Abstract
Runx2 regulates osteogenic differentiation and bone formation, but also suppresses pre-osteoblast proliferation by affecting cell cycle progression in the G1 phase. The growth suppressive potential of Runx2 is normally inactivated in part by protein destabilization, which permits cell cycle progression beyond the G1/S phase transition, and Runx2 is again up-regulated after mitosis. Runx2 expression also correlates with metastasis and poor chemotherapy response in osteosarcoma. Here we show that six human osteosarcoma cell lines (SaOS, MG63, U2OS, HOS, G292, and 143B) have different growth rates, which is consistent with differences in the lengths of the cell cycle. Runx2 protein levels are cell cycle-regulated with respect to the G1/S phase transition in U2OS, HOS, G292, and 143B cells. In contrast, Runx2 protein levels are constitutively expressed during the cell cycle in SaOS and MG63 cells. Forced expression of Runx2 suppresses growth in all cell lines indicating that accumulation of Runx2 in excess of its pre-established levels in a given cell type triggers one or more anti-proliferative pathways in osteosarcoma cells. Thus, regulatory mechanisms controlling Runx2 expression in osteosarcoma cells must balance Runx2 protein levels to promote its putative oncogenic functions, while avoiding suppression of bone tumor growth.
Osteosarcoma is the most common bone tumor in children and adolescents (Young and Miller, 1975). The highest incidence of osteosarcoma is in the second decade of life, which suggests a relationship between bone growth and tumor development (Fraumeni, 1967; Cotterill et al., 2004). One of the critical steps for normal skeletal development and bone formation is the proliferative expansion of mesenchymal cells, osteoprogenitors, and immature osteoblasts. Cell growth and differentiation of normal osteoprogenitors and pre-osteoblasts is tightly regulated by Runx2, which favors a quiescent state (Pratap et al., 2003; Galindo et al., 2005). The growth suppressive potential of Runx2 is controlled by modulation of its protein levels during the cell cycle (Galindo et al., 2005, 2007). Cell cycle dependent changes of Runx2 levels occur with respect to G1 progression at a cell cycle stage when normal osteoblasts monitor extra-cellular cues for competency to initiate cell cycle progression beyond the G1/S phase transition. Accordingly, transient Runx2 overexpression in synchronized cells delays cell cycle entry into S phase and significantly decreases cell proliferation in the MC3T3 pre-osteoblasts, Runx2 null calvarian osteoprogenitors, C2C12 pluripotent mesenchymal, and IMR-90 fibroblasts cell lines (Pratap et al., 2003; Galindo et al., 2005; Young et al., 2007a; Teplyuk et al., 2008, 2009a). The function of Runx2 as a negative regulator of cell proliferation is also reflected by linkage of Runx2 deficiency to cell immortalization and tumorigenesis (Kilbey et al., 2007; Zaidi et al., 2007a).
Apart from the growth suppressive potential that is evident during late G1 in osteoblasts (Pratap et al., 2003; Galindo et al., 2005), Runx2 may have mitogenic potential in early G1 (Teplyuk et al., 2008). Several studies indicate that Runx2-dependent control of proliferation is cell type-specific. Runx2 inhibits proliferation of osteoprogenitors and committed osteoblasts (Pratap et al., 2003; Galindo et al., 2005), but it may have distinct biological roles in chondrocytes (Galindo et al., 2005; Hinoi et al., 2006; Komori, 2008) and endothelial cells (Inman and Shore, 2003; Qiao et al., 2006). While immature osteoblasts from mice with Runx2 null mutations show accelerated proliferative potential, chondrocyte proliferation seems to be decreased in Runx2 null mice (Pratap et al., 2003; Yoshida et al., 2004), suggesting that Runx2 would also have opposites roles in different bone cell types. Moreover, ectopic expression of Runx2 in aortic endothelial cells increases cell proliferation (Sun et al., 2004), whereas Runx2 depletion inhibits cell proliferation in human marrow endothelial cells (Qiao et al., 2006). These findings support the concept that Runx2 protein can function as either a bona fide tumor suppressor or a classical oncoprotein depending on the cellular context (Blyth et al., 2005).
Current evidence indicates that Runx2 expression is a key pathological factor in osteosarcoma (Martin et al., 2011) by controlling a number of cancer-related genes (van der Deen et al., 2012). Moreover, osteosarcoma development may be associated with Runx2 overexpression and defects in osteogenic differentiation (Wagner et al., 2011). Over-expression of Runx2 in transgenic mice within the osteoblast lineage inhibits osteoblast maturation, increases bone resorption, and causes osteopenia with multiple fractures (Liu et al., 2001; Geoffroy et al., 2002). Runx2 is also clearly detected in clinical osteosarcoma samples (Andela et al., 2005; Lu et al., 2008; Sadikovic et al., 2009; Won et al., 2009; Kurek et al., 2010). Analysis of genomic DNA from osteosarcoma patients with amplication of the 6p12#x02013;p21 chromosomal interval, which spans the Runx2 locus, increases the Runx2 gene copy number and aberrantly elevates Runx2 expression (Lau et al., 2004; Lu et al., 2008; Sadikovic et al., 2009). Increased expression of Runx2 in osteosarcoma biopsies has been associated to increased tumorigenicity, tumor progression, metastases, lower survival, and poor prognosis (Won et al., 2009; Kurek et al., 2010; Sadikovic et al., 2010). Interestingly, osteosarcoma cell culture models may exhibit a similar variability of Runx2 gene expression, because Runx2 is expressed at different levels in a number of human osteosarcoma cell lines (Thomas et al., 2004; Lu et al., 2008; Luo et al., 2008; Kurek et al., 2010; Shapovalov et al., 2010). A subset of patient-derived osteosarcoma cell lines exhibit high levels of Runx2, whereas others show decreased Runx2 expression in accordance with the findings of Thomas and colleagues who suggested that Runx2 protein levels are negatively regulated in some types of osteosarcoma (Thomas et al., 2004; Nathan et al., 2009; Pereira et al., 2009; San Martin et al., 2009; Won et al., 2009; Kurek et al., 2010; Sadikovic et al., 2010).
Recently, we presented data indicating that cell cycle control of Runx2, which is readily observed in osteoblasts, is deregulated in osteosarcoma cells (Galindo et al., 2005; San Martin et al., 2009). Runx2 is constitutively expressed throughout the cell cycle in at least two osteosarcoma (human SaOS and rat ROS) cell lines (Young et al., 2007b; San Martin et al., 2009). Hence, the transcriptional and post-transcriptional mechanisms that mediate cell cycle control of Runx2 gene expression in osteoblasts could be compromised in osteosarcoma cells. The latter may occur in conjunction with abrogation of other molecular mechanisms cells that mediate normal osteoblast proliferation and that may bypass the growth suppressive properties of Runx2 in bone cancer cells (Nathan et al., 2009). In this article, we systematically examined human osteosarcoma cell lines with respect to Runx2 gene expression and cell cycle regulation to understand the biological functions of Runx2 in osteosarcoma cell proliferation. Our main finding is that forced expression of Runx2 suppresses growth in all cell lines, indicating that stimulation of Runx2 beyond its preestablished levels in osteosarcoma cells remains capable of triggering an anti-proliferative response. We propose that osteosarcoma cells in which Runx2 is present must balance prooncogenic functions of Runx2 with the requirement to maintain Runx2 at levels that avoid tumor suppression.
