Abstract
We carried out a comprehensive overview of inhibitory effects of selected antibiotics on planktonic and biofilm cells of Staphylococcus aureus (ATCC 29213) and Pseudomonas aeruginosa (ATCC 27853) strains. The possible involvement of protease activity and the lipopolysaccharide (LPS) profile of P. aeruginosa were also analyzed. Biofilm cells of both strains were more resistant to antibiotics than their planktonic counterparts. Protease activity was increased in both strains in the biofilm forms. Challenge with sublethal doses of antibiotics also increased proteolytic activity of biofilm cells. Additionally, the LPS profile of P. aeruginosa showed pattern alterations of the biofilm that can contribute to biofilm resistance and survival. These observations provide evidence for the involvement of bacterial proteolytic activity and LPS profile in the resistance of biofilm bacteria to antibiotics compared to their planktonic counterparts.
Keywords: biofilm, Pseudomonas aeruginosa, Staphylococcus aureus, proteolytic activity, lipopolysaccharide
Many pathogenic and commensal bacteria are capable of transitioning between lifestyles in the environment and the human host.1 These bacteria must be able to adapt to sudden shifts in availability of nutrients and to primary and secondary host immune defenses.2 One particularly important and clinically relevant example of bacterial adaptation is the ability to grow as biofilms.3–5
Biofilms, a surface-associated bacterial community, are complex and ordered bacterial societies that are capable of growing in connection with different biological or inert surfaces.1 The major clinical consequence of different disease-causing bacteria correlates with the problems of therapeutic killing of attached cells.6 Biofilms are commonly associated with many health problems, such as endocarditis, otitis media, periodontitis, prostatitis, and urinary tract infections.7–10 Several bacteria, such as Escherichia coli, Staphylococcus aureus, Haemophilus influenza, and Pseudomonas aeruginosa, can form biofilms in the body tissues, leading to different infections.10–12 It has been estimated that biofilms account for two-thirds of the bacterial infections that physicians encounter, particularly in immunocompromised patients.13
Antibiotics have been used to treat patients with infectious diseases. They target important bacterial structures and cellular pathways, such as the cell wall, DNA, RNA, protein synthesis machinery, and bacterial metabolism.14 However, uncontrolled or long-term use of antibiotics results in the adaptation and development of resistance leading to treatment failure, prolonged or additional hospitalization, increased costs of care, and increased mortality.11,15 The mechanism of resistance of microbial biofilms to antibiotics is not clear. However, it seems to be multifactorial and may vary from one organism to another.16 In this study we investigated the possible involvement of proteolytic activity and lipopolysaccharides (LPSs) in increased resistance to antibiotics during the biofilm state.
Materials and methods
Bacterial strains and culture
Pseudomonas aeruginosa (ATCC 27853) and S. aureus (ATCC 29213) strains were obtained from the American type culture collection and cultivated on Mueller Hinton agar (Becton Dickinson and Company, Cockeysville, MD, USA) for 24 hours at 37°C under standardized aseptic conditions.
Antimicrobial agents
The following antimicrobial agents were used for susceptibility testing against S. aureus: cefaclor (cephalosporins) at a concentration of (32 μg/mL), amoxicillin (aminoglycosides; 32 μg/mL), cotrimoxazole (sulfonamides/folic acid antagonists; 32 μg/mL), and ciprofloxacin (fluoroquinolones; 0.125 μg/mL). We used amikacin (aminoglycosides, 0.25 μg/mL) and cotrimoxazole (32 μg/mL), ciprofloxacin (0.0625 μg/mL), and ceftazidime (32 μg/mL) (cephalosporins) for susceptibility testing against P. aeruginosa. All antibiotics were used as raw material, and purchased from Sigma-Aldrich, MI, USA.
Bacterial culture
Staphylococcus aureus and P. aeruginosa biofilms were developed as previously described17 under standardized aseptic conditions. Briefly, 100 μL of bacterial suspension from each strain was cultivated in polypropylene tubes containing 2 mL of trypticase soy broth (TSB) supplemented with 1% glucose (Becton Dickinson and Company, Cockeysville, MD, USA) for 48 hours at 37°C. Culture media was refreshed after 24 hours of incubation. After 48 hours of incubation, biofilm cells were harvested by discarding the culture media and washing the tubes three times with phosphate buffer saline (PBS; pH 7.2) to remove nonadherent bacteria; the adhered cells were then harvested by vortex and centrifugation. The pellet was suspended in PBS (pH 7.2) to achieve the desired turbidity (comparable to a McFarland turbidity standard of 0.5). Screening for biofilm formation was achieved as previously described.18 Briefly, after being emptied from their content, culture tubes were stained with trypan blue or safranin. Biofilms were judged by the appearance of a visible film lining the walls of the tube. Observations were carried out by three independent observers. Biofilms were scored as absent (score 0), weak (score 1), moderate (score 2), or strong (score 3). Average scores were used.
