; U.S. Department of Health and Human Services, 2007).
WEEV strain CO92-1356, isolated from Culex tarsalis mosquitoes in Larimer County, Colorado on July 30, 1992, was purified and prepared for cryo-EM as described in (Sherman and Weaver, 2010).
RFVF vaccine strain MP12 purification and concentration was described in (Sherman, Freiberg, Holbrook et al., 2009).
Everglades virus strain Fe5-47et, isolated from Aedes (Ochlerotatus) taeniorhynchus mosquitoes collected in Florida on the 16th of January 1969, was purified and examined microscopically. The virus was passaged twice in suckling mouse brains, once in baby hamster kidney (BHK-21) cells, and then used to infect BHK-21 cells at a multiplicity of ca. one PFU/cell. Following the appearance of cytopathic effects, the cell culture medium containing 2% fetal bovine serum was harvested and clarified by centrifugation at 2,000 × g for 10 min. Next, polyethylene glycol and NaCl were added to 7% and 2.3% W/V concentrations, respectively, and the virus was allowed to precipitate overnight at 4°C. Following centrifugation at 4,000 × g for 30 min at 4°C, the pelleted precipitate was resuspended in TEN buffer [0.05M Tris-HCl (pH 7.4), 0.1 M NaCl, 0.001 M EDTA] and loaded onto a 20-70% continuous sucrose (W/V) gradient in TEN buffer. Following centrifugation at 270,000 × g for one hour, the visible virus band was harvested using a Pasteur pipette and centrifuged 4 times through an Amicon Ultra-4 100kDa filter (Millipore, Billerica, Massachusetts) with resuspension in 4 ml of TEN buffer at each step to remove residual sucrose.
Sindbis virus (SINV) strain AR339 was cultured in BHK-21 cells, purified and prepared for cryo-EM using the same methods described above for EVEV.
Grid preparation and freezing
Purified WEEV, EVEV, and RVFV were vitrified as reported previously (Freiberg, Sherman, Morais et al., 2008; Sherman, Guenther, Tama et al., 2006) on holey carbon film grids (R2×2 or R1.2/1.3 Quantifoil®; Micro Tools GmbH, Jena, Germany; or C-flat™, Protochips, Raleigh, North Carolina). Briefly, purified concentrated suspensions of viruses were applied to the holey films in a volume of ca. 3.5 μl, blotted with filter paper, and plunged into liquid ethane cooled in a liquid nitrogen bath. Frozen grids were stored under liquid nitrogen and transferred to a cryo-specimen 626 holder (Gatan, Inc., Pleasanton, California) under liquid nitrogen before loading into a JEOL 2200FS electron microscope, equipped with in-column energy filter (omega type) and a field emission gun (FEG), operating at 200 keV. Grids were maintained at near-liquid nitrogen temperature (−172 to −180°C) during imaging.
Particle imaging
Imaging was carried out either with the operator wearing all PPE inside of the containment laboratory, or from outside the containment lab using SIRIUS remote microscope control software (JEOL Inc.) through a computer network. WEEV (Sherman and Weaver, 2010) and EVEV images were acquired at 60,000x nominal microscope magnification, while RVFV particles (Sherman, Freiberg, Holbrook et al., 2009) were imaged at 40,000× magnification using a CCD camera (UltraScan 895, GATAN, Inc., Pleasanton, CA). Images were acquired with a ca. 20 electrons/Å2 dose; the CCD pixel size corresponded to 2 Å on the specimen scale for WEEV and EVEV, and to 3 Å for RVFV. We used a 0.3 – 1.64 μm defocus range for EVEV imaging. 4729 virus particle images were selected from 80 CCD frames. An in-column omega electron energy filter was used during imaging with a zero-loss electron energy peak selected with a 20 eV slit.
Image processing
WEEV and RVFV image processing was described in (Sherman and Weaver, 2010) and (Sherman, Freiberg, Holbrook et al., 2009), respectively.
Individual EVEV images were picked using EMAN2 package (Tang, Peng, Baldwin et al., 2007) (E2BOXER) and contrast transfer function (CTF) determination and correction was performed using (E2CTFCOR). Particle alignment and orientation determination were done using IMAGIC-5 package (van Heel, Harauz, Orlova et al., 1996). Three-dimensional (3D) reconstruction was calculated using BKPR (Orlov, Morgan and Cheng, 2006). Particle images were centered and the angular reconstitution technique (van Heel, 1987) was then used to determine the relative orientations of the images; these orientations were used to calculate 3D maps in consecutive iterations until the process converged. The final 3D map was reconstructed from the 1000 best virus images. The effective resolution of the map was 1 nm according to a 0.5 Fourier shell correlation (FSC) criterion (Rosenthal and Henderson, 2003). The 3D maps were surface-rendered and displayed with a one standard deviation (1σ) threshold in CHIMERA (Pettersen, Goddard, Huang et al., 2004), which accounted for ~100% particle volume. Homology modeling was done using SWISS-MODEL server (Arnold, Bordoli, Kopp et al., 2006) We performed fitting of E1 – E3 homology models into the cryo-EM density map in CHIMERA (Pettersen, Goddard, Huang et al., 2004).
Results
Architectural design of the containment
The W. M. Keck Center for Virus Imaging is part of cryo-EM Laboratory within the UTMB Sealy Center for Structural Biology and Molecular Biophysics. This Laboratory was designed to include both BSL-3 containment and BSL-2 open research space. While the open research component of the Laboratory is used to study non-infectious or BSL-2 biological agents, the Keck Center focuses on imaging of viruses and bacteria at BSL-3. The Laboratory houses 3 cryo-EM: JEM1400 LaB6 and JEM 2100 LaB6 outside of the containment, with GATAN Orius fast-scan- and Ultrascan 894 (JEM 1400) or Ultrascan 895 (JEM 2100) CCD cameras. A JEM 2200FS with a field emission gun, in-column omega-type electron energy filter and GATAN Orius and Ultrascan 895 CCD cameras is placed inside the containment and can operate under BSL-3 conditions. Additionally, it can be remotely controlled using JEOL SIRIUS software and a FasTEM knobset through the general computer network.
Cryo-EM allows for the preservation of viability of biological samples, including infectious agents; therefore studying them in a microscope should be performed with proper precautions to protect research and maintenance personnel from infection by the samples studied. Because we intended to study BSL-3 agents, a containment laboratory had to be built for the microscope. Requirements for BSL-3 containment are different from those for EM imaging, so we needed to design our containment taking into account both, BSL-3 conditions and recommendations for EM rooms with laminar air flow, very stable room temperature and air pressure, and low levels of floor vibrations and acoustical noise in the room.