Materials and Methods
Cell culture
Human osteosarcoma cell lines were maintained in culture medium (Invitrogen) supplemented with 10–15% fetal bovine serum (FBS) plus 2 mM l-glutamine and a penicillin–streptomycin cocktail at 37°C and 5% CO2 according to ATCC recomendations. SaOS cells were cultured in McCoy’s medium supplemented with 15% FBS. U2OS and G292 cells were cultured in McCoy’s medium with 10% FBS. MG-63 and HOS cells were grown in DMEM medium with 10% FBS. 143B cells were maintained in DMEM medium, 1 mM sodium piruvate, 100 µg/ml of bromodeoxiuridine and 10% FBS.
Cell growth analysis
Cells were plated in six-well plates (8 × 104 cells/well) and grown in supplemented medium at 37°C and 5% CO2. Alternatively, after 24 h, cells were infected with Adenovirus Runx2 or Adenovirus Vector as indicated below, and cultured for 6 days. The growth medium was changed every 2 days. Cell numbers were daily counted to determine growth curves.
Cell synchronization
Cells were seeded in 100-mm plates at 0.5 × 106 cells/plate and grown up to 60% of confluence. After that, cell cultures were treated for 24 h with 400 µM mimosine (Sigma–Aldrich, St. Louis, MO) to arrest cells in G1 phase (Galindo et al., 2005). Cells arrested in G1 were released by three washes in serum-free medium and stimulated to progress to S phase by the addition of fresh medium without drug containing FBS plus 2 mM l-glutamine and antibiotics (San Martin et al., 2009). After serum stimulation, cells were harvested at selected time points for Western blot and RT-PCR, as well as fluorescence-activated cell sorting (FACS) analysis.
Flow cytometric analysis
Distribution of cells at specific cell cycle stages was evaluated by assessment of DNA content by flow cytometry as previously described (Teplyuk et al., 2008). Cells were trypsinized, washed with phosphate-buffered saline (PBS), and fixed in 70% ethanol at −20°C overnight. Cells were then treated with RNase A (10 µg/ ml) at 37°C for 15 min. Subsequently, cells were stained with propidium iodide and subjected to FACS analysis based on DNA content. Samples (1 × 106 cells) were analyzed using the FACStar cell sorter and Consort 30 software (Becton–Dickinson, San Jose, CA).
Western blot analysis
Runx2, cyclin D, and β-actin were analyzed by Western blot analysis as described previously (Galindo et al., 2005). Briefly, equal amounts of total cellular proteins collected in the presence of the proteasome inhibitor MG132 (Calbiochem) and Complete® cocktail of protease inhibitor (Roche) were resolved in 10% SDS–PAGE and transferred to polyvinylidene difluoride membranes (Amersham Life Science, Buckinghamshire, UK). Blots were incubated with a 1:2,000 dilution of each primary antibody for 1 h. Runx2-specific mouse monoclonal antibody was the generous gift of Dr. Yoshiaki Ito (Cancer Science Institute, Singapore). Mouse monoclonal antibody (cyclin D1) and goat polyclonal antibody (actin) were acquired commercially (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.) for 1 h. Immunoreactive protein bands were visualized on a BioMax Light film (Kodak, Rochester, NY) using a chemiluminescence detection kit (PerkinElmer Life Sciences, Waltham, MA), and signal intensities were quantified by densitometry.
cDNA synthesis and PCR
Total RNA was isolated from osteosarcoma cells using TRIzol reagents (Invitrogen) according to the manufacturer’s specifications. Total RNA was separated in a 1% agarose-formaldehyde gel. Ethidium bromide staining of the gels was used to assess RNA quality of samples. Purified RNA was treated with RQ1RNase-Free DNase (Promega Corporation, Madison, WI) and subject to reverse transcription using random hexamer primers (Invitrogen Life Technologies, Carlsbad, CA) with M-MLV reverse transcriptase (Promega Corporation) according to the manufacturer’s recommendations. cDNA was amplified by PCR using PCR buffer 1 × (Promega), 0.2 mM dNTPs (Promega), 1.5 mM MgCL (Promega), 0.06 U/µl of Taq polymerase (Promega) with the following set of specific primers to human gene: Runx2 (forward primer: 5′-CCAGATGGGACTGTGGTTACC-3′, reverse primer: 5′-ACTT GGTGCAGAGTTCAGGG-3′), cyclin A (forward primer: 5′-GAAGACGAGACGGGTTGCAC-3′, reverse primer: 5′-GCAGTGCCCACAAGCTGAAG-3′), cyclin B (forward primer: 5′-GCTCCGAGT CACCAGGAACT-3′, reverse primer: 5′-TCCATTGGGCTTGGAGAGGC-3′), cyclin D1 (forward primer: 5′-CAGAAGAGCGCGAGGGAGCG-3′, reverse primer: 5′-CTTCTCGGCCGTCAGGGGG A-3′), cyclin E (forward primer: 5′-GGACAAGACCCTGGCCTCAG-3′, reverse primer: 5′-TCAGG TGTGGGGATCAGGGA-3′), and GADPH (forward primer: 5′-CCTTCATTGACCTCAACTA-3′, reverse primer: 5′-GGCCATCCACAGTCTTCT-3′). Aliquots of the resulting product (5 µl) were visualized in 1% agarose gels by ethidium bromide staining.
Adenovirus infections
Adenoviral delivery of vector containing cDNA of Runx2-IRES-GFP under the control of the CMV5 promoter was used as previously described (Pratap et al., 2003). Preparation and purification of virus were performed according to the manufacturer’s protocols (Promega). For control of infection, the same Adenovirus vector carrying GFP was used. The Adenovirus Runx2 (Adv-Runx2) contains both the GFP cassette and the Runx2 cDNA in forward orientation (+) and Adenovirus Vector (Adv-Vector) contains the GFP cassette in the forward orientation (+) and the Runx2 cDNA in reverse orientation (−) (Pratap et al., 2003).
Cells were plated for infections in 60 mm plates at a density of 30 × 104 cells/plate and cultured in DMEM with 10% FBS. After 24 h, cells were infected at 60–70% of confluence with 30 × 1010 OPU/ml (optical particle unit) of each virus in 900 µl of DMEM suplemented with 1% FBS for 4 h. Upon addition of 600 µl of media containing 1% FBS, cells were incubated for an additional 10 h. After adenoviral infection, cells were grown for 24–72 h. Infection efficiencies were assessed by expression analysis of a green fluorescent protein (GFP) under the control of an IRES signal, and images of GFP-expressing cells were taken using a fluorescence microscope with a CCD camera.