Determination of minimum inhibitory concentrations (MICs) of antibiotics for planktonic and biofilm cells
The MIC values of both S. aureus and P. aeruginosa planktonic and biofilm cells were tested against selected antibiotics. MICs were determined by using the broth macrodilution method.19 Briefly, 100 μL of adjusted bacterial suspensions equivalent to a 0.5 McFarland standard were added to a twofold serial dilution of selected antibiotics diluted in Mueller Hinton broth. The results were observed after 24 hours of incubation at 37°C. The lowest concentration of antibiotic needed to inhibit microbial growth compared to the control culture was defined as the MIC. Tests were performed in triplicate for each antibiotic.
Influence of sub-MICs of selected antibiotics on biofilm cells
To determine the effects of sub-MICs of antibiotics on S. aureus and P. aeruginosa biofilms, 100 μL of a bacterial biofilm suspension was added to TSB (supplemented with 1% glucose) containing sub-MICs of each antibiotic (for S. aureus: ciprofloxacin 32 μg/ml, cotrimoxazole 32 μg/ml, cefaclor 32 μg/ml, amoxicillin 32 μg/ml; and for P. aeruginosa: ciprofloxacin 8 μg/ml, amikacin 0.003 μg/ml, ceftazidime 32 μg/ml), and the suspension + antibiotic was then incubated at 37°C for 24 hours. After incubation, the antibiotics were removed by washing the tubes three times, and the cells were pelleted for further investigation.
Proteolytic activity assay
Total protease activity of S. aureus and P. aeruginosa in planktonic and biofilm cells was determined by the azocasein assay.20 Briefly, media from each bacterial strain (30 mL) was added to 50 mL azocasein substrate (2% azocasein (Sigma-Aldrich, MI, USA) in 10 mM Tris HCl, 8 mM CaCl2, pH 7.4). The reaction mixture was incubated for 20 hours. Thereafter, 240 mL 10% trichloroacetic acid was added, and the samples were mixed and allowed to stand for 15 minutes to ensure complete precipitation of undigested material. Tubes were centrifuged at 10,600 xg for 10 minutes, and 240 mL of the supernatant was transferred to tubes containing 280 mL 1.0 M NaOH. The absorbance at 440 nm was determined against a blank tube. One unit of enzyme activity corresponds to the absorbance at maximal digestion of 1 mg azocasein/hour.21 The protease activity was expressed as units/106 bacteria/hour.20
LPS extraction and analysis
We followed the LPS extraction kit guidelines (Intron Biotechnology, Kyungki-Do, Republic of Korea) to extract LPSs from P. aeruginosa planktonic and biofilm cells and biofilms induced with sub-MICs of antibiotics. The LPS profile was then determined using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) comprising a 4% stacking gel and a 12% separation gel.22 The LPS gel was then fixed and stained according to the method of Tsai and Frasch.23
Statistical analysis
Analysis was performed using GraphPad Prism software (version 4.0; GraphPad Software, Inc, La Jolla, CA). One-way analyses of variance followed by Dunnett’s post hoc test were used to determine any statistically significant difference. A P-value < 0.05 was considered significant.
Results
The MIC values of selected antibiotics against S. aureus and P. aeruginosa biofilm and planktonic cells were determined (Tables S1 and S2). The MIC values of biofilms were generally higher than their planktonic counterparts.
We determined protease activity of S. aureus and P. aeruginosa in order to evaluate the possible involvement of proteolytic activity in the resistance of the biofilm form of bacteria (Tables 1 and 2). Results demonstrated that control biofilm had significantly higher proteolytic activity than its planktonic counterpart. When biofilms cells were exposed to sub-MICs of selected antibiotics, most showed a slight but not significant increase in their proteolytic activity.
Table 1.
