Abstract
Background
The theory that short telomere length and genetic defects in maintaining telomere length are associated with familial nonmedullary thyroid cancer (FNMTC) is controversial. Thus, the aim of this study was to determine whether telomere length and genes involved in maintaining telomere length are altered in FNMTC.
Methods
Blood samples were collected from 44 members (13 affected and 31 unaffected) of six families with FNMTC and from 60 controls. Quantitative polymerase chain reaction (Q-PCR) and reverse transcription PCR were performed to analyze relative telomere length (RTL), gene copy number, and mRNA expression of telomerase reverse transcriptase (hTERT), telomere repeat binding factor 1 (TRF1), telomere repeat binding factor 2 (TRF2), repressor activator protein 1 (RAP1), TRF1 interacting nuclear factor 2 (TIN2), tripeptidyl peptidase 1 (TPP1), and protection of telomere 1 (POT1).
Results
Affected members had shorter RTL, as compared with unaffected members (0.98 vs. 1.23, p<0.01). There was no significant difference in hTERT, TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 gene copy number or mRNA expression between affected and unaffected members.
Conclusions
RTL is shorter in affected members with FNMTC but is not associated with altered copy number or expression in hTERT, TRF1, TRF2, RAP1, TIN2, TPP1, and POT1. The small differences in RTL preclude the utility of RTL as a marker for FNMTC in at-risk individuals.
Introduction
Thyroid cancer is the most common endocrine malignancy and is one of the fastest growing cancer diagnoses worldwide (1). Thyroid cancer may originate from follicular or parafollicular cells. The majority of thyroid cancer cases originate from follicular cells and are also referred to as nonmedullary thyroid cancer. Approximately 9% of nonmedullary thyroid cancer cases are familial (2–4). Some investigators have suggested that familial nonmedullary thyroid cancer (FNMTC) is associated with a younger age at presentation and more aggressive disease than is sporadic nonmedullary thyroid cancer (5). FNMTC has higher rates of multicentric tumors, lymph node metastasis, extra-thyroidal invasion, and persistent or recurrent disease. The risk of developing FNMTC in first-degree relatives of patients with differentiated thyroid cancer is significantly higher than in the general population (6,7). In addition, when comparing first and second generations of families with FNMTC, some investigators have found that the second generation had disease onset at an earlier age (8). However, no susceptibility gene(s) responsible for FNMTC has been identified.
Telomeres, which are important in maintaining chromosomal stability, are linear chromosomal ends formed by the tandem TTAGGG sequence. Telomere length is regulated by the telomerase complex consisting of the telomerase reverse transcriptase (TERT) catalytic subunit and the telomerase ribonucleic acid (RNA) complex (9). At the very end of a chromosome, a single-strand portion forms a loop, held together by a six-protein complex (telomere repeat binding factor 1 [TRF1], telomere repeat binding factor 2 [TRF2], repressor activator protein 1 [RAP1], TRF1 interacting nuclear factor 2 [TIN2], tripeptidyl peptidase 1 [TPP1], and protection of telomere 1 [POT1]), known as “shelterin.” This protein complex binds the TTAGGG sequence of telomere, protects chromosomes, and regulates telomerase activity (10–14). Dysregulated telomere length maintenance plays an important role in genomic stability, since shortened telomere length and increased telomerase activity result in facilitating malignant transformation (15,16).
Short telomere length and genetic defects in telomere maintenance have been associated with increased risk of cancers (breast, bladder, lung, and gastrointestinal); familial diseases, such as dyskeratosis congenital syndrome (17); and familial cancers, including papillary thyroid cancer (18). Some studies have also shown that telomere length is inherited and segregates in families, and that the decrease in telomere length may play a role in age-related genomic instability (19). In familial papillary thyroid cancer (also referred to as FNMTC), Capezzone et al. found shorter telomere length and increased telomerase activity in the lymphocytes of affected members with familial papillary thyroid cancer (FPTC), as compared with unaffected members and those with sporadic PTC, benign thyroid neoplasm, and healthy controls (18). In a follow-up study, this same group demonstrated short relative telomere length (RTL) in thyroid tumor tissue of patients with FPTC and sporadic disease but also showed shortened RTL in the contralateral normal thyroid tissue, and extra-thyroidal tissue only in affected members with FPTC and not in sporadic cases of papillary thyroid cancer (20). These findings suggest that telomere length may predispose an individual to FPTC. In contrast, Jendrzejewski et al. did not find affected members with FNMTC to have short RTL (21). Understanding the relative contribution of telomere length in FNMTC and what mechanism(s) may account for this alteration are important for determining what susceptibility gene(s) may exist.
Given these two conflicting observations of RTL in affected members with FNMTC, limitations in our understanding of the genetic basis of FNMTC, and the lack of genetic markers capable of risk-stratifying patients at risk for FNMTC, we investigated RTL, and the gene copy number and expression of genes involved in maintaining telomere length (hTERT, TRF1, TRF2, RAP1, TIN2, TPP1, and POT1) in the blood lymphocytes of a cohort screened for FNMTC and in controls.
Materials and Methods
Patients
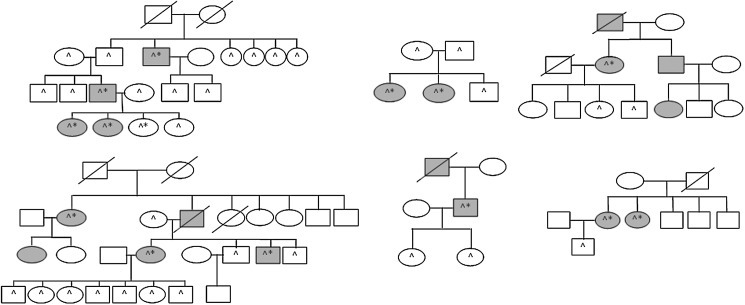
Patients were enrolled in a clinical protocol approved by the National Institutes of Health review board after written informed consent was obtained. All patients completed a family history questionnaire. All patients had a thyroid ultrasound to screen for a thyroid nodule. If a thyroid nodule was present, thyroid fine-needle aspiration biopsy was performed to exclude a thyroid cancer diagnosis. All participants (and their legal guardians) agreed to annual follow-up with blood tests and thyroid ultrasound. Blood samples were collected from study participants, and deoxyribonucleic acid (DNA) and total RNA were extracted. Six families with FNMTC (13 affected and 31 unaffected family members) were included in this study. The family pedigrees for the six families are summarized in Figure 1. FNMTC was defined when two or more first-degree relatives were affected (22). All histologic diagnoses were reviewed by an endocrine pathologist to confirm the diagnosis of thyroid cancer, which were all conventional papillary thyroid cancer. In addition to the blood samples from FNMTC cases, we analyzed blood samples from patients with sporadic papillary thyroid cancer (n=40), sporadic multinodular goiter (n=10), and sporadic primary hyperparathyroidism (n=10) as controls.
FIG. 1.
Pedigrees of six families in study cohort. □, male; ○, female;/, deceased; shaded, affected with thyroid cancer; *, underwent thyroidectomy; ^, blood sample included in analysis.
Blood DNA and RNA preparation
Both genomic DNA and total RNA were extracted from peripheral blood samples at the same time. Lymphocytes were extracted from whole blood using a PAXgene blood DNA kit (Qiagen; 761133). Genomic DNA and total RNA were extracted from lymphocytes with an RNeasy Mini kit (Qiagen; 74104) following the manufacturer's instructions. The quantity and quality of DNA and RNA were evaluated by spectrophotometry (NanoDrop 2000; Thermo Scientific). DNA stock (10 ng/μL) was prepared and used for experiments. Total RNA (400 ng) was reverse-transcribed to complementary DNA (cDNA) using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems [ABI]; 4374967). Stock cDNA at 1:10 dilution was prepared and used for subsequent experiments.
Quantitative polymerase chain reaction to detect RTL
The quantitative polymerase chain reaction (Q-PCR) assay was performed as previously described (18). Telomere length quantification was determined as the relative ratio of telomere (T) repeat copy number to a single copy gene (S), called the T/S ratio, in experimental samples using standard curves. The 36B4, encoding acidic ribosomal phosphoprotein P0, was used as the control single copy gene. All samples, including the endogenous control, were run on the same 384-well plate to eliminate the effect of inter-assay variability as a result of different PCR efficiency between plates. The PCR master mix included 5 μL of SYBR Green PCR master mix (2×; ABI), 1 μL of forward primer and 1μL of reverse primer, and 3 μL (30 ng) of stock DNA in a total volume of 10 μL. A standard curve and a negative control (no DNA template) were included in each experiment. For the standard curve, the reference DNA sample (pooled DNA samples) was diluted serially to produce five final concentrations (25, 3.13, 0.39, 0.049, and 0.0061 ng/μL). The PCR cycling condition for both amplicons was 95°C for 10 minutes, followed by 30 cycles at 95°C for 15 seconds, 54°C for 1 minute, and 60°C for 30 seconds for telomere PCR; and 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute for 36B4 PCR. To exclude the presence of nonspecific binding between Syber Green probes and template, a dissociation stage was included at the end of all PCR amplifications. To determine equal copy numbers per cell, the β-globin gene was amplified in all DNA samples. The primer sequences and concentrations of telomere, 36B4, and β-globin were as described by Capezzone et al. (18). All PCRs were performed on the 7900HT Fast Real-Time PCR System. ABI's SDS 2.3 software was used to quantify PCR products for each sample based on the standard curve.
hTERT copy number and mRNA expression
To determine hTERT copy number and mRNA expression, we performed DNA amplification using the SYBR Green method and mRNA expression using ABI predesigned TaqMan® primer and probe and TaqMan gene expression master mix. Thirty nanograms of genomic DNA was amplified using a final concentration of 300 nM of the following primers: sense, 5′-TGA CAC CTC ACC TCA CCC AC-3′ and antisense, 5′-CAC TGT CTT CCG CAA GTT-3′ in a final volume of 10 μL Syber Green reaction mix. A standard curve and negative control were included in each experiment. The thermal cycling condition was 95°C for 10 minutes followed by 30 cycles at 95°C for 15 seconds and 60°C for 30 seconds. A dissociation stage was included to exclude nonspecific binding between SYBR Green probes and template. The TaqMan assay was used to measure hTERT expression, using 3 μL of cDNA in a final volume of 10 μL reaction. hTERT TaqMan primer and probe were purchased from ABI (Hs00972652_g1). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control gene (Hs99999905_m1). The difference in cycle threshold (Ct) between hTERT and GAPDH [−ΔCt=−(Ct of hTERT−Ct of GAPDH)] was calculated to determine hTERT mRNA expression relative to GAPDH expression.
Copy number and expression of shelterin complex genes
The shelterin complex is composed of six core proteins: TRF1, TRF2, TIN2, POT1, TPP1, and RAP1. The copy number and mRNA expression level of these genes were evaluated using DNA and RNA from blood lymphocytes. The primers and probes for these genes for the TaqMan copy number and mRNA expression assay were purchased from ABI (TRF1: HS00783530_cn; TRF2: HS02635053_cn; RAP1: HS02244780_cn; TIN2: HS02040625_cn; TPP1: HS02844556_cn; POT1: HS02381501_cn). PCRs were performed in four biologic replicates in a duplex reaction with RNaseP as an endogenous control gene. Copy number was determined using the CopyCaller v 1.0 software (www.appliedbiosystems.com/absite/us/en/home/support/software/real-time-pcr/copycaller.html). Human GAPDH mRNA expression was used as an endogenous control. The expression level was determined by the −ΔCt method.
Data analysis
Statistical analysis was performed with GraphPad Prism 5.04 for Windows (1992–2010 GraphPad Software, Inc.) and IBM® SPSS® Statistics for Windows, version 16.0 (SPSS, Inc.). Student's t-test was used to determine statistical difference in RTL, and hTERT and shelterin complex gene copy number and expression between affected and unaffected FNMTC cases and other control groups. One-way analysis of variance (ANOVA) was used to account for multiple comparisons and differences among groups. Post hoc tests using least square difference (LSD) and Bonferroni's method were performed to compare the mean difference between groups. The Pearson correlation coefficient was calculated for RTL and age and other variables, using 95% confidence interval. Demographic and clinical data were analyzed by Fisher's exact test for categorical data. Student's t-test and Mann–Whitney test were used to compare parametric and nonparametric data, respectively. Data are shown as mean±standard deviation. A two-sided p-value of <0.05 was considered statistically significant.
Results
Relative telomere length
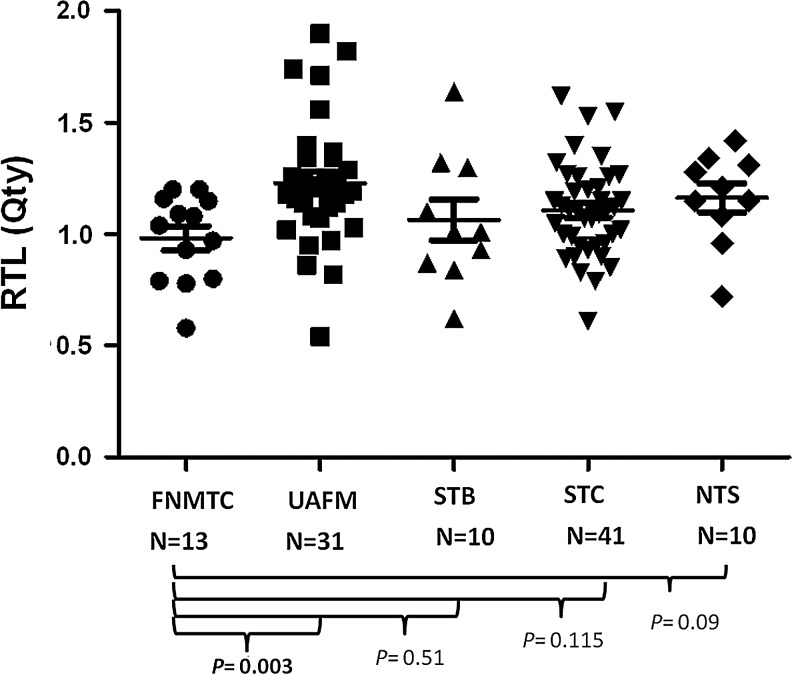
RTL was measured by Q-PCR in six families and other control groups. The demographic and clinical characteristics of the study cohort are summarized in Table 1. Affected members in families with FNMTC had a higher rate of advanced tumors (T3 and T4) than patients with sporadic PTC (46.2% vs. 12.5%, p=0.02). We found significantly shorter RTL in FNMTC affected members (0.98±0.20) as compared with unaffected members (1.23±0.29, p<0.01 by LSD method, with ANOVA p=0.03; Fig. 2). However, there was no significant difference in RTL between FNMTC affected members and other control groups (Fig. 2).
Table 1.
Study Cohort Clinical Characteristics
| |
Familial |
Sporadic |
Parathyroid |
|
||
|---|---|---|---|---|---|---|
| Characteristics | Affected (n=13) | Unaffected (n=31) | Sporadic thyroid cancer (n=40) | Multinodular goiter (n=10) | No thyroid disease (n=10) | p-Value |
| Sex (M/F) | 4/9 | 15/16 | 5/35 | 2/8 | 4/6 | 0.20a |
| Age (mean±SD) | 44.4±14.4 | 40.0±22.3 | 47.2±12.5 | 41.0±14.6 | 41.6±18.8 | 0.36 |
| TSHb (median±SD) (reference range: 0.4–4.0 mIU/L) | 0.11±0.3 | 0.85±2.5 | 2.02±1.4 | 2.00±1.9 | 1.41±1.2 | 0.01 |
| Pathology | ||||||
| Thyroid cancers | 0.70 | |||||
| Classical PTC | 11 (84.6%) | 30 (75%) | ||||
| Follicular variant PTC | 1 (7.7%) | 7 (17.5%) | ||||
| Other thyroid cancer subtypesc | 1 (7.7%) | 3 (7.5%) | ||||
| Multinodular goiter | 10 | |||||
| Parathyroid adenoma | 8 | |||||
| Parathyroid hyperplasia | 2 | |||||
| Tumor characteristics | ||||||
| T3 and T4 | 46.2% | 12.5% | 0.02 | |||
| Size (median±SD) in mm | 18±11.2 | 9±16.8 | 0.11 | |||
| Lymph node metastasis | 46.2% | 38.1% | 0.73 | |||
| Distant metastasis | 0% | 3.8% | 1.0 | |||
| Extrathyroidal extension | 30.8% | 7.5% | 0.05 | |||
| Lymphovascular invasion | 25% | 7.5% | 0.19 | |||
| Multifocality | 46.2% | 45% | 1.0 | |||
| Thyroiditis | 38% | 15% | 0.08 | |||
| Disease recurrence | 7.7% | 2.5% | 0.43 | |||
| Radioactive iodine treatment | 75% | 50% | 0.19 | |||
p-Value reflects a comparison of sex distribution between affected members of family with FNMTC and patients with sporadic thyroid cancer.
Postoperative values in affected member, sporadic thyroid cancer, and multinodular goiter groups.
Diffusing sclerosing and tall-cell variant PTC.
TSH, thyrotropin; FNMTC, familial nonmedullary thyroid cancer; SD, standard deviation.
FIG. 2.
Relative telomere length (RTL) measurement in affected and unaffected members with familial nonmedullary thyroid cancer (FNMTC) and other control groups. Affected members had significantly shorter RTL than unaffected members. The mean RTL of each group is indicated in the long black line, and standard error of the mean is shown. FNMTC, affected patients with FNMTC; UAFM, unaffected family members; STB, sporadic thyroid benign disease; STC, sporadic thyroid cancer; NTS, nonthyroid subjects.
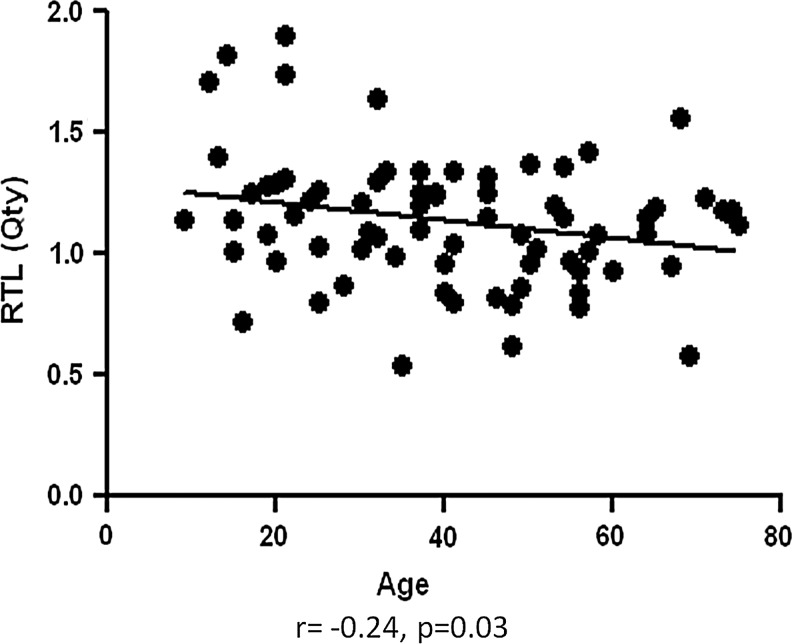
As expected, we found an inverse association of RTL and age in the entire cohort (r=− 0.24, p=0.03; Fig. 3). There was no significant difference in age between the groups. Also, there was no significant association between RTL and other demographics and clinical characteristics, such as sex, advanced T stage (T3 and T4 vs. T1 and T2), lymph node or distant metastasis, extrathyroidal extension, lymphovascular invasion, multifocality, recurrent disease, presence of benign thyroid nodules, thyroiditis, history of other cancers, and tobacco use (Table 2).
FIG. 3.
Association of RTL and age in cohort.
Table 2.
Relative Telomere Length and Study Cohort Clinical Characteristics in Patients with Sporadic Thyroid Cancer and Affected Members of Families with Familial Nonmedullary Thyroid Cancer
| Patient characteristics | Mean RTL±SD | p-Value |
|---|---|---|
| Sex | 0.06 | |
| Male | 0.96±0.18 | |
| Female | 1.11±0.20 | |
| Advanced T stage (T3–4) | 0.81 | |
| Yes | 1.07±0.14 | |
| No | 1.9±0.22 | |
| Lymph node metastasis | 0.82 | |
| Yes | 1.06±0.20 | |
| No | 1.07±0.16 | |
| Distant metastasis | 0.20 | |
| Yes | 0.79 | |
| No | 1.06±0.20 | |
| Extrathyroidal extension | 0.68 | |
| Yes | 1.05±0.13 | |
| No | 1.09±0.22 | |
| Lympho-vascular invasion | 0.46 | |
| Yes | 1.03±0.22 | |
| No | 1.11±0.21 | |
| Multifocality | 0.72 | |
| Yes | 1.07±0.22 | |
| No | 1.09±0.20 | |
| Disease recurrence | 0.57 | |
| Yes | 1.17±0.06 | |
| No | 1.08±0.21 | |
| Presence of benign thyroid nodules | 0.60 | |
| Yes | 1.10±0.21 | |
| No | 1.07±0.21 | |
| Thyroiditis | 0.53 | |
| Yes | 1.05±0.17 | |
| No | 1.10±0.22 | |
| History of other cancers | 0.84 | |
| Yes | 1.10±0.18 | |
| No | 1.08±0.21 | |
| Tobacco use | 0.70 | |
| Yes | 1.13±0.15 | |
| No | 1.10±0.21 | |
| Radioactive iodine treatment | 0.20 | |
| Yes | 1.05±0.20 | |
| No | 1.13±0.21 |
RTL, relative telomere length.
There were no significant differences between affected and unaffected members of FNMTC with regard to sex distribution (31% men vs. 48% men, p=0.33), body mass index (27.9 vs. 27.4, p=0.81), the rates of tobacco use (44% vs. 20%, p=0.20), benign thyroid nodules (20% vs. 57%, p=0.07), and prior history of other cancers (0% vs. 7%, p=1.0).
hTERT and shelterin complex copy number and gene expression
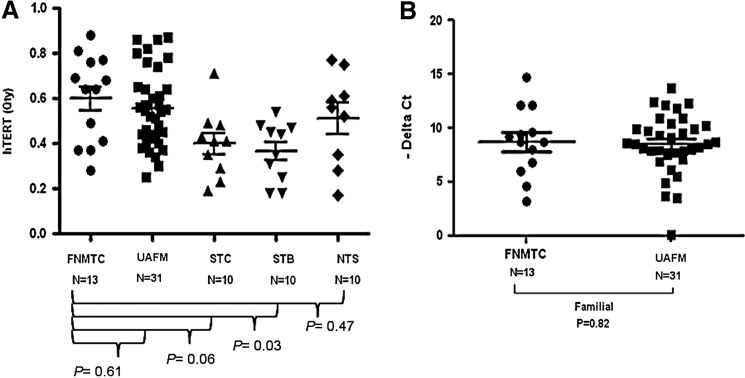
No differences in hTERT copy number were observed in affected versus unaffected members with FNMTC (0.60±0.18 and 0.55±0.17, respectively; p=0.61). We also found no significant difference in hTERT mRNA level in affected versus unaffected FNMTC members (8.77±2.92 vs. 8.34±5.12; p=0.82; Fig. 4). Affected members had significantly higher hTERT copy number than sporadic benign thyroid group (p=0.03). There was no significant difference in copy number and mRNA expression level of TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 in affected versus unaffected members with FNMTC.
FIG. 4.
Telomerase reverse transcriptase (hTERT) copy number and mRNA expression in affected and unaffected members. (A) hTERT gene copy number in affected members with FNMTC and control groups. (B) hTERT mRNA expression level in affected and unaffected family members. FNMTC, affected patients with FNMTC; UAFM, unaffected family members; STB, sporadic thyroid benign disease; STC, sporadic thyroid cancer; NTS, nonthyroid subjects.
Discussion
In this study, we determined whether telomere length was shorter in affected members with FNMTC as compared with unaffected members and other control groups. As previously reported, we found that affected members had shorter RTL in blood lymphocytes. To explore the possible mechanism of the shorter RTL observed in affected members with FNMTC, we studied genes involved in regulating telomere length for copy number and mRNA expression differences. We, however, found no significant difference in hTERT, TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 copy number and mRNA expression in affected and unaffected members with FNMTC and other control groups. This finding suggests that RTL may be associated with FNMTC.
Although we confirmed, in an independent cohort, the findings of Capezzone et al. (18) in regards to shorter RTL in affected members with FNMTC, the difference in RTL was much smaller than that observed by them with overlap in RTL between affected and unaffected members. Therefore, RTL is not likely to be a useful marker for FNMTC. Although patients who had a negative thyroid ultrasound with no prior history of thyroid cancer were defined as unaffected, it is possible that some may develop thyroid cancer in the future. These patients may contribute to shorter RTL in “unaffected” members. Follow-up of these families will be important to determine whether this is indeed the case. We did not find any significant differences in other factors (tobacco use or history of other cancers or rates of benign thyroid diseases) and RTL in the entire study cohort, or in affected and unaffected members. Affected members of families with FNMTC in our study had a significantly higher rate of locally invasive thyroid cancer (T3 and T4) than patients with sporadic thyroid cancer as has been observed in some studies (5). While our study is not the first to demonstrate shorter RTL in affected members with FNMTC, our study is important in that it demonstrates that this finding may be more applicable to a heterogeneous population of FNMTC studied in our analysis than to a more homogenous population from Italy and Portugal (21,23). Further, the lack of significant difference between affected members with FNMTC and those with sporadic and benign thyroid disease also suggests that short RTL is not necessarily specific to FNMTC.
In contrast to the study by Capezzone et al. (18), we found no difference in hTERT gene copy number and mRNA expression in affected members with FNMTC, as compared with unaffected members using the same methods. However, affected members with FNMTC in our study had a significantly higher hTERT gene copy number than patients with benign thyroid disease but this was not associated with a difference in RTL. A recent study suggested that telomere length may be inherited and that the telomere set-point may be reset, which provides indirect evidence to support the hypothesis that shorter RTL among affected members with FNMTC may be indeed inherited (24). However, it is unclear why shorter RTL may result in one specific familial cancer, such as thyroid cancer, since the shorter telomere length would be present in all cell types and thus should result in a higher risk of malignancies in other organs. Further, we observed a relatively small difference in RTL among affected members with FNMTC, compared with that observed by Capezzone et al. (18). Taken together, these observations suggest that shorter telomere length is likely not to be the sole predisposing genetic factor in FNMTC, since RTL differences may also occur due to a variety of factors, such as dietary and environmental factors (25,26).
To understand what possible mechanism(s) may account for the shorter RTL we found among affected members with FNMTC, we evaluated hTERT copy number and mRNA expression. However, we found no difference in hTERT copy number and expression, in contrast to the study by Capezzone et al. (18). In addition, because the shelterin complex proteins have also been implicated in maintaining telomere length and genomic stability, we also analyzed the copy number and mRNA expression of these genes and found no difference. These findings suggest that alterations in hTERT, TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 copy number and mRNA expression level are not likely to account for the shorter RTL we observed in our cohort of FNMTC.
In summary, telomere length is shorter in affected members with FNMTC but the significance to thyroid cancer development and the mechanism of this finding still remains unclear. The small difference and overlapping telomere length between unaffected and affected members preclude the utility of RTL as a marker for FNMTC in at-risk individuals. Additional studies in larger population with long-term follow-up to confirm this finding are needed.
Acknowledgments
This research was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, and National Institutes of Health.
Disclosure Statement
The authors declared that no competing financial interests exist.
References
- 1.Chen AY. Jemal A. Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer. 2009;115:3801–3807. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 2.Loh KC. Familial nonmedullary thyroid carcinoma: a meta-review of case series. Thyroid. 1997;7:107–113. doi: 10.1089/thy.1997.7.107. [DOI] [PubMed] [Google Scholar]
- 3.Stoffer SS. Van Dyke DL. Bach JV. Szpunar W. Weiss L. Familial papillary carcinoma of the thyroid. Am J Med Genet. 1986;25:775–782. doi: 10.1002/ajmg.1320250415. [DOI] [PubMed] [Google Scholar]
- 4.Moses W. Weng J. Kebebew E. Prevalence, clinicopathologic features, and somatic genetic mutation profile in familial versus sporadic nonmedullary thyroid cancer. Thyroid. 2011;21:367–371. doi: 10.1089/thy.2010.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vriens MR. Suh I. Moses W. Kebebew E. Clinical features and genetic predisposition to hereditary nonmedullary thyroid cancer. Thyroid. 2009;19:1343–1349. doi: 10.1089/thy.2009.1607. [DOI] [PubMed] [Google Scholar]
- 6.Goldgar DE. Easton DF. Cannon-Albright LA. Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86:1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 7.Hemminki K. Eng C. Chen B. Familial risks for nonmedullary thyroid cancer. J Clin Endocrinol Metab. 2005;90:5747–5753. doi: 10.1210/jc.2005-0935. [DOI] [PubMed] [Google Scholar]
- 8.Capezzone M. Marchisotta S. Cantara S. Busonero G. Brilli L. Pazaitou-Panayiotou K. Carli AF. Caruso G. Toti P. Capitani S. Pammolli A. Pacini F. Familial non-medullary thyroid carcinoma displays the features of clinical anticipation suggestive of a distinct biological entity. Endocr Relat Cancer. 2008;15:1075–1081. doi: 10.1677/ERC-08-0080. [DOI] [PubMed] [Google Scholar]
- 9.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 10.De Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 11.Hockemeyer D. Daniels JP. Takai H. de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 12.Wu L. Multani AS. He H. Cosme-Blanco W. Deng Y. Deng JM. Bachilo O. Pathak S. Tahara H. Bailey SM. Deng Y. Behringer RR. Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 13.He H. Multani AS. Cosme-Blanco W. Tahara H. Ma J. Pathak S. Deng Y. Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez P. Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 15.Masutomi K. Hahn WC. Telomerase and tumorigenesis. Cancer Lett. 2003;194:163–172. doi: 10.1016/s0304-3835(02)00703-6. [DOI] [PubMed] [Google Scholar]
- 16.Ma H. Zhou Z. Wei S. Liu Z. Pooley KA. Dunning AM. Svenson U. Roos G. Hosgood HD., 3rd Shen M. Wei Q. Shortened telomere length is associated with increased risk of cancer: a meta-analysis. PLoS One. 2011;6:e20466. doi: 10.1371/journal.pone.0020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vulliamy T. Marrone A. Szydlo R. Walne A. Mason PJ. Dokal I. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet. 2004;36:447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 18.Capezzone M. Cantara S. Marchisotta S. Filetti S. De Santi MM. Rossi B. Ronga G. Durante C. Pacini F. Short telomeres, telomerase reverse transcriptase gene amplification, and increased telomerase activity in the blood of familial papillary thyroid cancer patients. J Clin Endocrinol Metab. 2008;93:3950–3957. doi: 10.1210/jc.2008-0372. [DOI] [PubMed] [Google Scholar]
- 19.Granger MP. Wright WE. Shay JW. Telomerase in cancer and aging. Crit Rev Oncol Hematol. 2002;41:29–40. doi: 10.1016/s1040-8428(01)00188-3. [DOI] [PubMed] [Google Scholar]
- 20.Capezzone M. Cantara S. Marchisotta S. Busonero G. Formichi C. Benigni M. Capuano S. Toti P. Pazaitou-Panayiotou K. Caruso G. Carli AF. Palummo N. Pacini F. Telomere length in neoplastic and nonneoplastic tissues of patients with familial and sporadic papillary thyroid cancer. J Clin Endocrinol Metab. 2011;96:E1852–E1856. doi: 10.1210/jc.2011-1003. [DOI] [PubMed] [Google Scholar]
- 21.Jendrzejewski J. Tomsic J. Lozanski G. Labanowska J. He H. Liyanarachchi S. Nagy R. Ringel MD. Kloos RT. Heerema NA. de la Chapelle A. Telomere length and telomerase reverse transcriptase gene copy number in patients with papillary thyroid carcinoma. J Clin Endocrinol Metab. 2011;96:E1876–E1880. doi: 10.1210/jc.2011-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malchoff CD. Malchoff DM. Familial nonmedullary thyroid carcinoma. Cancer Control. 2006;13:106–110. doi: 10.1177/107327480601300204. [DOI] [PubMed] [Google Scholar]
- 23.Helgason A. Nicholson G. Stefansson K. Donnelly P. A reassessment of genetic diversity in Icelanders: strong evidence from multiple loci for relative homogeneity caused by genetic drift. Ann Hum Genet. 2003;67:281–297. doi: 10.1046/j.1469-1809.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- 24.Chiang YJ. Calado RT. Hathcock KS. Lansdorp PM. Young NS. Hodes RJ. Telomere length is inherited with resetting of the telomere set-point. Proc Natl Acad Sci USA. 2010;107:10148–10153. doi: 10.1073/pnas.0913125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L. Trimarchi JR. Navarro P. Blasco MA. Keefe DL. Oxidative stress contributes to arsenic-induced telomere attrition, chromosome instability, and apoptosis. J Biol Chem. 2003;278:31998–32004. doi: 10.1074/jbc.M303553200. [DOI] [PubMed] [Google Scholar]
- 26.Schoeftner S. Blasco MA. A ‘higher order’ of telomere regulation: telomere heterochromatin, telomeric RNAs. EMBO J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]