Results
Analysis of Runx2 expression and cell proliferation potential in human osteosarcoma cell lines
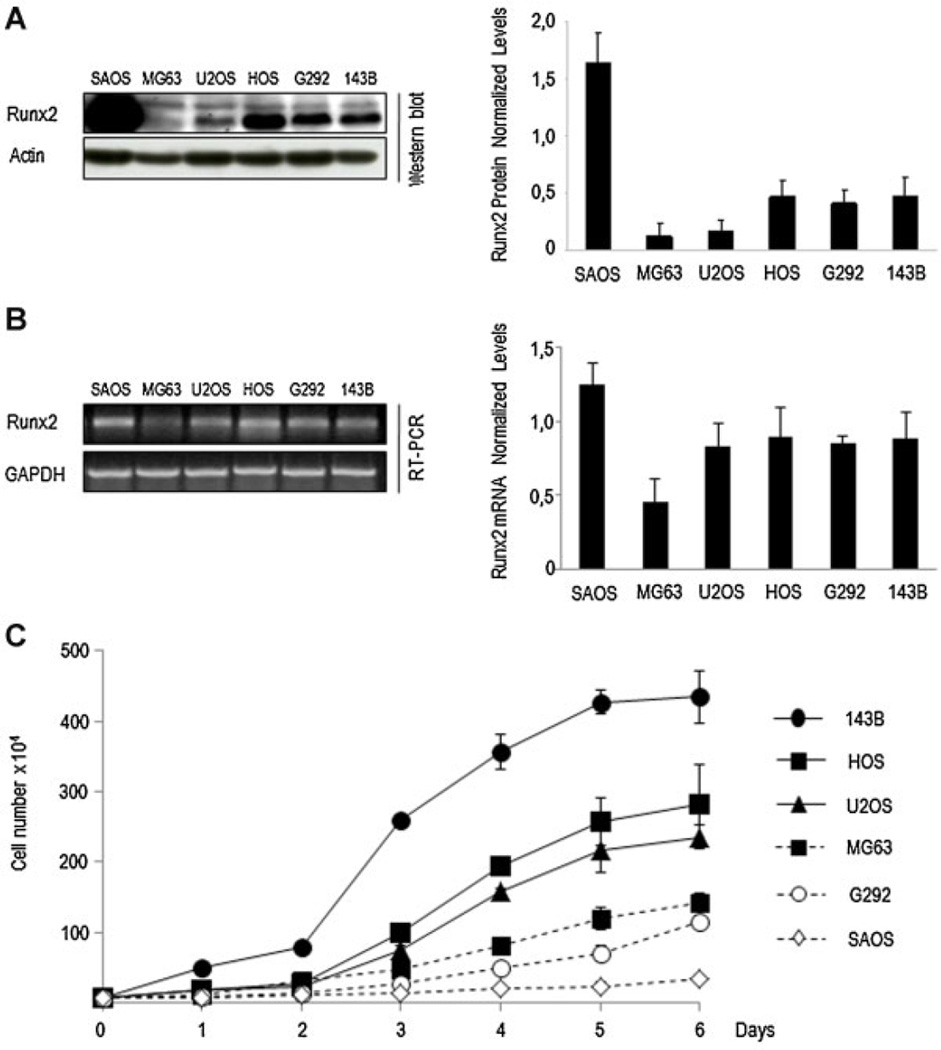
To better understand Runx2 function in osteosarcoma cell proliferation, we first examined Runx2 protein levels in six human osteosarcoma cell lines (SaOS, MG63, U2OS, HOS, G292, and 143B) (Fig. 1A). Western blot analysis revealed that SaOS cells exhibit the highest Runx2 protein levels compared to the other five osteosarcoma cell lines. Runx2 protein is also detected in HOS, G292, and 143B cells, but it is expressed at significantly lower levels than in SaOS cells. U2OS cells exhibit significantly lower Runx2 protein expression, and Runx2 protein expression is barely detected in MG63. Consistently, relatively high Runx2 mRNA levels are detected by RT-PCR in SaOS cells (Fig. 1B). Runx2 mRNA levels are also detected in U2OS, HOS, G292 and 143B cells, but Runx2 mRNA is only weakly detected in MG63 (Fig. 1B).
Fig. 1.
Runx2 expression and growth profile in human osteosarcoma cell lines. Runx2 expression was assessed in SaOS-2, G292, MG63, U2OS, HOS, and 143B osteosarcoma cells. Runx2 protein (A, right part) and mRNA (B, right part) levels were evaluated by Western blot analysis and RT-PCR, respectively (A). Runx2 protein and mRNA values were normalized to actin (A, left part) and GAPDH (B, left part), respectively. Proliferation was monitored by determining cell number at the indicated days (C). All data are presented as mean ± SEM.
To establish a correlation between cell proliferation and Runx2 expression in osteosarcoma, we assessed cell growth profiles. Cell growth was monitored daily by cell counting during a period of 6 days (Fig. 1C). SaOS cells display very slow growth rates compared to the other cell lines. MG63 and G292 cells exhibit moderate growth rates, U2OS and HOS cells have intermediate growth rates, while 143B cells have higher growth rates. Interestingly, SaOS cells show high Runx2 levels and lower growth rates, suggesting that presence of Runx2 may negatively correlated with cell proliferation in osteosarcoma cells.
Characterization of cell cycle profiles in human osteosarcoma cell lines
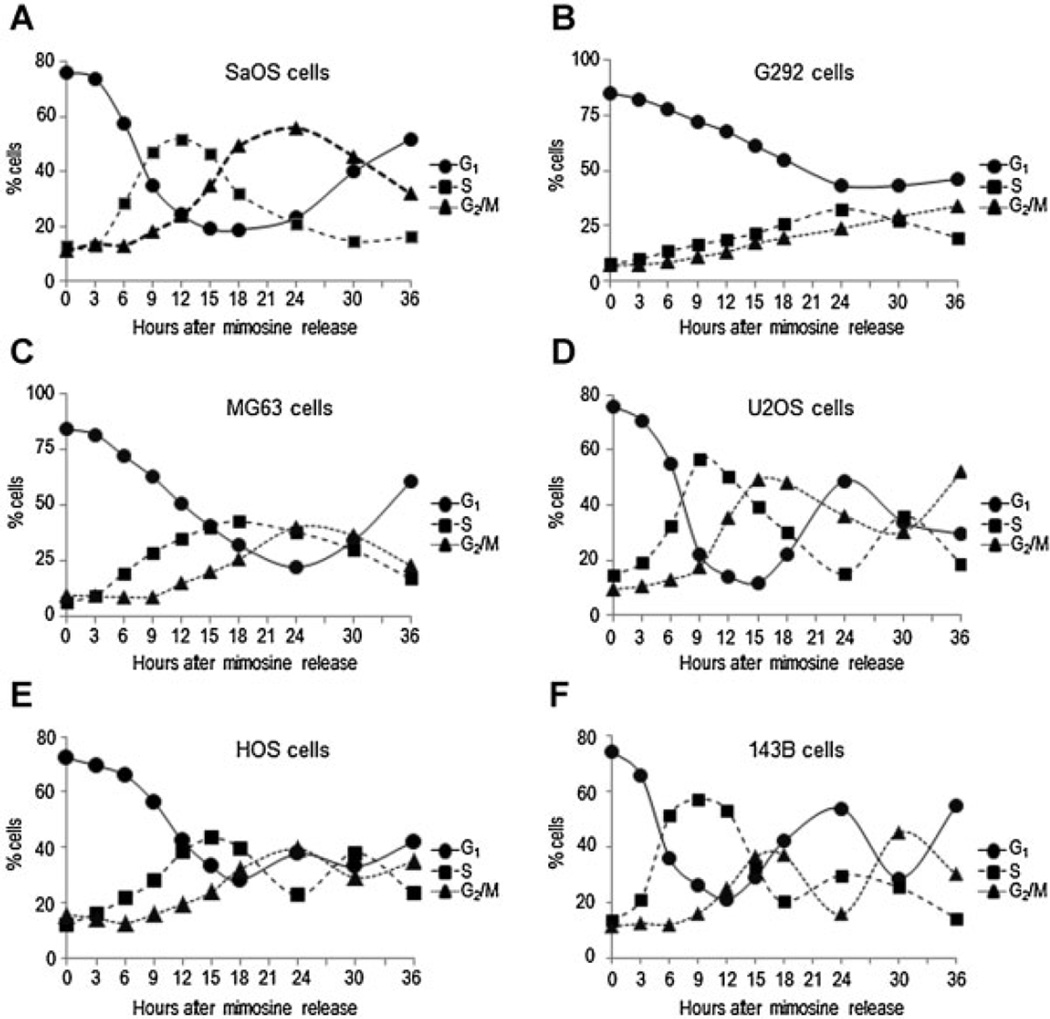
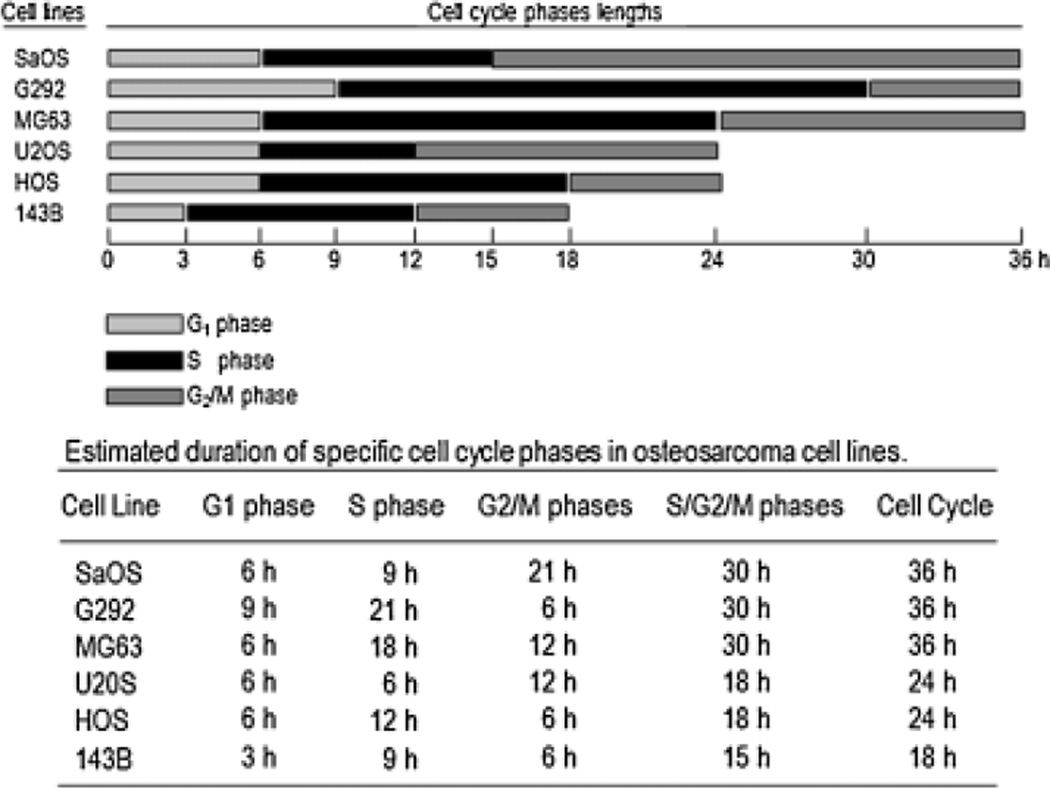
We characterized the temporal profiles of progression in different phases of the cell cycle (G1, S, and G2/M) in synchronized osteosarcoma cells by assessment of DNA content and cell cycle markers (cyclin D, E, A, and B) using flow cytometry as well as RT-PCR and Western blot analysis, respectively (Fig. 2). We estimated the cell cycle length and length of specifics cell cycle phases based on the percentage of cells distributed in G1, S and G2/M at different time points after stimulation of proliferation in mimosine-arrested cells (Figs. 3 and 4). Thus, the apparent duration of the cell cycle is 36 h for SaOS, G292, and MG63 cells; 24 for U2OS and HOS cells; and 18 h for 143B cells, approximately.
Fig. 2.
Human osteosarcoma cell lines exhibit different cell cycle profiles. SaOS-2, G292, MG63, U2OS, HOS, and 143B osteosarcoma cells were synchronized by incubation for 24 h with mimosine to generate a G1 phase block. Then, cells were released from G1 phase arrest by the addition of fresh culture medium without mimosine and harvested after 0, 3, 6, 9, 12, 15, 18, 24, 30, and 36 h (A–F). Progression through successive cell cycle phases (G1, S, and G2/M) was monitored by flow cytometry (top part). Expression of cell cycle markers cyclins A, B, D, and E mRNA (middle part) and cyclin D1 protein (bottom part) were analyzed by Western blot and RT-PCR, respectively.
Fig. 3.
Human osteosarcoma cell lines show different patterns of progression through the cell cycle. Percentage of osteosarcoma cells in different cell cycle phases (G1, S and G2/M) were obtained from data shown in Figure 2. Graphic representation of progression through successive cell cycle phases are showed for each osteosarcoma cell line (A–F).
Fig. 4.
Human osteosarcoma cell lines show different cell cycle lengths. Cell cycle lengths and lengths of specifics cell cycle phases (G1, S, and G2/M) were determined from data shown in Figure 3. Graphic representation of duration of specifics cell cycle phases are showed for each osteosarcoma cell line (A–F). The bottom portion shows the estimated duration of specific stages during the cell cycle in hours (h).
A detailed analysis showed that cell cycle progression was slower in SaOS, G292, and MG63 cells compared to 143B, indicating a dynamic differences in cell cycle kinetics between these cell lines (Figs. 3 and 4). The distinctions in cell cycle lengths observed in SaOS, G292, and MG63 cells is corroborated by a significant change in the lengths of G1 (from 3 to 6 h in SaOS and MG63, and from 3 to 9 h in G292), S (from 9 to 18 h in MG63, and from 9 to 21 h in G292), and G2/M (from 6 to 12 h in MG63, and from 6 to 21 h in SaOS) as compared to 143B (Fig. 4). We also observed a progressive reduction in the lengths of specific cell cycle phases in U2OS cells (S: 21 to 6 h and G2/M: 21 to 12 h) and HOS (S: 21 to 12 h and G2/M: 21 to 6 h), as compared to G292 and SaOS (Fig. 4). Based on these results, we conclude that changes in cell cycle kinetics are related to major changes in the length of specific cell cycle phases, including extension of the S and G2/M phases (Fig. 4). Interestingly, the estimated cell cycle lengths and kinetics of progression in different phases are consistent with the growth rates observed in each of the osteosarcoma cell lines (Figs. 1C and 4).
Cell cycle regulation of Runx2 expression in osteosarcoma
We examined Runx2 expression during cell cycle progression in synchronized osteosarcoma cells (143B, G292, HOS, U2OS, MG63, and SaOS) at multiple time points after stimulation of proliferation (Fig. 5). Runx2 protein levels decrease during G1/S phase transition in 143B, G292, HOS, and U2OS (Fig. 6B, D–F). Thus, cell cycle dependent changes in Runx2 protein expression observed in normal pre-osteoblasts cells synchronized in the G1 phase (Galindo et al., 2005) is also evident in 143B, G292, HOS, and U2OS osteosarcoma cells. However, we observed that Runx2 protein levels are rapidly up regulated at the end of S phase in U2OS cells. Furthermore, Runx2 protein levels are retained after mitosis during early G1. In contrast, Runx2 is constitutively expressed throughout the cell cycle (and beyond the G1 phase) in SaOS and MG63 cells (Figs. 5A and C and 6A and C). Yet, Runx2 mRNA levels do not exhibit any major fluctuations during the cell cycle in these osteosarcoma cell lines (Fig. 5). These data together indicate that Runx2 protein expression is modulated during the cell cycle in a sub-set of osteosarcoma-derived cells.
Fig. 5.
Runx2 expression is differentially regulated during the cell cycle in different human osteosarcoma cell lines. Runx2 gene expression was assessed during progression through the cell cycle to determine the specific transition stages when Runx2 levels are modulated in SaOS-2, G292, MG63, U2OS, HOS, and 143B osteosarcoma cell lines (A–F). Cells were synchronized at the G1 phase and stimulated to progress through the cell cycle as described in Figure 2. Cells were harvested after 0, 3, 6, 9, 12, 15, 18, 24, 30 and 36 h stimulation. This set of samples was the same that were used to asses cell cycle markers showed in Figure 2. Cell cycle-dependent modulations in Runx2 protein and mRNA levels were evaluated by Western blot analysis and RT-PCR. Actin and GAPDH showed in Figure 2 were also used as a loading control for Runx2 protein and mRNA, respectively. Cell cycle phases as determined by flow cytometry are indicated at the base of the parts.
Fig. 6.
Runx2 protein levels are differentially modulates during the cell cycle in human osteosarcoma cell lines. Runx2 expression in osteosarcoma cells were quantified from data shown in Figure 5. Cell cycle-related changes in Runx2 protein levels after serum stimulation were depicted in a graphic representation (A–F). Protein values were normalized to actin. Cell cycle phases as determined by flow cytometry are indicated at the base of the graphs.
Runx2 suppresses cell proliferation in osteosarcoma
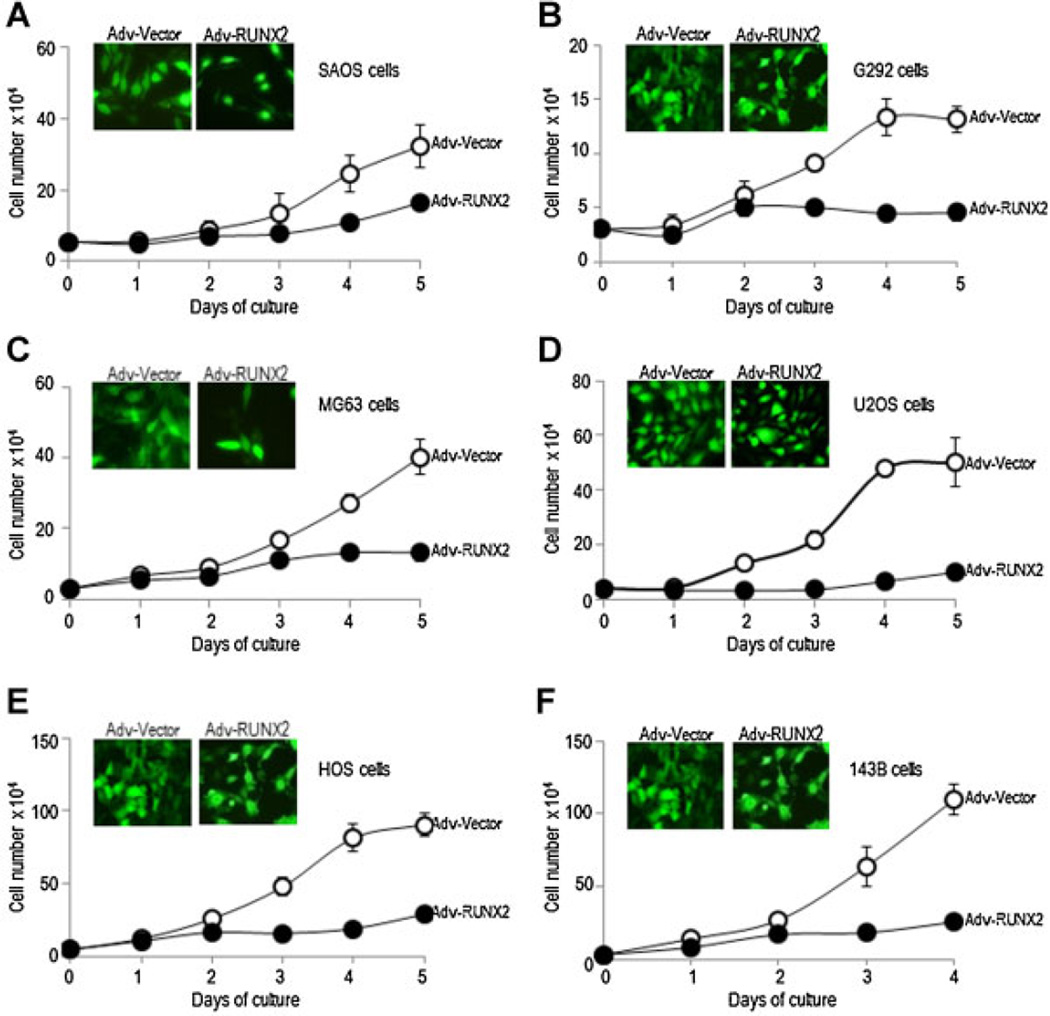
We conducted a systematic study on the function of Runx2 gene expression in osteosarcoma cell lines to define the role of Runx2 in osteosarcoma cell proliferation. We achieved 95% infection efficiency for adenoviral vectors based on fluorescence detection of co-expressed GFP proteins, as assessed by fluorescence microscopy (Fig. 7). As expected, exogenous expression of Runx2 into any of the osteosarcoma cell lines inhibits cell proliferation as reflected by a prominent decrease in the slope of the growth curve when compared with control cells expressing GFP alone (Fig. 7).
Fig. 7.
Runx2 modulates cell proliferation in human osteosarcoma cell lines. SaOS-2, G292, MG63, U2OS, HOS, and 143B osteosarcoma cells were infected with adenoviral vectors expressing Runx2 plus GFP (Adv-Runx2, closed circles) or GFP alone (Adv-Vector, open circles) at comparable efficiencies of infection. After transduction, cell growth was monitored at the indicated days. Growth curves for each osteosarcoma cell lines were obtained by cell counting at daily intervals until 5 days of culture after infection (A–F). Day 0 is the day of infection. Microscopic analysis of GFP expression shows efficient transduction (up to 90%) for each cell line (insert parts, cells infected with Adv-Runx2 or Adv-Vector). All data are presented as mean ± SEM.
Discussion
In osteoblast cells, Runx2 protein levels are dramatically upregulated in quiescence (G0) or proliferative arrest induced by serum deprivation or by contact inhibition. In contrast, Runx2 protein decreases to minimal levels when the cells are stimulated to proliferate, consistent with its function as a negative regulator of cell proliferation (Pratap et al., 2003; Galindo et al., 2005, 2007). More specifically, Runx2 protein levels decreases at G1/S transition and remains low during G2 and M phases, regaining higher levels postmitotically in early G1 (Galindo et al., 2005, 2007). Thus, in normal osteoblastic cells (e.g., MC3T3), Runx2 protein levels are modulated at three different cell cycle stages (i.e., the G1/G0, G1/S, and M/G1 transitions) (Galindo et al., 2005).
Previously, we showed that Runx2 is constitutively expressed at high levels throughout the cell cycle in osteosarcoma cells (i.e., human SaOS and rat ROS) (Young et al., 2007b; San Martin et al., 2009). However, these studies included only a single human cell line. Our current results advance our understanding of Runx2 expression during the cell cycle in a panel of six osteosarcoma cell lines. In osteosarcoma, Runx2 protein levels are down-regulated at the G1/S phase transition in 143B, G292, HOS, and U2OS cell lines and it is constitutively expressed through the cell cycle only in MG63 and SaOS cells. Down-regulation of Runx2 in late G1 (G1/S transition) in osteosarcoma may be mediated by CDK-related and ubiquitin/proteasome dependent protein degradation as suggested previously (Galindo et al., 2005; Shen et al., 2006; Rajgopal et al., 2007). Moreover, the proteasome inhibitor bortezomib suppresses growth and increases Runx2 protein levels in osteosarcoma cell lines (Shapovalov et al., 2010). The latter finding may be linked to our demonstration that stabilization of Runx2 protein levels by the proteasomal inhibitor MG132 is restricted to a stage in late G1 in osteosarcoma cells (San Martin et al., 2009). Runx2 gene expression during the cell cycle is also controlled at least in part at the transcriptional level, because Runx2 mRNA levels are also down-regulated in pre-osteoblastic cells progressing towards the G1/S transition (Galindo et al., 2005). However, we do not observed major variations in Runx2 mRNA levels during progression to S phase in osteosarcoma cells, which suggest that protein destabilization by the proteasome pathway could be one of the principal mechanisms that modulate Runx2 expression during cell cycle progression in osteosarcoma cells.
We have previously shown that Runx2 deficiency increases the proliferative potential of osteoprogenitors and that reintroduction of wild type Runx2 into primary calvarian cells restores stringent cell growth control (Pratap et al., 2003). Also, ectopic over-expression of Runx2 inhibits proliferation of osteoprogenitors and committed osteoblasts (Galindo et al., 2005; Young et al., 2007a; Teplyuk et al., 2008). Interestingly, exogenous Runx2 suppresses growth of G292 osteosarcoma cells, but other osteosarcoma cell lines, such as SaOS and U2OS, appears to be refractory to Runx2 anti-proliferative functions (Thomas et al., 2004). Our present results suggest that Runx2 may retain its ability to control osteosarcoma cell growth, based on the observation that Runx2 suppresses growth of all osteosarcoma cells when over-expressed. This latter finding complements studies using an osteosarcoma xenograft mouse model in which BMP-promoted tumor growth in MG63 cells is inhibited by exogenous Runx2 expression (Luo et al., 2008).
It has been suggested that lack of functional pRB may account for the inability of Runx2 to attenuate cell proliferation in osteosarcoma (Thomas et al., 2004). However, our results may indicate that Runx2 growth control in osteosarcoma involves other molecular pathway linked to the cell cycle. The growth regulatory functions of Runx2 are mediated in part through epigenetic mechanisms and the ability of Runx2 to regulate the expression of cell growth related genes that promote a nonproliferative state (Pratap et al., 2003; Galindo et al., 2005; Kilbey et al., 2007; Young et al., 2007a,b; Zaidi et al., 2007b, 2010; Teplyuk et al., 2008, 2009a,b). Runx2 target genes linked to cell cycle may contribute to the control of osteosarcoma tumor growth (Manara et al., 2006; Young et al., 2007a,b; Sadikovic et al., 2009; van der Deen et al., 2012).
From a broader perspective, Runx1, Runx2, and Runx3 transcription factors are specific master regulators for hematopoiesis, bone and gastric differentiation, respectively (Stein et al., 2010). Inactivating mutations or epigenetic silencing of Runx1 and Runx3 are associated to leukemia and gastric cancer, respectively (Song et al., 1999; Li et al., 2002; Cameron and Neil, 2004). However, Runx2 mutations have been related mainly to skeletal defects but not to bone cancer, suggesting that this factor is not inactivated in osteosarcoma. Previous studies have reported that Runx2 is expressed in distinct cancer types. Runx2 has been involved in tumor progression and bone metastasis in breast and prostate cancer (Pratap et al., 2005, 2006, 2008), as well as tumorigenesis in lymphomas (Blyth et al., 2005). In normal mammary epithelial cells Runx2 is expressed at low levels, but it is expressed at high levels in metastatic mammary cancer cells (Selvamurugan and Partridge, 2000; Inman and Shore, 2003; Barnes et al., 2004) and promotes bone metastatic properties of breast and prostate cancer cells (Brubaker et al., 2003; Javed et al., 2005; Pratap et al., 2005, 2006, 2008). Runx2 enhanced expression is also associated to lymphoma development where it oncogenic potential has been well established (Blyth et al., 2006). Moreover, Runx2 is strongly expressed in human malignant melanoma, thyroid papillary carcinoma, glioma, and pituitary tumor, which suggests that this cancer gene could promote tumorigenesis in a broad spectrum of tumors (Riminucci et al., 2003; Endo et al., 2008; Vladimirova et al., 2008; Zhang et al., 2009). Regarding to bone cancer, several studies have shown that Runx2 is differentially expressed in osteosarcoma patient specimens where high expression has been associated with tumor progression, poor response to chemotherapy, metastasis, and lower survival (Won et al., 2009; Kurek et al., 2010; Sadikovic et al., 2010). The findings presented here indicate unambiguously that Runx2 protein controls osteosarcoma cell proliferation when over-expressed beyond its normal pre-established levels in a given osteosarcoma cell type. Therefore, we propose that Runx2 expression is biologically and functionally linked to tumor growth control in human osteosarcoma.
Acknowledgments
We thank the members of our research groups for stimulating discussions, including Francisco Villanueva, Karina Villegas, Hector Araya, and Nelson Varela (University of Chile), as well as Nadiya Teplyuk, Margaretha van der Deen, and Hanna Taipaleenmaki (University of Massachusetts Medical School). We are also grateful to Sergio Lavanderos and Mario Chiong (Facultad de Ciencias Química y Farmacéuticas, University of Chile) for advice and assistance in adenovirus purification. This study was supported by Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT), grant number 1095234 (to M.G.) and Iniciativa Cientifico Milenio. Ministerio de Economía, Fomento y Turismo, grant number P09/016-F (to M.G.). This work was also supported by National Institutes of Health Grants R01AR049069 (to A.J.v.W.) and P01 CA082834 (to G.S.S.).
Contract grant sponsor: Iniciativa Científica Milenio. Ministerio de Economía, Fomento y Turismo;
Contract grant number: P09/016-F.
Contract grant sponsor: Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT);
Contract grant number: 1095234.
Contract grant sponsor: National Institutes of Health;
Contract grant numbers: R01AR049069, P01 CA082834.
Literature Cited
- Andela VB, Siddiqui F, Groman A, Rosier RN. An immunohistochemical analysis to evaluate an inverse correlation between Runx2/Cbfa1 and NFκB in human osteosarcoma. J Clin Pathol. 2005;58:328–330. doi: 10.1136/jcp.2004.017640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, Stein GS, Gerstenfeld LC. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases associated osteolytic disease. Cancer Res. 2004;64:4506–4513. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The RUNX genes: Gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, Jenkins A, Neil JC, Cameron ER. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 2006;66:2195–2201. doi: 10.1158/0008-5472.CAN-05-3558. [DOI] [PubMed] [Google Scholar]
- Brubaker KD, Vessella RL, Brown LG, Corey E. Prostate cancer expression of runt-domain transcription factor Runx2, a key regulator of osteoblast differentiation and function. Prostate. 2003;56:13–22. doi: 10.1002/pros.10233. [DOI] [PubMed] [Google Scholar]
- Cameron ER, Neil JC. The Runx genes: Lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23:4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- Cotterill SJ, Wright CM, Pearce MS, Craft AW. Stature of young people with malignant bone tumors. Pediatr Blood Cancer. 2004;42:59–63. doi: 10.1002/pbc.10437. [DOI] [PubMed] [Google Scholar]
- Endo T, Ohta K, Kobayashi T. Expression and function of Cbfa-1/Runx2 in thyroid papillary carcinoma cells. J Clin Endocrinol Metab. 2008;93:2409–2412. doi: 10.1210/jc.2007-2805. [DOI] [PubMed] [Google Scholar]
- Fraumeni JF., Jr Stature and malignant tumors of bone in childhood and adolescence. Cancer. 1967;20:967–973. doi: 10.1002/1097-0142(196706)20:6<967::aid-cncr2820200606>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M, Kahler RA, Teplyuk NM, Stein JL, Lian JB, Stein GS, Westendorf JJ, van Wijnen AJ. Cell cycle related modulations in Runx2 protein levels are independent of lymphocyte enhancer-binding factor 1 (Lef1) in proliferating osteoblasts. J Mol Histol. 2007;38:501–506. doi: 10.1007/s10735-007-9143-0. [DOI] [PubMed] [Google Scholar]
- Geoffroy V, Kneissel M, Fournier B, Boyde A, Matthias P. High bone resorption in adult aging transgenic mice overexpressing cbfa1/runx2 in cells of the osteoblastic lineage. Mol Cell Biol. 2002;22:6222–6233. doi: 10.1128/MCB.22.17.6222-6233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E, Bialek P, Chen YT, Rached MT, Groner Y, Behringer RR, Ornitz DM, Karsenty G. Runx2 inhibits chondrocyte proliferation and hypertrophy through its expression in the perichondrium. Genes Dev. 2006;20:2937–2942. doi: 10.1101/gad.1482906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman CK, Shore P. The osteoblast transcription factor Runx2 is expressed in mammary epithelial cells and mediates osteopontin expression. J Biol Chem. 2003;278:48684–48689. doi: 10.1074/jbc.M308001200. [DOI] [PubMed] [Google Scholar]
- Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbey A, Blyth K, Wotton S, Terry A, Jenkins A, Bell M, Hanlon L, Cameron ER, Neil JC. Runx2 disruption promotes immortalization and confers resistance to oncogene-induced senescence in primary murine fibroblasts. Cancer Res. 2007;67:11263–11271. doi: 10.1158/0008-5472.CAN-07-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and maintenance by Runx2. Front Biosci. 2008;13:898–903. doi: 10.2741/2730. [DOI] [PubMed] [Google Scholar]
- Kurek K, Del Mare S, Salah Z, Abdeen S, Sadiq H, Lee S, Gaudio E, Zanesi N, Jones KB, De Young B, Amir G, Gebhardt M, Warman M, Stein GS, Stein JL, Lian JB, Aqeilan RI. Frequent attenuation of the WWOX tumor suppressor in osteosarcoma is associated with increased tumorigenicity and aberrant RUNX2 expression. Cancer Res. 2010;70:5577–5586. doi: 10.1158/0008-5472.CAN-09-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CC, Harris CP, Lu XY, Perlaky L, Gogineni S, Chintagumpala M, Hicks J, Johnson ME, Davino NA, Huvos AG, Meyers PA, Healy JH, Gorlick R, Rao PH. Frequent amplification and rearrangement of chromosomal bands 6p12-p21 and 17p11.2 in osteosarcoma. Genes Chromosomes Cancer. 2004;39:11–21. doi: 10.1002/gcc.10291. [DOI] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Liu W, Toyosawa S, Furuichi T, Kanatani N, Yoshida C, Liu Y, Himeno M, Narai S, Yamaguchi A, Komori T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155:157–166. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XY, Lu Y, Zhao YJ, Jaeweon K, Kang J, Xiao-Nan L, Ge G, Meyer R, Perlaky L, Hicks J, Chintagumpala M, Cai WW, Ladanyi M, Gorlick R, Lau CC, Pati D, Sheldon M, Rao PH. Cell cycle regulator gene CDC5L, a potential target for 6p12-p21 amplicon in osteosarcoma. Mol Cancer Res. 2008;6:937–946. doi: 10.1158/1541-7786.MCR-07-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Chen J, Song WX, Tang Ni, Luo J, Deng ZL, Sharff KA, He G, Bi Y, He BC, Bennett E, Huang J, Kang Q, Jiang W, Su Y, Zhu GH, Yin H, He Y, Wang Y, Souris JS, Chen L, Zuo GW, Montag AG, Reid RR, Haydon RC, Luu HH, He TC. Osteogenic BMPs promote tumor growth of human osteosarcomas that harbor differentiation defects. Lab Invest. 2008;88:1264–1277. doi: 10.1038/labinvest.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manara MC, Bernard G, Lollini PL, Nanni P, Zuntini M, Landuzzi L, Benini S, Lattanzi G, Sciandra M, Serra M, Colombo MP, Bernard A, Picci P, Scotlandi K. CD99 acts as an oncosuppressor in osteosarcoma. Mol Biol Cell. 2006;17:1910–1921. doi: 10.1091/mbc.E05-10-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JW, Yoshimoto M, Ludkovski O, Thorner PS, Zielenska M, Squire JA, Nuin PA. Analysis of segmental duplications, mouse genome synteny and recurrent cancer-associated amplicons in human chromosome 6p21-p12. Cytogenet Genome Res. 2010;128:199–213. doi: 10.1159/000308353. [DOI] [PubMed] [Google Scholar]
- Martin JW, Zielenska M, Stein GS, van Wijnen AJ, Squire JA. The role of RUNX2 in osteosarcoma oncogenesis. Sarcoma 2011. 2011:13. doi: 10.1155/2011/282745. Article ID 282745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan SS, Pereira BP, Zhou YF, Gupta A, Dombrowski C, Soong R, Pho RW, Stein GS, Salto-Tellez M, Cool SM, van Wijnen AJ. Elevated expression of Runx2 as a key parameter in the etiology of osteosarcoma. Mol Biol Rep. 2009;36:153–158. doi: 10.1007/s11033-008-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira BP, Zhou Y, Gupta A, Leong DT, Aung KZ, Ling L, Pho RW, Galindo M, Salto-Tellez M, Stein GS, Cool SM, van Wijnen AJ, Nathan SS. Runx2, p53, and pRB status as diagnostic parameters for deregulation of osteoblast growth and differentiation in a new pre-chemotherapeutic osteosarcoma cell line (OS1) J Cell Physiol. 2009;221:778–788. doi: 10.1002/jcp.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Galindo M, Zaidi SK, Vraidii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of pre-osteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25:589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M, Shapiro P, Fosbrink M, Rus H, Kumar R, Passaniti A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281:7118–7128. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- Rajgopal A, Young DW, Mujeeb KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J Cell Biochem. 2007;100:1509–1517. doi: 10.1002/jcb.21137. [DOI] [PubMed] [Google Scholar]
- Riminucci M, Corsi A, Peris K, Fisher LW, Chimenti S, Bianco P. Coexpression of bone sialoprotein (BSP) and the pivotal transcriptional regulator of osteogenesis, Cbfa1/Runx2, in Malignant Melanoma. Calcif Tissue Int. 2003;73:281–289. doi: 10.1007/s00223-002-2134-y. [DOI] [PubMed] [Google Scholar]
- Sadikovic B, Yoshimoto M, Chilton-MacNeill S, Thorner P, Squire JA, Zielenska M. Identification of interactive networks of gene expression associated with osteosarcoma oncogenesis by integrated molecular profiling. Hum Mol Genet. 2009;18:1962–1975. doi: 10.1093/hmg/ddp117. [DOI] [PubMed] [Google Scholar]
- Sadikovic B, Thorner P, Chilton-MacNeill S, Martin JW, Cervigne NK, Squire J, Zielenska M. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer. 2010;10:202. doi: 10.1186/1471-2407-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin IA, Varela N, Gaete M, Villegas K, Osorio M, Tapia JC, Antonelli J, Mancilla E, Pereira BP, Nathan SS, Lian JB, Stein JL, Stein GS, van Wijnen AJ, Galindo M. Impaired cell cycle regulation of the osteoblast-related heterodimeric transcription factor Runx2-Cbfbeta in osteosarcoma cells. J Cell Physiol. 2009;221:560–571. doi: 10.1002/jcp.21894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamurugan N, Partridge NC. Constitutive expression and regulation of collagenase-3 in human breast cancer cells. Mol Cell Biol Res Commun. 2000;3:218–223. doi: 10.1006/mcbr.2000.0215. [DOI] [PubMed] [Google Scholar]
- Shapovalov Y, Benavidez D, Zuch D, Eliseev RA. Proteasome inhibition with bortezomib suppresses growth and induces apoptosis in osteosarcoma. Int J Cancer. 2010;127:67–76. doi: 10.1002/ijc.25024. [DOI] [PubMed] [Google Scholar]
- Shen R, Wang X, Drissi H, Liu F, O’Keefe RJ, Chen D. Cyclin D1-Cdk4 induce Runx2 ubiquitination and degradation. J Biol Chem. 2006;281:16347–16353. doi: 10.1074/jbc.M603439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- Stein GS, Stein JL, van Wijnen AJ, Lian JB, Montecino M, Croce CM, Choi JY, Ali SA, Pande S, Hassan MQ, Zaidi SK, Young DW. Transcription factor-mediated epigenetic regulation of cell growth and phenotype for biological control and cancer. Adv Enzyme Regul. 2010;50:160–167. doi: 10.1016/j.advenzreg.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Vitolo MI, Qiao M, Anglin IE, Passaniti A. Regulation pof TGFb1-mediated growth inhibition and apoptosis by Runx2 isoforms in endothelial cells. Oncogene. 2004;23:4722–4734. doi: 10.1038/sj.onc.1207589. [DOI] [PubMed] [Google Scholar]
- Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Runx2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J Biol Chem. 2008;283:27585–27597. doi: 10.1074/jbc.M802453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyuk NM, Zhang Y, Lou Y, Hawse JR, Hassan MQ, Teplyuk VI, Pratap J, Galindo M, Stein JL, Stein GS, Lian JB, van Wijnen AJ. The osteogenic transcription factor Runx2 controls genes involved in sterol/steroid metabolism, including Cyp11a1 in osteoblasts. Mol Endocrinol. 2009a;23:849–861. doi: 10.1210/me.2008-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyuk NM, Haupt LM, Ling L, Dombrowski C, Mun FK, Nathan SS, Lian JB, Stein JL, Stein GS, Cool SM, van Wijnen AJ. The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J Cell Biochem. 2009b;107:144–154. doi: 10.1002/jcb.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Johnson S, Sims N, Trivett M, Slavin J, Rubin B, Waring P, McArthur G, Walkley C, Holloway A, Diyagama D, Grim J, Clurman B, Bowtell D, Lee J, Gutierrez G, Piscopo D, Carty S, Hinds P. Terminal osteoblast differentiation, mediated by Runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Deen M, Akech J, Lapointe D, Gupta S, Young DW, Montecino MA, Galindo M, Lian JB, Stein JL, Stein GS, van Wijnen AJ. Genomic promoter occupancy of runt-related transcription factor RUNX2 in osteosarcoma cells identifies genes involved in cell adhesion and motility. J Biol Chem. 2012;287:4503–4517. doi: 10.1074/jbc.M111.287771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirova V, Waha A, Luckerath K, Pesheva P, Probstmeier R. Runx2 is expressed in human glioma cells and mediates the expression of galectin3. J Neurosci Res. 2008;86:2450–2461. doi: 10.1002/jnr.21686. [DOI] [PubMed] [Google Scholar]
- Wagner ER, Luther G, Zhu G, Luo Q, Shi Q, Kim SH, Gao JL, Huang E, Gao Y, Yang K, Wang L, Teven C, Luo X, Liu X, Li M, Hu N, Su Y, Bi Y, He BC, Tang N, Luo J, Chen L, Zuo G, Rames R, Haydon RC, Luu HH, He TC. Defective osteogenic differentiation in the development of osteosarcoma. Sarcoma 2011. 2011:12. doi: 10.1155/2011/325238. Article ID 325238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won KY, Park HR, Park YK. Prognostic implication of immunohistochemical Runx2 expression in osteosarcoma. Tumori. 2009;95:311–316. doi: 10.1177/030089160909500307. [DOI] [PubMed] [Google Scholar]
- Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T. Runx2 and Runx3 are essencial for chondrocyte maduration, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JL, Jr, Miller RW. Incidence of malignant tumors in US children. J Pediatr. 1975;86:254–258. doi: 10.1016/s0022-3476(75)80484-7. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Pande S, Pratap J, Gaur T, Grigoriu S, Ali SA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci USA. 2007a;104:19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007b;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic book marking of genes: A novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Jin L, Stilling GA, Ruebel KH, Coonse K, Tanizaki Y, Raz A, Lloyd RV. RUNX1 and RUNX2 upregulate Galectin-3 expression in human pituitary tumors. Endocrine. 2009;35:101–111. doi: 10.1007/s12020-008-9129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]