Protease activity of Staphylococcus aureus cells
| Samples | Proteolytic activity (units/106 bacteria/hour) |
|---|---|
| Planktonic | 2.00 ± 0.33 |
| Biofilm | 3.34 ± 0.55* |
| Biofilm treated with (1/4) MIC of ciprofloxacin | 2.44 ± 0.40 |
| Biofilm treated with (1/32) MIC of cefaclor | 2.88 ± 0.43 |
| Biofilm treated with (1/8 ) MIC of cotrimoxazole | 3.56 ± 0.65* |
| Biofilm treated with (1/16) MIC of amoxicillin | 6.44 ± 0.57* |
Notes: n = 4 experiments.
indicates significant difference from the planktonic group at P < 0.05.
Abbreviation: MIC, minimum inhibitory concentration.
Table 2.
Protease activity of Pseudomonas aeruginosa cells
| Samples | Proteolytic activity (units/106 bacteria/hour) |
|---|---|
| Planktonic | 2.89 ± 0.47 |
| Biofilm | 4.44 ± 0.38* |
| Biofilm treated with (1/8) MIC of ciprofloxacin | 5.33 ± 0.46* |
| Biofilm treated with (1/8) MIC amikacin | 5.78 ± 0.61* |
| Biofilm treated with (1/8) MIC ceftazidime | 5.10 ± 0.44* |
Notes: n = 4 experiments.
indicates significant difference from the planktonic group at P < 0.05.
Abbreviation: MIC, minimum inhibitory concentration.
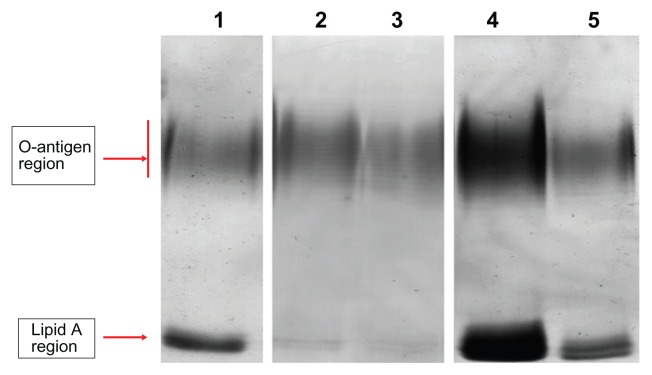
LPSs of the P. aeruginosa cell membrane also have an essential barrier function and directly affect bacterial susceptibility for antibiotics.24 We therefore analyzed the LPS profile by SDS–PAGE and silver stain. LPSs displayed a ladder-like pattern of bands with the slower migrating band of the LPS extract in the O-antigen region and the faster band in the lipid A region (Figure 1). In comparison to planktonic cells, biofilm-forming cells showed a different LPS profile; the faster migrating band (lipid A) had an increased staining intensity and a slightly decreased number of bands in the O-antigen region. In the presence of (1/8) MIC of ceftazidime, the number of bands in the O-antigen region increased and the faster migrating band (lipid A) decreased to being barely observable when compared with the control biofilm. For (1/4) MIC of ciprofloxacin and (1/8) MIC of amikacin, the number of bands in the O-antigen region decreased slightly and lipid A intensity increased.
Figure 1.
Electrophoretic profile of LPS of Pseudomonas aeruginosa.
Notes: Lane 1, LPS extracted from biofilm cells; lane 2, LPS extracted from planktonic cells; lane 3, LPS extracted from biofilm cells treated with (1/8) MIC of ceftazidime; lane 4, LPS extracted from biofilm cells treated with (1/4) MIC of ciprofloxacin; lane 5, LPS extracted from biofilm cells treated with (1/8) MIC of amikacin.
Abbreviations: LPS, lipopolysaccharide; MIC, minimum inhibitory concentration.
Discussion
Biofilm forms of bacteria are responsible for a variety of life-threatening infections. They have the ability to resist attack by host defenses and show resistance to most antibiotics.25,26 A wide range of pathogens, such as P. aeruginosa and S. aureus, are capable of forming biofilms. Both bacterial types are medically significant microbes and can cause implant and prosthetic device infections. Thus, assessment of possible mechanisms for antibiotic resistance in their biofilm form is critical.
Results of this study showed that proteolytic activity increases when bacteria switch from a planktonic to biofilm phenotype. This indicates that biofilms are more virulent and have a greater ability to cause tissue destruction, which correlates with the conclusions of previous studies.27–29 Additionally, the proteolytic potential slightly increased when biofilms were exposed to sublethal concentrations of selected antibiotics. This possibly explains results of clinical studies that show increased severity of disease when subtherapeutic doses or inadequate duration of antibiotics are used.30–33
LPSs are a major constituent of the P. aeruginosa membrane, and changes observed in membrane structure may result in changes to the antibiotic permeability barrier.34,35 For example, the presence of full-length O-antigen renders the LPS smooth, whereas absence or reduction of O-antigen makes the LPS rough. This represents a bacterial shift from an acute to chronic lifestyle, leading to increased persistence of bacteria and a consequent high relapse of disease.36 Results of our study showed decreased O-antigen and increased lipid A in biofilm-forming cells compared to planktonic cells, indicating a phenotypic switch in the LPSs from a smooth form to a rough form.37
Apart from an LPS role in resistance, LPSs are generally considered endotoxins.38 Accordingly, the increased virulence of P. aeruginosa biofilms compared to the planktonic form could be related to an increase in lipid A. In the LPS pattern of P. aeruginosa-treated biofilms, lipid A expression in biofilms exposed to amikacin and ciprofloxacin was up-regulated compared to untreated biofilms. These changes in LPS expression indicate that antibiotic-exposed biofilms had more virulence potential than untreated biofilms. Further studies are required to elucidate the mechanisms by which these antibiotics induce changes in LPSs.
In this study we investigated the effect of certain antibiotics on proteolytic activity of P. aeruginosa and S. aureus and/or membrane LPSs of P. aeruginosa. We chose antibiotics that are most commonly used for the treatment of infections by these two bacterial strains. Future work could cover other important antibiotics and also commonly used antibiotics, such as vancomycin and aztreonam. Studies should also address the possibility of membrane protein involvement in increased virulence of biofilms, especially when challenged with sublethal concentrations of antibiotics.
Collectively, the antibiotic susceptibility results presented in this study showed that biofilms are more tolerant to antimicrobial agents than planktonic forms. Biofilms (control and treated strains) revealed changes in proteolytic activity and LPS patterns that may result in antibiotic resistance. A decrease in O-antigen bands of LPSs could be a mechanism that helps biofilms evade the immune system, while increased lipid A contents may indicate an increase in biofilm endotoxicity. These LPS changes along with increased protease activity indicate that biofilms are more virulent than their planktonic counterparts.
Supplementary tables
Table S1.
Minimum inhibitory concentration values of Staphylococcus aureus planktonic and biofilm cells
| Antibiotics | Planktonic cells | Biofilm cells |
|---|---|---|
| Ciprofloxacin | 0.5 ± 0.1 μg/mL | 128 ± 25 μg/mL |
| Amoxicillin | 4 ± 0.9 μg/mL | 512 ± 110 μg/mL |
| Cotrimoxazole | 4 ± 0.0 μg/mL | 256 ± 60 μg/mL |
| Cefaclor | 8 ± 1.8 μg/mL | >1024 μg/mL |
Table S2.
Minimum inhibitory concentration values of Pseudomonas aeruginosa planktonic and biofilm cells
| Antibiotics | Planktonic cells | Biofilm cells |
|---|---|---|
| Ciprofloxacin | 0.125 ± 0.02 μg/mL | 64 ± 13 μg/mL |
| Ceftazidime | 2 ± 0.4 μg/mL | 256 ± 80 μg/mL |
| Cotrimoxazole | 256 ± 60 μg/mL | 512 ± 100 μg/mL |
| Amikacin | 2 ± 0.0 μg/mL | 0.02 ± 0.004 μg/mL |
Acknowledgment
This project was supported by a grant (No 37/2010) from the Deanship of Research at the Jordan University of Science and Technology.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Johnjulio W, Fuge LH, Kad M, Post C. Introduction to biofilms in family medicine. South Med J. 2012;105(1):24–29. doi: 10.1097/SMJ.0b013e31823c3ee4. [DOI] [PubMed] [Google Scholar]
- 2.Wolcott R, Dowd S. The role of biofilms: are we hitting the right target? Plast Reconstr Surg. 2011;127(Suppl 1):28S–35S. doi: 10.1097/PRS.0b013e3181fca244. [DOI] [PubMed] [Google Scholar]
- 3.Brooks JL, Jefferson KK. Staphylococcal biofilms: quest for the magic bullet. Adv Appl Microbiol. 2012;81:63–87. doi: 10.1016/B978-0-12-394382-8.00002-2. [DOI] [PubMed] [Google Scholar]
- 4.Aparna MS, Yadav S. Biofilms: microbes and disease. Braz J Infect Dis. 2008;12(6):526–530. doi: 10.1590/s1413-86702008000600016. [DOI] [PubMed] [Google Scholar]
- 5.Lynch AS, Robertson GT. Bacterial and fungal biofilm infections. Annu Rev Med. 2008;59:415–428. doi: 10.1146/annurev.med.59.110106.132000. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77(16):5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson SK, Costerton JW. Biofilm and penile prosthesis infections in the era of coated implants: a review. J Sex Med. 2012;9(1):44–53. doi: 10.1111/j.1743-6109.2011.02428.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang R, Li M, Gregory RL. Bacterial interactions in dental biofilm. Virulence. 2011;2(5):435–444. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlassova N, Han A, Zenilman JM, James G, Lazarus GS. New horizons for cutaneous microbiology: the role of biofilms in dermatological disease. Br J Dermatol. 2011;165(4):751–759. doi: 10.1111/j.1365-2133.2011.10458.x. [DOI] [PubMed] [Google Scholar]
- 10.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyd JM, McGrath J, Krishnamurthy A. Mechanisms of bacterial resistance to antibiotics in infections of COPD patients. Curr Drug Targets. 2011;12(4):521–530. doi: 10.2174/138945011794751519. [DOI] [PubMed] [Google Scholar]
- 12.Jensen PO, Givskov M, Bjarnsholt T, Moser C. The immune system vs Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2010;59(3):292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 13.Sawhney R, Berry V. Bacterial biofilm formation, pathogenicity, diagnostics and control: An overview. Indian J Med Sci. 2009;63(7):313–321. [PubMed] [Google Scholar]
- 14.Gaynor M, Mankin AS. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr Top Med Chem. 2003;3(9):949–961. doi: 10.2174/1568026033452159. [DOI] [PubMed] [Google Scholar]
- 15.Canton R, Morosini MI. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev. 2011;35(5):977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Mah TF. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol. 2008;190(13):4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cernohorska L, Votava M. Antibiotic synergy against biofilm-forming Pseudomonas aeruginosa. Folia Microbiol (Praha) 2008;53(1):57–60. doi: 10.1007/s12223-008-0008-z. [DOI] [PubMed] [Google Scholar]
- 18.Christensen GD, Simpson WA, Younger JJ, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22(6):996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute (CLSI) Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard. ninth edition. Villanova, PA: 2012. [Google Scholar]
- 20.Schmidtchen A, Wolff H, Hansson C. Differential proteinase expression by Pseudomonas aeruginosa derived from chronic leg ulcers. Acta Derm Venereol. 2001;81(6):406–409. doi: 10.1080/000155501317208336. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto T, Akaike T, Suga M, et al. Activation of human matrix metalloproteinases by various bacterial proteinases. J Biol Chem. 1997;272(9):6059–6066. doi: 10.1074/jbc.272.9.6059. [DOI] [PubMed] [Google Scholar]
- 22.Duan ZG, Yan XJ, He XZ, et al. Extraction and protein component analysis of venom from the dissected venom glands of Latrodectus tredecimguttatus. Comp Biochem Physiol B Biochem Mol Biol. 2006;145(3–4):350–357. doi: 10.1016/j.cbpb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 24.Hoekstra JL, de Neeling AJ, van Klingeren V, Stobberingh EE, van Boven CP. Resistant strains of Pseudomonas aeruginosa isolated after exposure to several beta-lactam antibiotics. Eur J Clin Microbiol. 1987;6(1):22–27. doi: 10.1007/BF02097185. [DOI] [PubMed] [Google Scholar]
- 25.Cos P, Tote K, Horemans T, Maes L. Biofilms: an extra hurdle for effective antimicrobial therapy. Curr Pharm Des. 2010;16(20):2279–2295. doi: 10.2174/138161210791792868. [DOI] [PubMed] [Google Scholar]
- 26.Khan W, Bernier SP, Kuchma SL, Hammond JH, Hasan F, O’Toole GA. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int Microbiol. 2010;13(4):207–212. doi: 10.2436/20.1501.01.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Hoiby N, Ciofu O, Johansen HK, et al. The clinical impact of bacterial biofilms. Int J Oral Sci. 2011;3(2):55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simoes M. Antimicrobial strategies effective against infectious bacterial biofilms. Curr Med Chem. 2011;18(14):2129–2145. doi: 10.2174/092986711795656216. [DOI] [PubMed] [Google Scholar]
- 30.Fluit AC, Schmitz FJ. Bacterial resistance in urinary tract infections: how to stem the tide. