Abstract
The complete mitochondrial genome (mitogenome) of the rice moth, Corcyra cephalonica Stainton (Lepidoptera: Pyralidae) was determined as a circular molecular of 15,273 bp in size. The mitogenome composition (37 genes) and gene order are the same as the other lepidopterans. Nucleotide composition of the C. cephalonica mitogenome is highly A+T biased (80.43%) like other insects. Twelve protein-coding genes start with a typical ATN codon, with the exception of coxl gene, which uses CGA as the initial codon. Nine protein-coding genes have the common stop codon TAA, and the nad2, cox1, cox2, and nad4 have single T as the incomplete stop codon. 22 tRNA genes demonstrated cloverleaf secondary structure. The mitogenome has several large intergenic spacer regions, the spacer1 between trnQ gene and nad2 gene, which is common in Lepidoptera. The spacer 3 between trnE and trnF includes microsatellite-like repeat regions (AT)18 and (TTAT)3. The spacer 4 (16 bp) between trnS2 gene and nad1 gene has a motif ATACTAT; another species, Sesamia inferens encodes ATCATAT at the same position, while other lepidopteran insects encode a similar ATACTAA motif. The spacer 6 is A+T rich region, include motif ATAGA and a 20-bp poly(T) stretch and two microsatellite (AT)9, (AT)8 elements.
Keywords: Galleriinae, mitogenome
Introduction
Animal mitogenomes are typically enclosed circular molecules of 14–20 kb in length with 37 genes, 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), and two ribosomal RNA (rRNA). It also contains an A+T rich non-coding area (also called control region) responsible for regulating transcription and replication of the mitogenome (Boore 1999; Taanman 1999). Mitogenomes have a simple structure, undergo fast evolution, are normally maternal inherited, and have been broadly applied in phylogenetic reconstruction, phylogeography, population structure and dynamics, and molecular evolution (Zhang et al. 1995; Nardi et al. 2003; Arunkumar et al. 2006). Recent advancements in sequencing technology have lead to rapid growth of mitogenome data in Genbank. To date, the complete mitogenome sequences of more than 140 species have been determined for insects, including 31 species of Lepidoptera that have been entirely or nearly entirely sequenced (Coates et al. 2005; Kim et al. 2006; Lee et al. 2006; Cameron et al. 2007; Cha et al. 2007; Cameron and Whiting 2008; Liu et al. 2008; Jiang et al. 2009; Hong et al. 2009; Pan et al. 2008; Salvato et al. 2008; Kim MI et al. 2009; Hu et al. 2010; Liao et al. 2010; Li et al. 2010; Zhao et al. 2010; Margam et al. 2011).
Lepidoptera is the second largest order after Coleoptera within in Insecta and includes moths and butterflies. Most of them are agricultural and forestry pests, pollinators, and resources insects (Li et al. 2009). Corcyra cephalonica Stainton (Lepidoptera: Pyralidae) is in a small subfamily of Galleriinae with 261 species of Pyralidae, which contains more than 330 species of 70 genera (Heppner 1991). The genus Corcyra contains only two species, C. nidicolella and C. cephalonica; the latter is known to be a stored product pest, and is controlled with botanical insecticides and trapped with sex pheromone (Türkera 1998; Allotey and Azalekor 2000; Coelho et al. 2007). Corcyra cephalonica is used as the host for cultivating Trichogramma and other parasitoid wasps (Muthukrishnan et al. 2003; Jalali et al. 2007). Moreover, it is lately being used as an experimental model insect. A group of the functional genes have been identified (Nagamanju et al. 2003; Chaitanya and Dutta-Gupta, 2010; Damara et al. 2010; Gullipalli et al. 2010), but information regarding the mitochondrial genome is lacking. The availability of the mitogenome sequence will definitely be beneficial in the basic and applied studies on C. cephalonica.
In this paper, the mitogenome of C. cephalonica was sequenced and analyzed. So far, there are four species within Pyraloidea with known mitogenome: Diatraea saccharalis (Li et al. 2010, Ostriniafurnacalis and O. nubilalis (Coates et al. 2005), and Chilo suppressalis [unpublished, JF339041].
Materials and Methods
DNA samples extraction
Corcyra eggs were collected from Guangdong Province of China and raised in the laboratory in Beijing. The hatched adults were collected, preserved in 100% ethanol, and stored at —20 °C. Total DNA was extracted and isolated from single specimens using the DNeasy Tissue kit (QIAGEN, www.qiagen.com) according to manufacturer instructions.
Primer design, PCR, and sequencing
The short fragment amplifications were performed using the universal PCR primers from Simon et al. (1994). The degenerate and specific primer pairs were designed based on the known mitochondrial sequences in Lepidoptera, or designed by Primer 5.0 software on the fragments that we previously sequenced (Table 1). All the primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd, www.sangon.com. For fragments of length less than 2 kb, PCR conditions were as follows: 95 °C for five min, 34 cycles of 94 °C for 30 sec, 50–55 °C (depending on primer combinations), 1–3 min (depending on putative length of the fragments) at 68 °C, and a final extension step of 72 °C for 10 min. For fragments of length longer than 2 kb, PCR conditions were as follows: 92 °C for two min, 40 cycles of 92 °C for 30 sec, 50–55 °C for 30 sec (depending on primer combinations), 60 °C for 12 min, and a final extension step of 60 °C for 20 min.