The containment rooms are physically separated from the rest of the laboratory, have negative air pressure and a clean change vestibule room with PPE storage. In addition to the general requirements for a microscope room to be free of vibrations and stray electromagnetic fields, the microscope operation and maintenance issues included temperature and air pressure stability considering the required by BSL-3 standards movement of air through the facility that could result in column instability. The walls and the ceiling were specially designed with resilient channels embedded in 3-layered drywall to dampen acoustic noise from outside sources. This was essential because no porous wall coverings usually used as acoustic dampers are allowed in BSL-3 containment due to the need for surface decontamination in the event of an incident. A drop ceiling was installed to act as an air damper to ensure the supply of slow, laminar air flow. The air exhaust grilles were lowered and distributed around the perimeter of the room to minimize air movement around the microscope. The air ducts in the facility incorporated inner acoustic-dampening liners and flexible joints to minimize vibrations originating from the building air handlers, and acoustic transmittance was also minimized by the shape and lengths of the ducts. The general BSL-3 containment organization was described in (Sherman, Freiberg, Razmus et al., 2010) including entry and exit procedures from the containment, PPE required for persons inside the containment, mechanical design of the BSL-3 suite and the microscope room, etc.
Specimen Preparation
Owing to the excellent preservation of flash-frozen biological samples, they remain potentially infectious when thawed. This freezing method for cryo-EM leaves the agents immobilized in a solid water matrix, reducing the risk of aerosol exposure as long as the grids remain frozen. Although the volume of an agent on the grid is extremely small and the likelihood of producing infectious doses of aerosols is thus very limited, this risk cannot be excluded given the unique conditions required for grid preparation. The research personnel must therefore be protected at all times from possible exposure.
To protect researchers, grid freezing was performed in a biological safety cabinet (BSC). Before a plunging session, all necessary tools, bottles, solutions, stands, and other materials used during the session were placed inside the BSC before opening a vial containing an agent. After each session all the instruments, tools, etc. were surface-decontaminated before removing them from the BSC. In case of an incident, the BSC provides a contained air space, preventing the spread of the agent and contamination of the entire room.
Cryo-plunger
Most of the standardly used in cryo-EM cryo-plungers are not suitable for BSL-3 operation because they do not have a specimen chamber that is safe to operate in containment. In addition, they are usually too large to fit into a standard BSC and are very difficult to decontaminate after usage. We therefore developed a special, pneumatically operated cryo-plunger (Figure 1), which fits into a BSC, to flash-freeze samples. The plunger has a pneumatic latch circuit with two control valves that control a double-acting cylinder with a 6-inch long stroke (Clippard Instrument Laboratory, Inc. Cincinnati, Ohio). One control valve commanding the shaft to extend is attached to a foot pedal while the other valve (hand-operated) retracts the shaft. A tweezers holder is attached to the end of the shaft. A foam-insulated plastic box with metal insert holds liquid nitrogen and has a grid box holder inside as well as an insert for liquid ethane. The base of the plunger has a slot serving as a guide for the box to align it with the rest of the plunger. A safety shield around the tips of the tweezers is to protect the operator’s hands from being punctured by the sharp tips. This is especially important in BSL-3 containment where the required PPE includes double gloves that affect dexterity.
Figure 1.
Pneumatically operated cryo-plunger to vitrify samples for Cryo-EM in a BSL-3 containment of the W. M. Keck Center for Virus Imaging at UTMB. Note that the plunger is inside of a BSC to protect researchers from exposure to samples. Pneumatic cylinder, tweezers guard, container for waste with disinfectant, and other materials and tools used for freezing grids are indicated in the Figure.
The BSC air flow was adjusted so that air movement around the plunger was minimal to prevent ice contamination of the samples. To further reduce the chance of ice contamination during grid freezing, the freezing box has a lid that remains closed until the box is cooled and the grids are ready to be frozen. Frozen EM grids containing samples are either stored in a liquid nitrogen dewar in the sample preparation room, or are used for imaging immediately after freezing. The specimen transfer to the cryo-holder is performed in a dedicated cryo-transfer station.
Automation of data collection
Automation of data collection is desirable for BSL-3 operation to minimize the time researchers spend inside of containment and thus to reduce risk of their exposure to an agent. In addition, data collection automation broadens the user base by allowing persons without high containment or select agent access to engage in data collection. To collect automatically single particle data, we use the JADAS software package (JEOL) that can be set up and run remotely, even without using the Sirius program to control the microscope. This facilitates collaboration with remote users who send samples to freeze and load into the microscope, but wish to collect data themselves without being physically present in the Laboratory. SerialEM developed by Dr. D. N. Mastronarde (Mastronarde, 2005) is used to collect tilted data for EM tomography (cryo-EM tomography in particular) with minimal user intervention, and can also be run on the microscope through the general computer network.
Decontamination protocols
Any operation in high biosafety containment should be accompanied by an adequate disinfection procedure for use in the event of an incident or as a preventive measure for use prior to routine maintenance requiring opening of the microscope column or vacuum system. We therefore implemented several policies to decontaminate equipment and rooms within the containment. Some of them are used routinely after each EM session when using BSL-3 agents; others are used only in case of an incident involving infectious agents. As an example, the cryo-specimen holder is decontaminated after each use, as well as cryo-EM grids and cryo-transfer station and tools used for the transfer. In contrast, the inside of the microscope column and vacuum system would be decontaminated only if an incident had occurred, e.g. a specimen grid was lost or thawed inside the specimen airlock or column. This approach does not preclude surface decontamination of the outside of the microscope after EM sessions and before the biosafety level is lowered to allow service personnel to enter the facility.
Decontamination of the plunger
The plunger was designed to be easily serviceable in BSL-3 containment. It has minimal number of parts and its components can withstand chemical disinfectants. Most parts are made of chemically resistant stainless steel, and there are no electrical components. It was surface decontaminated after each session with CAVICIDE (Metrex Research Corporation, Romulus, MI). All tools used for specimen preparation and transfer to the microscope after each use are decontaminated first with CAVICIDE or 70% alcohol followed by heating them in an oven at 60-70°C for at least one hour.
Cryo-EM grids
Once an imaging session is complete, the cryo-holder is removed from the microscope and placed into the pre-cooled cryo-transfer station. The grid (still frozen) is removed from the holder and immediately immersed in CAVICIDE. A new grid is then inserted into the holder and transferred to the microscope for the next imaging session. In some cases the grids remain in the holder for heat decontamination in the pumping station, followed by immersion in CAVICIDE.
The cryo-transfer station and all tools used during the transfer of frozen grids to and from the cryo-holder are surface decontaminated by spraying them with 70% v/v ethanol or immersing them in CAVICIDE. Used cryo-grid storage boxes are stored in 70% v/v ethanol for at least one day followed by washing in deionized water and thoroughly drying them before the next use.
Specimen holder disinfection
Although the specimen holder must be decontaminated after use, harsh chemicals that would destroy the holder cannot be used. We therefore use heat decontamination at ca. 100°C for at least 2 hours in the GATAN 655 turbo pumping station (GATAN, Inc.) to ensure proper decontamination of the tip that contacted the grids. For decontamination, the cryo-holder is transferred with the shield closed while still at cryo-temperatures into the vacuum chamber, where it is warmed under high vacuum and maintained at 100°C for several hours. It can also be decontaminated chemically in a mixing box of the ClO2 generator (see below).
Decontamination of the microscope
Potential incidents affecting personnel safety
One of the major biosafety concerns was microscope contamination in case of an incident involving the thawing or loss of a grid inside the microscope column or in the specimen airlock during specimen transfer and pre-pumping. We use a GATAN 626 single tilt cryo-transfer holder with a 70° tip. A cryo-grid clamped in the holder by a clip-ring, if not secured properly, could be dislodged resulting in poor thermal contact with the holder and potential thawing. The grid could also fall from the holder and contact warm parts of the microscope column, resulting in instant thawing and the potential aerosolization of the agent inside the microscope column. Although the volume of the agent suspension is small (less than 2 – 3 nL), it would be difficult to locate a grid or droplet containing an agent inside the microscope to perform surface decontamination using typical protocols for infectious agents. An additional hazard that was considered was a vacuum leak that could cause airflow within the vacuum system to aerosolize a thawed agent. That, in turn, could create a hazard when servicing the microscope.
Choosing sterilants
Standard chemical surface decontamination procedures typically implemented in a BSL-3 environment are not possible within the microscope column because the use of liquid sterilants would compromise the ultra-high vacuum system. Also, most chemicals used for decontamination are corrosive to the microscope components even after thorough removal with a secondary cleaning solution. In addition, the required disassembly of the microscope column before decontamination would be unrealistic. Although our original heat decontamination protocol developed was shown to be effective against heat-labile virus strains, it required prolonged heating of the entire microscope column and other parts of the vacuum system, rendering the microscope nonoperational for several days. Also, heat decontamination would be ineffective for many bacteria, especially for spore-forming species. A gaseous chemical sterilant that kills biological agents quickly with minimal effects on the microscope components, and that penetrates all cavities inside the microscope’s vacuum system, would be ideal. Several sterilants widely used in medicine and biology were considered for the microscope decontamination (their properties listed in Table 1):
Formaldehyde, one of the most commonly used space decontaminating agents, has been available for a long time (Nordgren, 1939). It is a gas and a very effective disinfectant with good penetration and distribution. It cross-links proteins and lipids, preventing them from functioning and consequently leading to death of the exposed organisms. The gas is generated by heating paraformaldehyde powder to 450°F, causing its depolymerization and yielding the formaldehyde gas (NSF International/ANSI Standard). After the exposure, the gas must be neutralized with ammonium bicarbonate. The neutralization process creates a surface residue consisting of polymerized formaldehyde and methenamine (Luftman, 2005) within the vacuum system. Formaldehyde is a carcinogen , allergen and toxic substance.
Ethylene oxide is strong alkylating agent used for medical and scientific equipment and instruments sterilization. It is extremely flammable and explosive when mixed with air. In addition, it also leaves residual film on treated surfaces, and is carcinogenic (http://www.inchem.org/documents/hsg/hsg/hsg016.htm).
Vaporized hydrogen peroxide (VHP) is a strong oxidizing agent and therefore is a very effective sterilant produced by boiling/vaporizing 31%-35% liquid hydrogen peroxide. It is used for decontamination of laboratory and hospital rooms and small chambers (Akers, Agalloco and Kennedy, 1995; Herd and Warner, 2005; Meszaros, Antloga, Justi et al., 2005). Since the vapor is produced from an aqueous hydrogen peroxide solution, it is difficult if not impossible to avoid condensation that is undesirable in treating vacuum systems. VHP could corrode metals, especially in the presence of condensation (Malmborg, Wingren, Bonfield et al., 2001).
Chlorine dioxide (CD) is also strong oxidizer like VHP and a gas similar to formaldehyde. It is used for decontamination in BSC (isolators) (Czarneski and Lorcheim, 2005; Eylath, Wilson, Thatcher et al., 2003), processing vessels (Eylath, Madhogarhia, P. et al., 2003), rooms (Leo, Poisson, Sinclair et al., 2005), and large facilities (Czarneski, 2009). It easily penetrates intricate piping/cavities inside the microscope column/vacuum system. Although >50% humidity is required to kill microbes (Agalloco, Carleton and Frederick, 2008; Spotts Whitney, Beatty, Taylor et al., 2003; Westphal, Price, Leighton et al., 2003) it does not have an inherent condensation problem unlike VHP. It does not require neutralization or a post-exposure cleaning making CD an attractive decontaminating agent for a TEM.
Table 1.
| Sterilant | Needs cleanup |
gas | vapor | Carcinogen | corrosive | Equipment cost |
|---|---|---|---|---|---|---|
| Formaldehyde | Yes | Yes | No | Yes | No | Low |
| Ethylene oxide | Yes | Yes | No | Yes | No | Medium |
| Vaporized hydrogen peroxide | No | No | Yes | No | Yes | Medium |
| Chlorine dioxide | No | Yes | No | No | No | High |
In summary, formaldehyde is carcinogenic and leaves residual film on surfaces, which is unacceptable for ultra-high vacuum systems used in modern microscopes, and would require very complicated post-exposure cleaning of microscope parts, which is not realistic. Ethylene oxide is a very effective sterilant but leaves residues on treated surfaces, is highly flammable, even explosive when mixed with air, toxic and also carcinogenic similar to formaldehyde. Consequently, two candidates were selected for testing: VHP and ClO2. Both are widely used sterilants to decontaminate hospital rooms, BSC and other chambers and equipment exposed to pathogens. Hydrogen peroxide is a strong oxidant and therefore kills a wide range of pathogens. On the other hand, because of its oxidizing ability, it could damage some microscope materials. Our early attempts to use VHP with JEOL microscopes were not successful because of unacceptable level of corrosion of some parts inside the microscope column. Various parts were tested in a chamber filled with VHP and some showed visible discoloration and corrosion after the level of exposure necessary for a single decontamination cycle. In addition, VHP is a vapor and not a gas; it requires nearly saturated humidity and decomposes easily. Saturated humidity would lead to water droplet formation inside the microscope, which should be avoided at all times in electron microscopes. We therefore selected ClO2 (CD) for the microscope decontamination.