Table 1.
Region, primers and sequences for PCRs in this study.
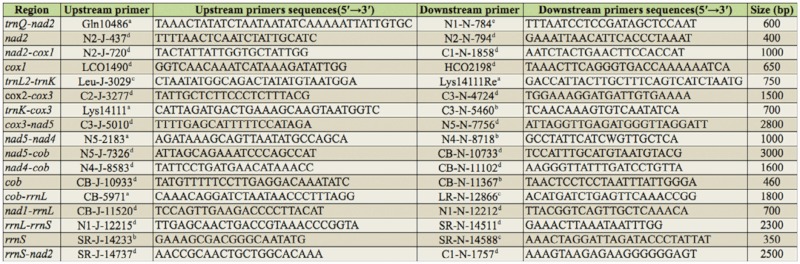
The entire mitogenome of the Corcyra was amplified in 17 fragments. For most fragments, 2× Taq PCR MasterMix (Tiangen Biotech Co., Ltd., www.tiangen.com) was used in the amplification; fragments longer than 2 kb (e.g., rrnL-rrnS and nad4-cob) and with higher AT contents (e.g., rrnS-nad2 and cox3-nad5) were amplified using Takara LA Taq (Takara Co. www.takara-bio.com). All amplifications were performed on an Eppendorf Mastercycler and Mastercycler gradient in 50 µL reaction volumes. The reaction volume of 2 × Taq PCR MasterMix contained 22 µL sterilized distilled water, 25 µL 2× Master Mix, 1 µL of each primer (10 uM), and 1 µL of DNA template; the reaction volume of Takara LA Taq consisted of 26.5 µL sterilized distilled water, 5 µL 10×LA PCR Buffer II (Takara), 5 µL 25 mM MgCl2, 8 µL of dNTPs Mixture, 2 µL of each primer (10 µM), 1 µL of DNA template, and 0.5 µL (1.25 U) of Takara LA Taq polymerase (Takara).
The PCR products were detected via electrophoresis in 1% agarose gel, purified using the 3 S Spin PCR Product Purification Kit, and sequenced directly with ABI-377 automatic DNA sequencer. All fragments were sequenced from both strands. Short amplified products were sequenced directly by internal primers, and long amplified products were sequenced completely by primer walking. The rrnS-nad2 region was sequenced after cloning. The purified PCR products were ligated to the pEASY-T3 Cloning Vector (Beijing TransGen Biotech Co., Ltd., transgen.com.cn) and then sequenced by M13F and M13-R primers and walking. Sequencing was performed using ABI BigDye ver 3.1 dye terminator sequencing technology and run on ABI PRISM 3730×1 capillary sequencers.
Analysis and annotation
Sequence annotation was performed using the DNAStar package (DNAStar Inc., www.dnastar.com). The sequence was checked manually for consistency by alignment, and tRNA genes were found using tRNAscan-SE software v. 1.21 (Lowe and Eddy 1997) with manual editing. The undermined putative tRNAs were identified by sequence alignment with other insects of Pyralidae (Diatraea, O. furnacalis, and O. nubilalis) using Bioedit (Hall 1999). Secondary structure was inferred using DNASIS v.2.5. The trnS1(AGN) secondarystructure was developed as proposed by Steinberg and Cedergren (1994). PCGs and rRNAs were identified by similarity to other lepidopteran sequences. The nucleotide sequences of the PCGs were translated based on the invertebrate mtDNA genetic code. Since the Corcyra does not utilize the AGG codon, use of the variant arthropod genetic code (Abascal et al. 2006) was unnecessary. Nucleotide composition and codon usage were calculated using MEGA4.0 (Tamura et al. 2007).
Results
Genome structure and organization
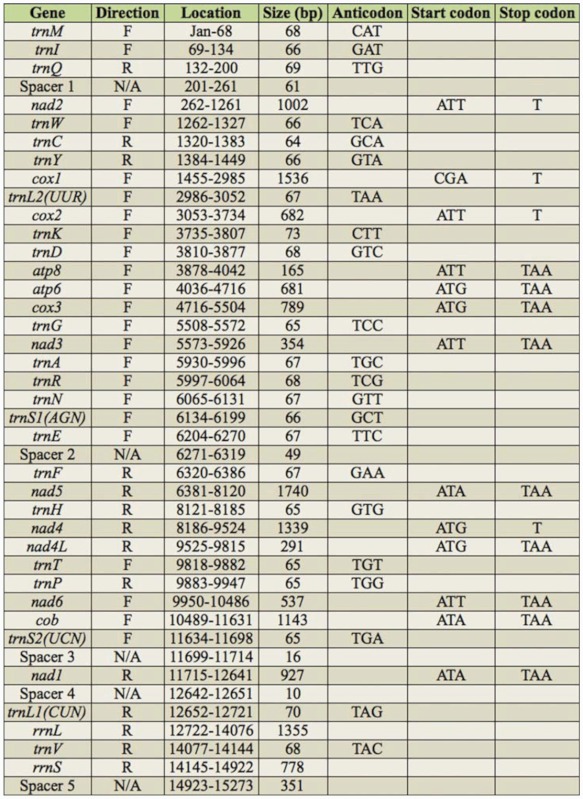
The Corcyra mitogenome is a circular molecule 15,273 bp in length; data were uploaded to Genbank (HQ897685). The Corcyra mitogenome showed the standard gene complement containing 13 PCGs, 2 rRNAs, 22 tRNAs, and non-coding regions typical for lepidopterans. The trnM is coded between the A+T rich region and tRNA-Ile (order is A+T region-trnM-trnl-trnQ), which was different from the ancestral gene order of insects (A+T region-trnl-trnQ-trnM). Since the trnS2(UCN) was not found by tRNAScan-SE, it was later determined by sequence comparison with other lepidopteran insects.
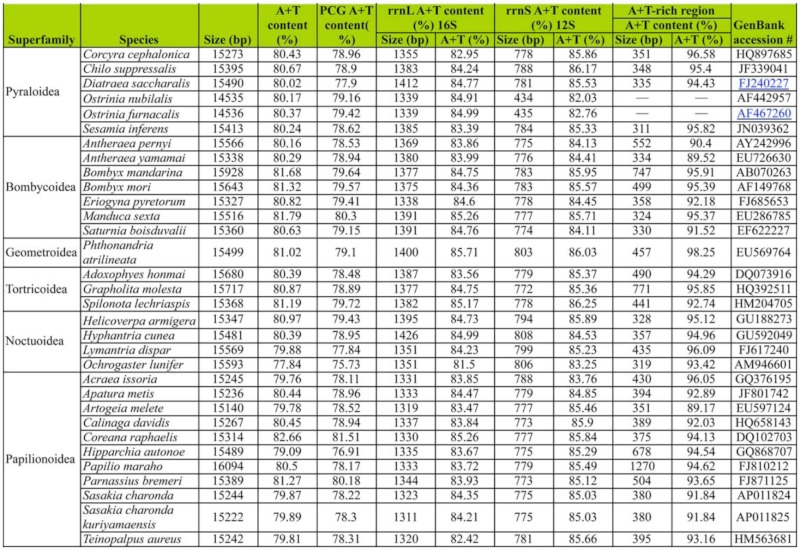
The Corcyra mitogenome was biased toward A+T content (80.43%) with the value falling into the lepidopteran range of 77.84% in Ochrogaster lunifer (Salvato et al. 2008) to 82.66% in Coreana raphaelis (Kim et al. 2006). Additionally, the A+T content was 78.96% in PCGs, 82.95%, in rrnL genes, and 85.86% in rrns genes. These values were also well within the range reported for other lepidopterans. The A+T content (96.58%) of A+T rich region was the highest value among the known lepidoteran MtDNA sequences (Table 3).
Table 3.
Characteristics of the lepidopteran mitogenomes.
Protein-coding genes
The initial and termination codons of thirteen PCGs are shown in Table 2. Twelve PCGs started with a typical ATN codon (ATT for nad2, cox2, atp8, nad3, nad6; ATA for nad5, cob, nad1; ATG for atp6,cox3, nad4, nad4l). One exception is cox1gene, which used CGA as a start codon.
Table 2.
Summary of the mitogenome of the Corcyra.
The putative start codon CGA is common across insects (Fenn et al. 2007) such as Bombyx mori (Yukuhiro et al. 2002), O. nubilalis and O. furnacalis (Coates et al. 2005), Adoxophyes honmai (Lee et al. 2006), Coreana (Kim et al. 2006), Antheraeapernyi (Liu et al. 2008), B. mandarina (Pan et al. 2008), Ochrogaster (Salvato et al. 2008), Artogeia melete (Hong et al. 2009), Eriogyna pyretorum (Jiang et al. 2009), and Hyphantria cunea (Liao et al. 2010).
Nine PCGs had the common stop codon TAA, while the nad2, cox1, cox2, nad4 have single T as an incomplete stop codon, also found in other animal mitochondrial genes (Clary and Wolstenholme 1985). The common interpretation of this phenomenon is that the TAA terminator is created via post– transcriptional polyadenylation (Ojala et al. 1981).
Transfer and ribosomal RNA genes
The 22 tRNA genes ranging from 64 to 73 nucleotides were spread over the mitogenome. Fourteen tRNAs were coded on the J-strand and eight on the N-strand, which is the same organization observed in other lepidopteran mitogenomes. Complete cloverleaf secondary structures could be inferred for 21 of the 22 tRNAs with the exception of trnS1(AGN), which lacks the DHU arm (Figure 1). A total of 43 unmatched base pairs were scattered in 20 tRNA genes, including 20 pairs in the DHU stems, eight pairs in the amino acid acceptor stems, nine pairs in the TψC stems, and six pairs in the anticodon stems. 24 of them are G-U pairs, which form a weak bond. The remaining were A-A, C-A, C-U, G-A, G-G, and U-U mismatches.
Figure 1.
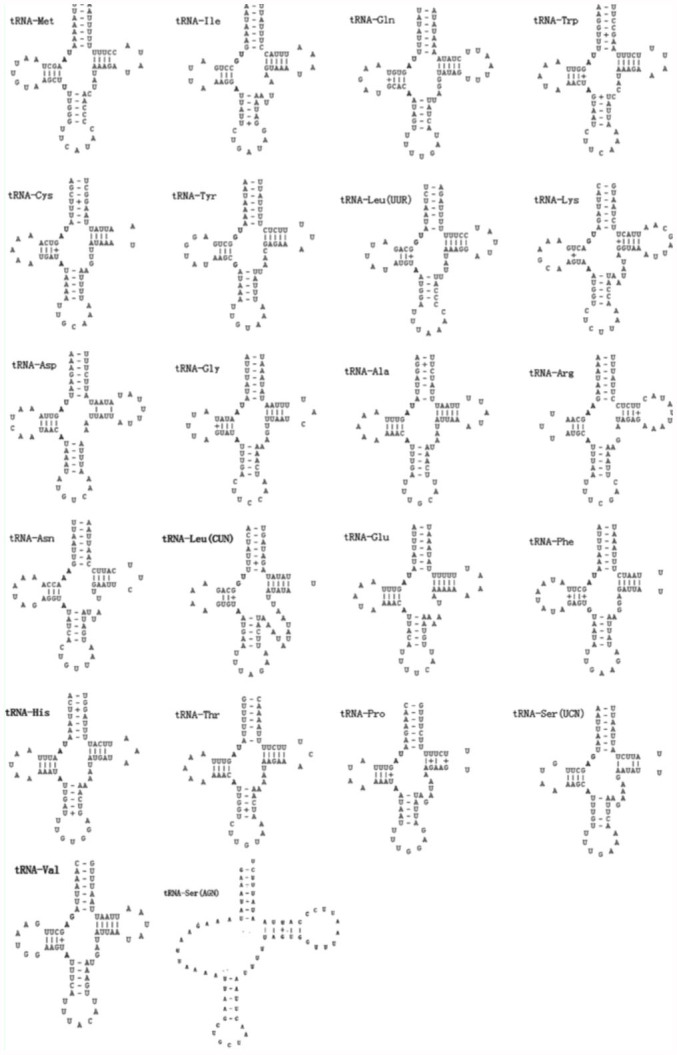
Putative secondary structures for the tRNA genes of the Corcyra cephalonica mitogenome. High quality figures are available online.
As in the other insect mitogenome sequences, two rRNA genes were present in Corcyra. The rrnL were found between trnL(CUN) and trnV, and the rrnS between trnV and the A+T rich region, respectively.
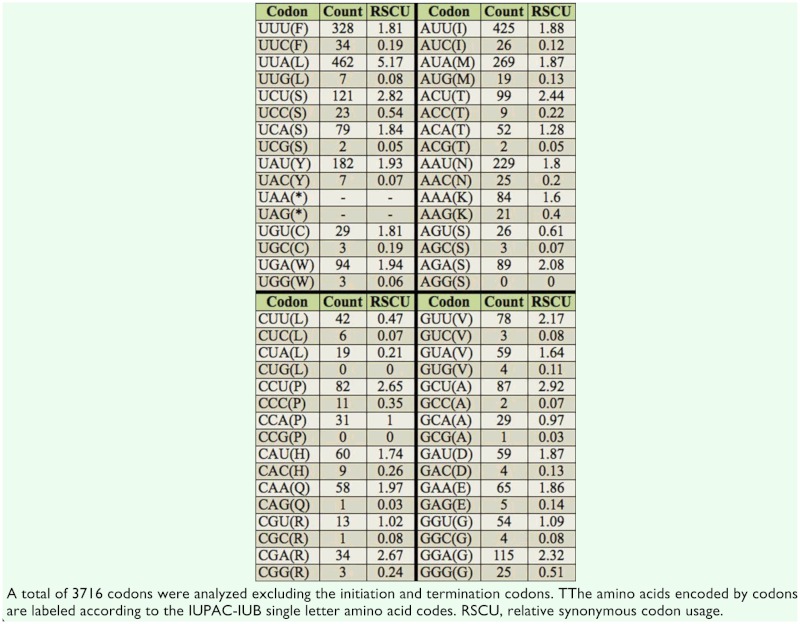
Codon usage
Relative synonymous codon usage values of Corcyra mitogenome are summarized in Table 4. The codons CTG, CCG, and AGG were not represented in the coding sequences. Leucine (14.42%), isoleucine (12.14%), phenylalanine (9.74%), and serine (9.23%) were the most common amino acids in Corcyra mitochondrial proteins (45.53%). These amino acids are abundant in other insects, averaging 45.08% (Lessinger et al. 2000).
Table 4.
Codon usage in the Corcyra mitochondrial genome.
Non-coding and overlapping region
The Corcyra mitogenome harbored 15 non-coding regions, from 1 to 351 bp to 512 bp. Intergenic spacer sequences covered four major regions of length more than 10 bp. The remaining intergenic spacer were less than 5 bp.
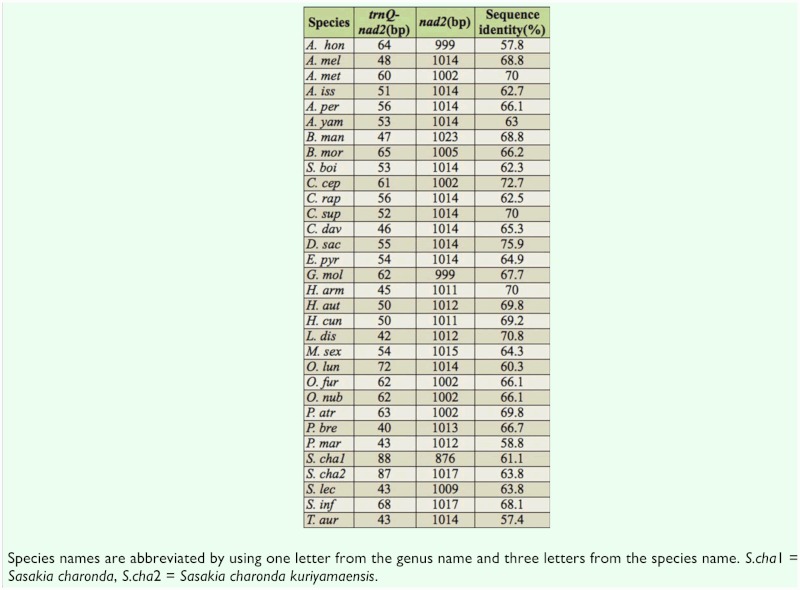
Spacer 1 (61 bp), located between trnQ gene and nad2 gene, is a common intergenic spacer rich in AT nucleotides (96.72%). The location of this spacer is fixed in lepidopterans, but varied in length from 40 bp (Parnassius bremeri) (Kim MI 2009) to 88 bp (Sasakia charonda) (Unpublished, AP011824). This spacer can be taken as a lepidopteran mitogenome marker not found in other insect mitogenomes. Kim MI (2009) found that the intergenic spacer sequences and the nad2 gene had higher sequence identity than other fragments of the mitogenome. There were 29 species with more than 60% identity of 32 total lepidopteran species sequenced (Table 5), suggesting that this spacer sequence originated from a partial duplication of the nad2 gene.
Table 5.
Sequence Identity of Spacer1 and nad2 in 32 Lepidoptera species.
Spacer 2 (49 bp) was found between trnE and trnF genes, including two microsatellite-like regions, (TA) 18 and (TTAT)3, similar to other lepidopterans. The spacer in Adoxophyes (Lee et al. 2006) is 222 bp and contains a different motif (TATTA)31. The spacer in Ochrogaster (Salvato et al. 2008) is 70 bp, contains a microsatellite (TA)23, and shows triplication of a 10-nucleotide motif with some changes. In other lepidoptera insects it is shorter than 10 bp.
Spacer 3 (16 bp) was between the trnS2(UCN) and nad1 genes, commonly detectable in lepidopteran insects, and measured 16–38 bp. This intergenic spacer sequence of most lepidopterans harbored the motif (ATACTAA), except for ATACTAT in Corcyra and ATCATAT in Sesamia (Figure 2). Similarly, in Hymenoptera there is a 6 bp conserved motif (THACWW) (Wei et al. 2010), and in Coleoptera the motif is 5 bp (TACTA). Such conservation suggests that the motif is functional in Lepidoptera. This motif is possibly fundamental to site recognition by the transcription termination peptide (Taanman 1999).
Figure 2.
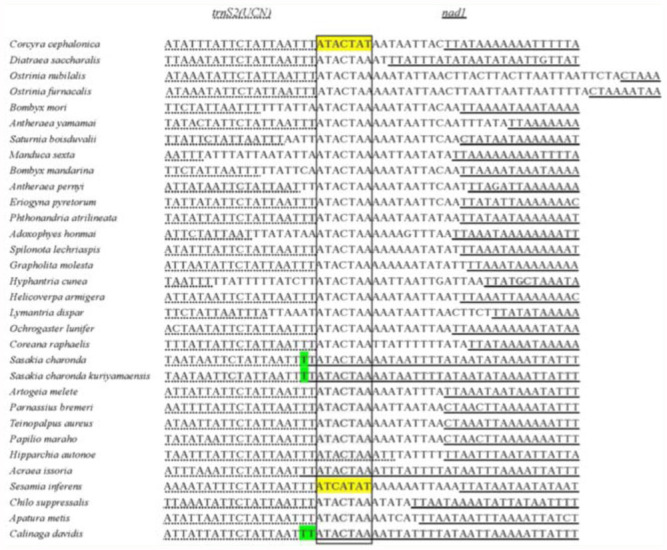
Alignment result of Spacer 4 tRNA-Ser(UCR)-NADI in 28 lepidopteran species. High quality figures are available online.
Spacer 4 (10 bp) was between nad1 and trnL(CUN). Ostrinia furnacalis and O. nubilalis also showed 10 bp spacers, while other lepidopteran spacers measured 1–6 bp.
Spacer 5 (351 bp) was A+T rich and found between rrnS and trnM with AT nucleotides (96.58%). There was a motif ATAGA followed by a 20 bp poly-T stretch downstream of rrnS, and two microsatellitelike regions (TA)9 and (TA)8. Finally, a 10 bp poly-A was present upstream of trnM. The feature was found to be common for other lepidopterans sequenced to date.
Overlapping sequences had a total of 35 bp spread over eight regions. Like other insect species (Adoxophyes) (35), atp8 and atp6 had a seven-nucleotide overlap (ATGATAA), known to be translated from the same cistronic mRNAs. The longest overlapping sequence (8 bp) was between trnW gene and trnC genes. The remaining overlapping sequences were all less than 6 bp.
Discussion
The Corcyra mitogenome is shorter than most lepidoteran mitogenomes previously reported. The shortest mitogenome is 15,140 bp (Artogeia) (Hong et al. 2009), and the longest is 15,928 bp (B. mandarina) (Pan et al. 2008). The Corcyra mitogenome had gene content and organization similar to other lepidopterans, which suggests that the mitochondrial gene arrangement in lepidopterans evolved independently after splitting from its stem lineage (Kim et al. 2006).
The most frequent amino acids in the Corcyra mitochondrial proteins were leucine, isoleucine, phenylalanine, and serine, all with high AT mutational bias that is a seemingly common feature in lepidopterans. Abascal et al. (2006) indicated that several arthropods have a new genetic code that translates the codon AGG as lysine instead of serineor arginine, these AGG reassignments may be events of parallel and correlated evolution between the arthropod genetic codes and the trnK/trnS. However, the variant codon, AGG, was not used by Corcyra.
The putative start codons of PCGs of the Corcyra mitogenome are ATNs, except for the CGA start codon of the cox1 gene. Although tetranucleotides TTAG and hexanucleotide TATTAG have also been proposed as start codons for the cox1 gene (Yukuhiro et al. 2002; Kim et al. 2006; Liu et al. 2008; Salvato et al. 2008; Kim SR et al. 2009), the TTAG lacks absolute conservation and may be of alternative function, not as an initiation codon (Margam et al. 2011). There are studies using ESTs (expressed sequence tags) to determine the cox1 start codon. For example, some dipterans have an unorthodox UCG serine initiation codon, which was confirmed through mitogenome EST data (Morlais and Severson 2002; Krzywinski et al. 2006; Stewart and Beckenbach 2009). Mitogenome ESTs and alignment of the mitogenome sequence from all lepidopterans had shown that arginine (CGR) functions as the start codon of the cox1 gene (Margam et al. 2011). These observations suggest that the use of EST data is valuable for the annotation of mitogenomes. The success of mitogenome sequencing will serve as the basis of the mating of EST and functional mitochondrial genome annotations.
Acknowledgements
Professor Qi-lian Qin and his lab members (Institute of Zoology, Chinese Academy of Sciences) kindly provided helpful advice and facilities for sequence cloning. We also thank Fu-qiang Chen and Xiao-he Wang (Institute of Zoology, Chinese Academy of Sciences) for their comments on the manuscript and help in specimen identification and data analysis. This project was supported mainly by the Public Welfare Project from the Ministry of Agriculture, China (Grant no. 201103024), Beijing Municipal Natural Science Foundation (6081002), and partially by the Innovation Program in the Chinese Academy of Sciences (KSCX2-EW-B-02/03). Grants from the National Science Foundation, China (30870268, J0930004) were awarded to Chaodong Zhu. Funding from the Shanxi Science and Technology Fund (2007031040-1) and the Academy of Agriculture Sciences (YGG0930) was given to Jie Li for screening and mass-culture of pest natural enemies.
Abbreviations
- mitogenome,
mitochondrial genome;
- PCGs,
protein-coding genes
References
- Abascal F, Posada D, Knight RD, Zardoya R. Parallel Evolution of the Genetic Code in Arthropod Mitochondrial Genomes. PLoS Biol. 2006;4(5):e127. doi: 10.1371/journal.pbio.0040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allotey J, Azalekor W. Some aspects of the biology and control using botanicals of the rice moth, Corcyra cephalonica (Stainton), on some pulses. Journal of Stored Product Research. 2000;36:235–243. doi: 10.1016/s0022-474x(99)00045-4. [DOI] [PubMed] [Google Scholar]
- Arunkumar KP, Metta M, Nagaraju J. Molecular phylogeny of silkmoths reveals the origin of domesticated silkmoth, Bombyx mori from Chinese Bombyx mandarina and paternal inheritance of Antheraea proylei mitochondrial DNA. Molecular Phylogenetics andEvolution. 2006;40:419–427. doi: 10.1016/j.ympev.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Research. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron SL, Johnson KP, Whiting MF. The Mitochondrial genome of the screamer Louse Bothriometopus (Phthiraptera: Ischnocera): effects of extensive gene rearrangements on the evolution of the genome. Journal of Molecular Evolution. 2007;65:589–604. doi: 10.1007/s00239-007-9042-8. [DOI] [PubMed] [Google Scholar]
- Cameron SL, Whiting MF. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 2008;408:112–123. doi: 10.1016/j.gene.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, Jin BR, Han YS, Kim I. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene. 2007;392:206–220. doi: 10.1016/j.gene.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Chaitanya RK, Dutta-Gupta A. Light chain fibroin and P25 genes of Corcyra cephalonica: Molecular cloning, characterization, tissue-specific expression, synchronous developmental and 20hydroxyecdysone regulation during the last instar larval development. General and Comparative Endocrinology. 2010;167:113–121. doi: 10.1016/j.ygcen.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. The mitochondrial DNA molecular of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. Journal of Molecular Evolution. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Coates BS, Sumerford DV, Hellmich RL, Lewis LC. Partial mitochondrial genome sequence of Ostrinia nubilalis and Ostrinia furnicalis. International Journal of Biological Sciences. 2005;1:13–18. doi: 10.7150/ijbs.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MB, Marangoni S, Macedo MLR. Insecticidal action of Annona coriacea lectin against the flour moth Anagasta kuehniella and the rice moth Corcyra cephalonica (Lepidoptera: Pyralidae). Comparative Biochemistry and Physiology, Part C: Toxicology and Pharmacology. 2007;146:406–414. doi: 10.1016/j.cbpc.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Damara M, Dutta-Gupta A. Identification of 86 kDa protein as methionine rich hexamerin in the rice moth, Corcyra cephalonica. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2010;157:229–237. doi: 10.1016/j.cbpb.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Damara M, Gullipalli D, Dutta-Gupta A. Cloning and expression of fat body hexamerin receptor and its identification in other hexamerin sequestering tissue of rice moth, Corcyra cephalonica. Journal of Insect Physiology. 2010;56:1071–1077. doi: 10.1016/j.jinsphys.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Damara M, Gullipalli D, Dutta-Gupta A. Ecdysteroid-mediated expression of hexamerin (arylphorin) in the rice moth, Corcyra cephalonica. Journal of Insect Physiology. 2010;56:1224–1231. doi: 10.1016/j.jinsphys.2010.03.026. [DOI] [PubMed] [Google Scholar]
- Fenn JD, Cameron SL, Whiting MF. The complete mitochondrial genome of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Molecular Biology. 2007;16:239–252. doi: 10.1111/j.1365-2583.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- Gullipalli D, Arif A, Aparoy P, Svenson GJ, Whiting MF, Reddanna P, Dutta-Gupta A. Identification of a developmentally and hormonally regulated Delta-Class glutathione S-transferase in rice moth Corcyra cephalonica. Comparative Biochemistry and Physiology, Part B: Biochemistry and Molecular Biology. 2010;156:33–39. doi: 10.1016/j.cbpb.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Heppner JB. Faunal regions and the diversity of Lepidoptera. Tropical Lepidoptera. 1991;2(1):1–85. [Google Scholar]
- Hong GY, Jiang ST, Yu M, Yang Y, Li F, Xue FS, Wei ZJ. The complete nucleotide sequence of the mitochondrial genome of the cabbage butterfly, Artogeia melete (Lepidoptera: Pieridae). Acta Biochimica et Biophysica Sinica. 2009;41:446–455. doi: 10.1093/abbs/gmp030. [DOI] [PubMed] [Google Scholar]
- Hu J, Zhang DX, Hao JS, Huang DY, Cameron S, Zhu CD. The complete mitochondrial genome of the yellow coaster, Acraea issoria (Lepidoptera: Nymphalidae: Heliconiinae: Acraeini): sequence, gene organization and a unique tRNA translocation event. Molecular Biology Reports. 2010;37(7):431–438. doi: 10.1007/s11033-009-9934-3. [DOI] [PubMed] [Google Scholar]
- Jalali SK, Venkatesana T, Murthya KS, Rabindraa RJ, Lalitha Y. Vacuum packaging of Corcyra cephalonica (Stainton) eggs to enhance shelf life for parasitization by the egg parasitoid Trichogramma chilonis. Biological Control. 2007;41(1):64–67. [Google Scholar]
- Jiang S, Hong G, Yu M, Li N, Yang Y, Liu Y, Wei Z. Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogynapyretorum (Lepidoptera: Saturniidae). International Journal of Biological Sciences. 2009;5:351–365. doi: 10.7150/ijbs.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Lee EM, Seol KY, Yun EY, Lee YB, Hwang JS, Jin BR. The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Molecular Biology. 2006;15(2):217–225. doi: 10.1111/j.1365-2583.2006.00630.x. [DOI] [PubMed] [Google Scholar]
- Kim SR, Kim MI, Hong MY, Kim KY, Kang PD, Hwang JS, Han YS, Jin BR, Kim I. The complete mitogenome sequence of the Japanese oak silkmoth, Antheraea yamamai (Lepidoptera: Saturniidae). Molecular Biology Reports. 2009;36(7):1871–1880. doi: 10.1007/s11033-008-9393-2. [DOI] [PubMed] [Google Scholar]
- Kim MI, Baek JY, Kim MJ, Jeong HC, Kim KG, Bae CH, Han YS, Jin BR, Kim I. Complete Nucleotide Sequence and Organization of the Mitogenome of the RedSpotted Apollo Butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and Comparison with Other Lepidopteran Insects. Molecules and Cells. 2009;28(31):347–363. doi: 10.1007/s10059-009-0129-5. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Molecular Phylogenetics and Evolution. 2006;39:417–423. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Lee ES, Shin KS, Kim MS, Park H, Cho S, Kim CB. The mitochondrial genome of the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae). Gene. 2006;373:52–57. doi: 10.1016/j.gene.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Lessinger AC, Junqueira AC, Lemos TA, Kemper EL, da Silva FR, Vettore AL, Arruda P, Azeredo-Espin AM. The mitochondrial genome of the primary screwworm fly Cochliomyia hominivorax (Diptera: Calliphoridae). Insect Molecular Biology. 2000;9:521–529. doi: 10.1046/j.1365-2583.2000.00215.x. [DOI] [PubMed] [Google Scholar]
- Li QQ, Duan YQ, Li DY, Liu XF, Xu HL, Zhou RM, Cao N, Li FL. Research Progress on Mitochondrial DNA of Lepidoptera Insect. Journal of Yunnan Agricultural University. 2009;24(5):746–753. [Google Scholar]
- Li W, Zhang X, Fan Z, Yue B, Huang F, King E, Ran J. Structural Characteristics and Phylogenetic Analysis of the Mitochondrial Genome of the Sugarcane Borer, Diatraea saccharalis (Lepidoptera: Crambidae). DNA and Cell Biology. 2010;30(1):3–8. doi: 10.1089/dna.2010.1058. [DOI] [PubMed] [Google Scholar]
- Liao F, Wang L, Wu S, Li YP, Zhao L, Huang GM, Niu CJ, Liu YQ, Li MG. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). International Journal of Biological Sciences. 2010;6:172–186. doi: 10.7150/ijbs.6.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YQ, Li YP, Pan MH, Dai FY, Zhu XW, Lu C, Xiang ZH. The complete mitochondrial genome of the Chinese oak silkmoth, Antheraeapernyi (Lepidoptera: Saturniidae). Acta Biochimica et Biophysica Sinica. 2008;40(8):693–703. [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margam VM, Coates BS, Hellmich RL, Agunbiade T, Seufferheld MJ, Sun W, Ba MN, Sanon A, Binso-Dabire CL, Baoua I, Ishiyaku MF, Covas FG, Srinivasan R, Armstrong J, Murdock LL, Pittendrigh BR. Mitochondrial Genome Sequence and Expression Profiling for the Legume Pod Borer Maruca vitrata (Lepidoptera: Crambidae). PLoS One. 2011;6(2):e16444. doi: 10.1371/journal.pone.0016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlais I, Severson DW. Complete mitochondrial DNA sequence and amino acid analysis of the cytochrome coxidase subunit I (COI) from Aedes aegypti. DNA Research. 2002;13:123–127. doi: 10.1080/10425170290030051. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan N, Porchezhian T, Venugopal MS, Janarthanan R. Recycling spent larval food of Corcyra cephalonica Stainton as a broiler feed ingredient. Bioresource Technology. 2003;86:39–44. doi: 10.1016/s0960-8524(02)00105-0. [DOI] [PubMed] [Google Scholar]
- Nagamanju P, Hansen IA, Burmester T, Meyer SR, Seheller K, Dutta-Gupta A. Complete sequence, expression and evolution of two members of the hexamerin protein family during the larval development of the rice moth, Corcyra cephalonica. Insect Biochemistry and Molecular Biology. 2003;33:73–80. doi: 10.1016/s0965-1748(02)00178-9. [DOI] [PubMed] [Google Scholar]
- Nardi F, Spinsanti G, Boore JL, Carapelli A, Dallai R, Frati F. Hexapod origins: Monophyletic or paraphyletic? Science. 2003;299:1887–1889. doi: 10.1126/science.1078607. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Pan MH, Yu QY, Xia YL, Dai FY, Liu YQ, Lu C, Zhang Z, Xiang ZH. Characterization of mitochondrial genome of Chinese wild mulberry silkworm, Bomyx mandarina (Lepidoptera: Bombycidae). Science in China Series C: Life Sciences. 2008;51(8):693–701. doi: 10.1007/s11427-008-0097-6. [DOI] [PubMed] [Google Scholar]
- Salvato P, Simonato M, Battisti A, Negrisolo E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics. 2008;9:331–345. doi: 10.1186/1471-2164-9-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Frati F, Bekenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain-reaction primers. Annals of the Entomological Society of America. 1994;87:651–701. [Google Scholar]
- Stewart JB, Beckenbach AT. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene. 2009;445:49–57. doi: 10.1016/j.gene.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Steinberg S, Cedergren R. Structural compensation in atypical mitochondrial tRNAs. Nature Structural Biology. 1994;1:507–510. doi: 10.1038/nsb0894-507. [DOI] [PubMed] [Google Scholar]
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochimica et Biophysica Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Türkera L. Theoretical study on the Sex-pheromones of the Rice Moth, Corcyra cephalonica Stainton. Turkish Journal of Biology. 1998;22:229–232. [Google Scholar]
- Wei SJ, Tang P, Zheng LH, Shi M, Chen XX. The complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A + T content and a long intergenic spacer between atp8 and atp6. Molecular Biology Reports 3. 2010;7:1931–1942. doi: 10.1007/s11033-009-9640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Molecular Biology and Evolution. 2002;19:1385–1389. doi: 10.1093/oxfordjournals.molbev.a004200. [DOI] [PubMed] [Google Scholar]
- Zhang DX, Szymura JM, Hewitt GM. Evolution and structural conservation of the control region of insect mitochondrial DNA. Journal of Molecular Evolution. 1995;40:382–391. doi: 10.1007/BF00164024. [DOI] [PubMed] [Google Scholar]
- Zhao JL, Zhang YY, Luo AR, Jiang GF, Cameron SL, Zhu CD. The complete mitochondrial genome of Spilonota lechriaspis Meyrick (Lepidoptera: Tortricidae). Molecular Biology Reports. 2010;38(6):3757–3764. doi: 10.1007/s11033-010-0491-6. [DOI] [PubMed] [Google Scholar]