Decontamination protocol. Design and testing
Preliminary studies of CD decontamination of an electron microscope
Since CD has never been used before to decontaminate a TEM, we needed first to determine the compatibility of the microscope materials with the treatment and to establish basic parameters of a decontamination cycle. We therefore performed a set of both visual and functional tests at the ClorDiSys facility using an old JEM 2000 microscope. These tests not only established feasibility of using CD for TEM decontamination, but also provided data for the design of an automated microscope decontamination system. Longevity tests were performed at the same time demonstrating long-term survival of the microscope after periodical CD treatment.
The tests confirmed the ability of CD gas to kill biological indicators [Bacillus Atrophaeus spores, (Czarneski and Lorcheim, 2008)], and also demonstrated a low damage rate induced by the gas and good preservation of the microscope parts during and after the tests. We showed that with regular maintenance the microscope should be able to function properly even after 25-30 years of usage with monthly CD treatment of the microscope column and vacuum system. The proof-of-principle tests at the ClorDiSys Facility were followed by battery of tests using the JEM 2200FS at UTMB. Although most of the parts inside the microscope survived these tests, the scintillator screen in the JEM 2000FS was damaged. However, because it is an easy replaceable and inexpensive part of the machine, it was decided to consider it as a consumable.
Interfacing CD generator with an electron microscope
The 2200FS microscope with the CD generator in the W. M. Keck Center for Virus Imaging (UTMB) is shown in Figure 2a and one of the CD injection ports is shown in Figure 2b. It was necessary to integrate decontamination procedures with the microscope, which required extensive modifications of both the CD generator (Minidox M, ClorDiSys, http://www.clordisys.com/site.php?index.php&20) and the JEM 2200FS. We therefore developed an interface between the Minidox-M, and the microscope so that both pieces of equipment would be mechanically and electronically coupled and would communicate, providing a semi-automated system easy to program, control and monitor (Figure 3a). A controller/interface was developed by JEOL to operate vacuum valves in the proper sequence to allow venting and continuous gas flow through the intended path of the microscope. In addition, it communicated with the Minidox M generator to start and end a decontamination cycle and to perform some other functions. Minidox M sent signals to the new microscope controller, initiated the valve sequence, and maintained the proper flow path. In turn, the controller informed the Minidox M of microscope’s readiness for a cycle. The Minidox M prepared a CD mixture with nitrogen and humidified it as needed, maintaining proper concentration and humidity of the CD. The overall scheme of the interface is shown in Figure 3b. A set of electronic valves was added to Minidox M to allow to custom design a decontamination cycle controlled by the generator without manual intervention through the cycle. After a decontamination cycle was complete, the whole system was purged using dry nitrogen gas to remove CD and residual moisture from the vacuum lines and the generator before restarting the normal operation of the microscope. To prevent accidental CD exposure of the vacuum system/other parts of the microscope and personnel, a safety interlock was installed in the microscope, precluding an accidental decontamination run from starting while the microscope was operational. Only after the microscope was turned off and its diffusion pumps were cooled down would the interlock allow a cycle to start.
Figure 2.
General view of the JEM 2200FS EM with Minidox-M CD generator (A). One of the CD injection ports into the microscope for decontamination of the specimen chamber (B). The microscope is interfaced with the CD generator with automated CD cycles to disinfect sections of the EM column.
Figure 3.
A. Schematic view of CD flow through the system. Minidox-M generator supplies CD to the mixing chamber at required concentration where it is humidified to facilitate the disinfection. Once the mixture is ready, a close loop is established between the microscope and the mixing chamber, a small diaphragm pump is used to pump the mixture through the loop. B. CD decontamination cycle. CD is injected into the mixing chamber until required concentration is reached followed by humidifying the mixture in the mixing chamber. Once both parameters (CD concentration and humidity) are in a require range, control valves open to establish a close loop between part of EM column to be decontaminated and the mixing chamber and a pump is activated to pump the mixture through. CD concentration and humidity are monitored and maintained during the cycle. After the cycle is finished, the system is purged with dry nitrogen gas while residual CD is scrubbed
The current system was designed to perform multiple decontamination runs semi-automatically using the Minidox-M ClO2 generator interfaced with the microscope. The design was aimed at minimal human intervention during runs. A multipoint gas injection system with full control of the microscope vacuum valves was developed during the system design, setup and testing, and proved to be effective in killing bacterial spore indicators (BIs) commonly used to test disinfection protocols (Czarneski and Lorcheim, 2008). The microscope was interfaced with the Minidox-M generator; an additional valve controller was installed allow for independent manipulation of the microscope vacuum system when the microscope was turned off. The microscope vacuum system was partitioned into several compartments because of its complexity, and each compartment was separately decontaminated with CD; an example is presented in Figure 2b where an injection port is shown to introduce CD into specimen chamber of the microscope. The system is fully contained and is safe to run even without using PPE (gas masks, gloves, chemical suites, etc.). A ClO2 monitor was permanently mounted on the wall in the microscope room to monitor possible CD leaks and to alert researchers about potentially dangerously high level of the gas in the room. It was necessary to add water vapor to the decontamination loop to allow spores in BIs to swell facilitating CD penetration into the spores to kill them. A sufficiently high humidity level (above 50% RH) was maintained in the loop while running a decontamination cycle. The CD concentration during a run was maintained at ~1 mg/L level. A small diaphragm pump (Grainger, SHURFLO 4UN55) was used to pump the gas through the loop. There was a mixing box in which properly humidified CD was prepared first and then injected into the microscope at the desired entry ports. Gas then traveled through the microscope column with its intricate chambers and pipes via opening preset for the cycle microscope valves. CD left the microscope column either through diffusion pump(s) and reservoir tank or through turbo-molecular pump by vacuum lines to the mechanical pumps and further through their exhaust lines. The exhaust of the mechanical vacuum pumps was HEPA filtered to prevent any possible hazardous discharge from the pumps themselves. Just before the HEPA filters a combination of normally open bypass valves (closed during the decontamination process) and normally closed valves (opened during decontamination) were installed thus allowing the gas to return to the Minidox CD generator. Thus, a closed loop for continuous gas circulation was established, allowing us to monitor and to maintain the necessary gas concentration and relative humidity in the loop during a run. At the end of a run when the preset CD exposure was achieved, the loop was purged with dry nitrogen to remove ClO2 from the system using a neutralizing device (scrubber). After all necessary decontamination runs were finished, the microscope was turned on and pumped to high vacuum normally followed by an overnight “bake out” to remove all the residual moisture and to outgas the interior of the column. This concluded the decontamination of the entire microscope.
Compartmentalization of the EM for reliable decontamination
When designing the decontamination protocol we considered several possible contamination scenarios and decided to perform the decontamination in several stages. One location in the microscope considered at highest risk of contamination was the specimen chamber, where a cryo-EM grid could fall out of the holder if not clipped securely. Therefore that area can be separately decontaminated if the incident did not involve any other parts of the microscope. Another scenario included a grid thawing in the holder if it’s dewar maintaining specimen at the cryo-temperature ran out of liquid nitrogen and warmed. In that case both upper (above the omega filter) and lower (below the omega filter but above the valve separating the column and the camera chamber) parts of the column would need to be decontaminated. The film camera chamber could be decontaminated separately from the rest of the microscope, as well as the FEG. Of course both CCD cameras would be decontaminated when the camera chamber is exposed to CD. An additional cycle was designed to decontaminate the turbomolecular vacuum pump (TMP) and it’s backing mechanical pump. The rest of the microscope vacuum system not exposed in the previously described cycles would be decontaminated in an additional cycle. All 3 mechanical pumps were part of the corresponding cycles.
Decontamination tests with biological indicators
BIs for the test runs were placed in several critical locations in the microscope column and the camera chamber to validate the elimination of potential contamination of the vacuum system by infectious agents. In general, the critical and most difficult locations for the gas flow were selected for BI placement: (a) the pipe in the specimen airlock in the goniometer; (b) a deep pocket right behind the door in the camera chamber of JEM 2200FS (CD was introduced from the back of the chamber), (c) in the condenser aperture holder, (d) in the objective lens pole-piece, (e) at the entrance aperture of the omega filter, and in several other locations. Since the microscope compartments intended for a run were vented with the CD gas, many of these hard-to-reach places were flooded with the humid gas at that time and then gas concentration and humidity were maintained by the continuous gas flow through the compartments and the rest of the vacuum system pathway for that particular loop.
A typical decontamination run included 2 hours of CD exposure and total run time was usually ca. 3 hours including gas mixture preparation and system purge time. The microscope can be restarted immediately afterward. With the most likely place of contamination in the specimen airlock, the microscope would be ready for operation the next day after an incident, while whole microscope decontamination including all the other necessary decontamination cycles would take 2 days in total. The Minidox-M continuously sampled several parameters in the loop during a run, e.g. the gas concentration, humidity, temperature, etc.; it produced logs that were printed during the cycle, and archived them on a hard drive as well.
Although the amount of infectious agents in the containment was less than a few microliters, so that the chance of an incident requiring whole-room decontamination was minimal, if necessary the microscope and specimen preparation rooms can still be decontaminated by using chlorine dioxide gas.
Structures of Agents Studied in the Center
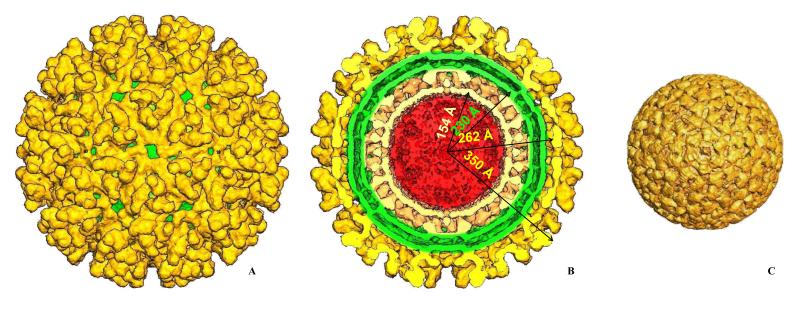
Several agents were studied in the Laboratory, including western equine encephalitis virus (WEEV), an alphavirus that causes highly debilitating and often fatal disease in humans and horses (Smith, Mackenzie and Weaver, 2009). WEEV was classified as a NIAID high priority Category B biothreat agent. To our knowledge, this was the first BSL-3 virus ever imaged in BSL-3 containment using Cryo-EM.. We used IMAGIC 5 (van Heel, Harauz, Orlova et al., 1996), EMAN (Ludtke, Baldwin and Chiu, 1999) and PFT (Baker, Olson and Fuller, 1999) to process images and to obtain 3D reconstruction of WEEV (Sherman and Weaver, 2010). Figure 4 shows the 3D reconstruction of WEEV. Particles were ~ 700 Å in diameter, and exhibited T = 4 quasi-symmetry within the icosahedral lattice on the virus surface. E1 and E2 glycoproteins formed heterodimers that in turn oligomerized into 80 trimeric spikes protruding perpendicular to the virus surface. The spikes were ca. 88 Å high and 120 Å in diameter. In Figure 4B the front half of the map is removed to reveal the inner virion organization. The outer glycoprotein shell (gold color) of the virus was formed by the E1/E2 glycoproteins. The positions and orientations of E1 in the map were derived using a homology model based on the E1 structure from Semliki Forest virus (Lescar, Roussel, Wien et al., 2001) and were similar to E1 in SINV (Zhang, Mukhopadhyay, Pletnev et al., 2002a; Zhang, Mukhopadhyay, Pletnev et al., 2002b). The rest of the spike density was attributed to E2 glycoproteins, forming the “stem” of the spikes. The E2 protein apparently interacts with cell receptors and conceals the fusogenic peptide of the E1 (Kuhn, 2007a; Kuhn, 2007b). The glycoprotein shell was separated from the nucleocapsid core by a lipid bilayer derived from the host cell (Figure 4B, green). The E2 cytoplasmic domain penetrated through the bilayer and appeared to bind the capsid protein, organizing the latter into well-ordered icosahedral particles with T = 4 quasi-symmetry. The nucleocapsid core had pronounced capsomers protruding by 40 Å from its surface. Hexons were skewed with a diameter of ~116 Å; pentons were 104 Å across. The outer glycoprotein WEEV shell was similar to that of SINV, which was expected since WEEV is a recombinant virus derived from two distinct alphavirus lineages, namely ancestral eastern equine encephalitis virus (EEEV) and SINV. WEEV inherited the E1 and E2 genes from SINV and the capsid protein from EEEV lineages. Surprisingly, the capsomers in the WEEV nucleocapsids were slightly rotated (~7 deg) relative to those in Venezuelan equine encephalitis virus (VEEV), a member of EEEV cluster.
Figure 4.
A 1.3 nm resolution Cryo-EM reconstruction of western equine encephalitis virus (WEEV), a level 3 alphavirus. A. Surface rendering of the reconstruction. E1/E2 glycoprotein shell is shown in gold; a green membrane envelope is seen through the holes in the E1/E2 shell. E1/E2 heterodimers form trimer spikes with E2 glycoprotein being up and inside the spike while E1 forming the floor of the shell. B. Same as A, but with the front half of the map removed. Major virus dimensions are shown for nucleocapsid, membrane envelope and the glycoprotein shell. Overall size of the virus is 70 nm with nucleocapsid protecting the viral RNA of 46 nm in diameter. C. The nucleocapsid computationally extracted from the map. It has the same icosahedral organization as the outer glycoprotein shell with T=4 symmetry. WEEV E1/E2 shell is very similar to that of Sindbis virus, another alphavirus, while the nucleocapsid pentamers and hexamers are slightly rotated relative to those in Sindbis virus (Sherman and Weaver, 2010).
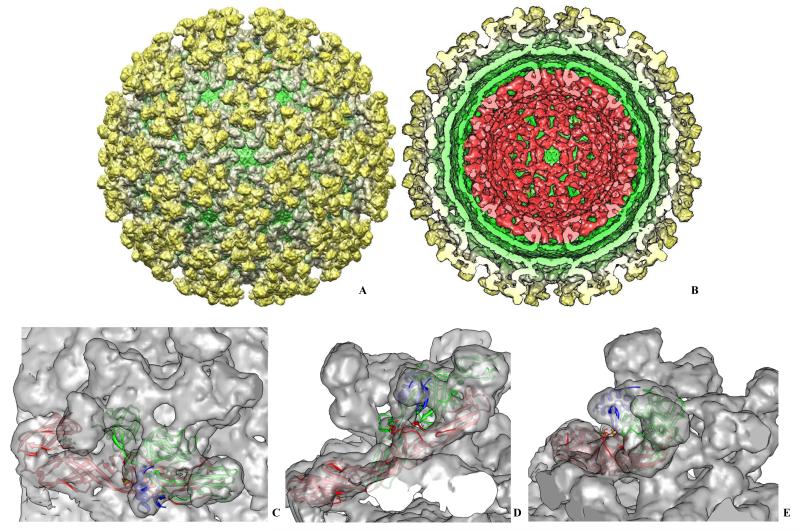
Another example of a BSL-3 alphavirus studied in the Laboratory was Everglades virus (EVEV), a close relative of VEEV (Coffey, Crawford, Dee et al., 2006). It was recently imaged and reconstructed to 1 nm resolution using IMAGIC 5 (van Heel, Harauz, Orlova et al., 1996) and BKPR (Orlov, Morgan and Cheng, 2006) (Figure 5).
Figure 5.
A 1 nm resolution Everglades virus cryo-EM map. A. Surface rendering of EVEV showing overall organization of E1/E2/E3 glycoprotein spikes on the surface. E1 forms the lower part of the glycoprotein shell (floor) and extends to the upper part of the spikes making contacts with E2 glycoprotein. Through holes in shell the membrane envelope is seen (shown in green). The rendering was done at 1 standard deviation of the density to include 100 % of the virus mass. B. Same as A. but with front half of the map removed to show internal organization of the EVEV. Gold and gray colors represent E1/E2/E3 glycoprotein shell, the host-derived lipid membrane envelope is shown in green, and the capsid is shown in red. The viral genomic RNA was computationally removed from the map to show inner surface of the capsid shell. C. Top view of one of the trimeric glycoprotein spikes is shown with pseudo-atomic models of E1, E2 and E3 fitted into the reconstruction as rigid bodies. . D. Side view of the same spike shown in C. E1 is shown in red, E2 is in green and E3 is shown in blue. E. Another side view of the spike shown in D. with the same color scheme. The rigid body fit was performed in Chimera (Pettersen, Goddard, Huang et al., 2004).
Our reconstruction is quite similar to the published higher resolution VEEV cryo-EM map (Zhang, Hryc, Cong et al., 2011) and other alphaviruses (Mancini, Clarke, Gowen et al., 2000; Mukhopadhyay, Zhang, Gabler et al., 2006; Zhang, Mukhopadhyay, Pletnev et al., 2002b; Zhang, Fisher, Olson et al., 2002), demonstrating that alphaviruses in general have very similar 3D organization. EVEV particles were ca. 70 nm in diameter, similar to WEEV and other alphaviruses with glycoprotein spikes and internal nucleocapsid features. An unusual density was found in the spikes that cannot be attributed to E1/E2 heterodimers; it appears to belong to the E3 glycoprotein typically present in PE2 (E2 precursor polyprotein) but normally cleaved off during spike maturation and generally not present in mature alphavirions (Figure 5C-E, blue). We obtained homology models of E1, E2 and E3 based on the high-resolution structures of chikungunya virus glycoproteins (Voss, Vaney, Duquerroy et al., 2010). The occupancy of E3 glycoprotein in our map was less than 100% but was still present in our reconstruction similar to (Zhang, Hryc, Cong et al., 2011). We performed rigid body fitting of the EVEV E1 – E3 homology models in Chimera (Pettersen, Goddard, Huang et al., 2004) with E1 model giving 0.74 cross-correlation coefficient (CC), E2 had CC = 0.84, and E3 fitted into the map with CC = 0.74 (Figure 5D). That again demonstrated similarity of the alphavirus structures across the different lineages.
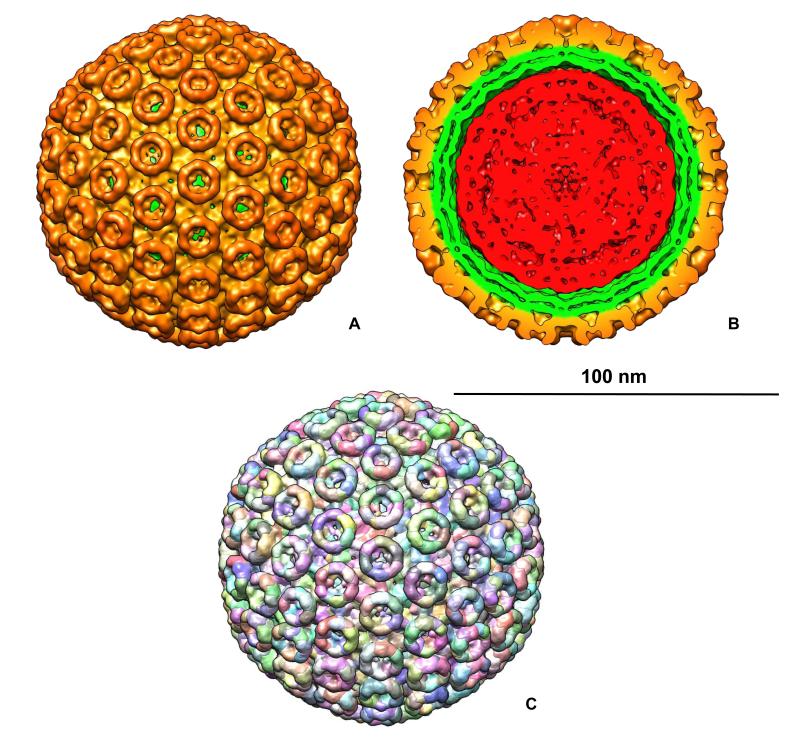
Yet another example of 3D reconstruction obtained in the Center is the structure of Rift Valley fever virus (RVFV) (Freiberg, Sherman, Morais et al., 2008; Sherman, Freiberg, Holbrook et al., 2009), a prototypical bunyavirus associated with major disease outbreaks in livestock and humans throughout Africa and the Arabian Peninsula. RVFV is classified as a NIAID priority Category A biothreat agent. RVFV is a spherical virus with a lipid-membrane envelope and surface protrusions composed of two viral transmembrane glycoproteins (GN and GC). RVFV contains a tripartite, single stranded, negative-sense RNA genome. The three RNA segments each bind N and L proteins to form ribonucleoprotein (RNP) complexes that are contained within the interior of the virus particle. The L, M, and S RNP complexes are necessary for genome transcription, replication and virus infectivity (Schmaljohn and Hooper, 2001; Schmaljohn and Nichol, 2007). The 3D map of RVFV (Figure 6) (Sherman, Freiberg, Holbrook et al., 2009) clearly showed T=12 icosahedral symmetry. The two glycoproteins, GN and GC, were organized in 122 distinct capsomers on the virus surface, extending by ca. 96 Å above the lipid envelope. Twelve of them were pentons and the rest (110) formed hexons. Capsomers were hollow cylinders. The pentons and hexons were interconnected by ridges located just above the lipid envelope. The hexons had outer and inner diameters of 125 Å and 84 Å, respectively with a constriction at 65 Å radially from the lipid membrane. Pentons were more compact than hexons with outer and inner diameters of 120 Å and of 65 Å, respectively. The space inside pentons was blocked by plugs at 58 Å from the penton base. Inside the ridges connecting the capsomers channels were found running between adjacent capsomers penetrating their walls into the inner cavities. The ridges interconnected the capsomers were thought to be essential for the virus assembly and stability.
Figure 6.
A. Surface rendering of Rift Valley fever virus (RVFV) T = 12 icosahedral reconstruction (Sherman, Freiberg, Holbrook et al., 2009) showing surface spikes composed of viral glycoproteins. RVFV particles were ca. 100 nm in diameter; B. Similar to A. but with front part of the reconstruction removed to show surface spikes (gold), viral envelope (green) and ribonucleoprotein core (red) containing viral RNA. C. Segmentation of the EM map showing tentative Gn/Gc glycoprotein distribution in the reconstruction. The scale bar is 100 nm.
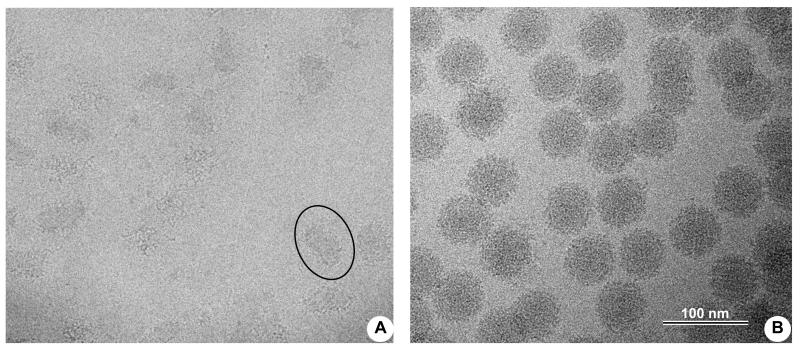
To demonstrate changes in viruses caused by irradiation treatment that is a standard UTMB decontamination practice before releasing them from BSL-3 containment, we purified and concentrated SINV and imaged samples before and after irradiation. The typical gamma-irradiation dose used in our BSL-3(4) labs is about 7 rad, which is reported to render infectious agents safe to handle in BSL-2 labs. We used the same dose for SINV particles before imaging them with cryo-EM (Figure 7A). No intact virus particles were observed in the sample; only small membrane patches and free RNA were present, although virus particles before irradiation looked perfectly “healthy” (Figure 7B). The experiment confirmed the efficacy of irradiation for decontaminating virus samples before removing them from BSL3 containment.
Figure 7.
Sindbis virus was cultured and purified as described in Materials and Methods. The sample was divided in two parts, with one subjected to gamma-irradiation and the other left untreated. A. Cryo-EM of Sindbis virus particles after irradiation with 7 rads of gamma-rays. Only small membrane patches (encircled) and some free RNA and protein aggregates were visible with no intact virus particles present. B. Same virus preparation frozen before gamma-irradiation. All particles look similar to other untreated alphaviruses with characteristic spiky appearance.
Acknowledgements
The Center was established through generous gifts from W. M. Keck Foundation, and the Kleberg Foundation, and by a grant from Health Resources and Services Administration. The Center operation was supported in part by grants to MS and SCW from NIAID through the Western Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research, NIH grant number U54 AI057156.
Footnotes
Accession codes The three-dimensional density maps have been deposited into the EBI-MSD EMD database with accession codes EMD-5210 for the structure of WEEV, EMD-5124 for the structure of RVFV, and EMD-XXX for EVEV.
Disclosure Statement JEOL is the manufacturer of JEM 2200FS and ClorDiSys manufactures Minidox-M CD generator
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- National Toxicology Program P. H. S. Department of Health and Human Services, National Toxicology Program, editor. [Retrieved June 10, 2011];Report on Carcinogens. 2011 Jun; from: http://ntp.niehs.nih.gov/go/roc12.
- NSF International/ANSI Standard 49-2007 . Class II (laminar flow) biosafety cabinetry. American National Standards Institute (ANSI); Ann Arbor (MI): [Google Scholar]
- Agalloco J, Carleton P, Frederick J. Validation of pharmaceutical processes. Informa Healthcare USA Inc.; New York: 2008. [Google Scholar]
- Akers JE, Agalloco JP, Kennedy CM. Experience in the design and use of isolator systems for sterility testing. PDA Journal Pharmaceutical Science Technology. 1995;49:140–144. [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Downing KH, Dubochet J, Glaeser RM, Heide HG, et al. Cryoprotection in electron microscopy. J. Microsc. 1986;141:385–391. [Google Scholar]
- Coffey LL, Crawford C, Dee J, Miller R, Freier J, et al. Serologic evidence of widespread everglades virus activity in dogs, Florida. Emerging infectious diseases. 2006;12:1873–1879. doi: 10.3201/eid1212.060446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarneski MA. Microbial decontamination of a 65-room new pharmaceutical research facility. Applied Biosafety: Journal of the American Biological Safety Association. 2009;14:81–88. [Google Scholar]
- Czarneski MA, Lorcheim P. Isolator decontamination using chlorine dioxide gas. Pharmaceutical Technology. 2005;29:124–133. [Google Scholar]
- Czarneski MA, Lorcheim P. Validation of chlorine dioxide sterilization. In: Agalloco J, Carleton F, editors. Validation of pharmaceutical processes. Inform Healthcare, Inc.; New York: 2008. pp. 281–287. [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Eylath A, Wilson D, Thatcher D, Pankau A. Successful sterilization using chlorine dioxide gas: Part one-Sanitizing an aseptic fill isolator. BioProcess International. 2003;1:52–56. [Google Scholar]
- Eylath AS, Madhogarhia ER, P. L, Czarneski MA. Successful sterilization using chlorine dioxide gas: Part two-Cleaning process vessels. Bio-Process International. 2003;1:54–56. [Google Scholar]
- Freiberg AN, Sherman MB, Morais MC, Holbrook MR, Watowich SJ. Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J Virol. 2008;82:10341–10348. doi: 10.1128/JVI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herd ML, Warner A. Hydrogen peroxide vapor biodecontamination of the Jackson Laboratory’s new animal facility. Animal Lab News. 2005;4:31–39. [Google Scholar]
- Kuhn RJ. Togaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields’ Virology. New York: 2007a. pp. 1001–1022. [Google Scholar]
- Kuhn RJ. Togaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields’ Virology. Fifth Edition Lippincott, Williams and Wilkins; New York: 2007b. pp. 1001–1022. [Google Scholar]
- Leo F, Poisson P, Sinclair CS, Tallentire A. Design, development, and qualification of a microbiological challenge facility to assess the effectiveness of BFS aseptic processing. PDA Journal of Pharmaceutical Science and Technology. 2005;59:33–48. [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, et al. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Luftman H. Neutralization of formaldehyde gas by ammonium bicarbonate and ammonium carbonate. Applied Biosafety: Journal of the American Biological Safety Association. 2005;10:101–106. [Google Scholar]
- Malmborg A, Wingren M, Bonfield P, McDonnell G. VHP takes its Place in Room Decontamination. Clean Rooms. 2001:11. [Google Scholar]
- Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Meszaros JE, Antloga K, Justi C, Plesnicher C, McDonnell G. Area fumigation with hydrogen peroxide vapor. Applied Biosafety: Journal of the American Biological Safety Association. 2005;10:91–100. [Google Scholar]
- Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren G. Investigations on the sterilization efficacy of gaseous formaldehyde. Journal of the American Medical Association. 1939;113:1759–1760. [Google Scholar]
- Orlov IM, Morgan DG, Cheng RH. Efficient implementation of a filtered back-projection algorithm using a voxel-by-voxel approach. J Struct Biol. 2006;154:287–296. doi: 10.1016/j.jsb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Schmaljohn CS, Hooper JW. Bunyaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott, Williams & Wilkins; Philadelphia, PA: 2001. pp. 1581–1602. [Google Scholar]
- Schmaljohn CS, Nichol ST. Bunyaviridae. In: K. DM, Howley PM, editors. Fields Virology. Wolters Kluwer; Philadelphia, PA: 2007. pp. 1741–1790. [Google Scholar]
- Sherman MB, Weaver SC. Structure of the recombinant alphavirus Western equine encephalitis virus revealed by cryoelectron microscopy. J Virol. 2010;84:9775–9782. doi: 10.1128/JVI.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MB, Freiberg AN, Holbrook MR, Watowich SJ. Single-particle cryo-electron microscopy of Rift Valley fever virus. Virology. 2009;387:11–15. doi: 10.1016/j.virol.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, et al. Removal of divalent cations induces structural transitions in red clover necrotic mosaic virus, revealing a potential mechanism for RNA release. Journal of virology. 2006;80:10395–10406. doi: 10.1128/JVI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MB, Freiberg AN, Razmus D, Yazuka S, Koht C, et al. A Unique BSL-3 Cryo-Electron Microscopy Laboratory at UTMB. Applied biosafety : journal of the American Biological Safety Association. 2010;15:130–136. doi: 10.1177/153567601001500305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DW, Mackenzie JS, Weaver SC. Alphaviruses. In: Richman DD, et al., editors. Clinical Virology. ASM Press; Washington, D.C.: 2009. pp. 1241–1274. [Google Scholar]
- Spotts Whitney EA, Beatty ME, Taylor TH, Jr., Weyant R, Sobel J, et al. Inactivation of Bacillus anthracis spores. Emerging infectious diseases. 2003;9:623–627. doi: 10.3201/eid0906.020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Transmission electron microscopy of frozen hydrated biological material. Electron Microsc Rev. 1989;2:117–121. doi: 10.1016/0892-0354(89)90012-9. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services . Biosafety in Microbiological and Biomedical Laboratories. 5th edition U.S. Government Printing Office; Washington, D.C.: 2007. [Google Scholar]
- Unwin N. The use of cryoelectron microscopy in elucidating molecular design and mechanisms. Ann N Y Acad Sci. 1986;483:1–4. doi: 10.1111/j.1749-6632.1986.tb34485.x. [DOI] [PubMed] [Google Scholar]
- van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–124. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Westphal AJ, Price PB, Leighton TJ, Wheeler KE. Kinetics of size changes of individual Bacillus thuringiensis spores in response to changes in relative humidity. Proceedings of the National Academy of Sciences. 2003;100:3461–3466. doi: 10.1073/pnas.232710999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, et al. 4. A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. Embo J. 2011;30:3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, et al. Placement of the structural proteins in Sindbis virus. Journal of virology. 2002a;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, et al. Placement of the structural proteins in Sindbis virus. J Virol. 2002b;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fisher BR, Olson NH, Strauss JH, Kuhn RJ, et al. Aura virus structure suggests that the T=4 organization is a fundamental property of viral structural proteins. J Virol. 2002;76:7239–7246. doi: 10.1128/JVI.76.14.7239-7246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]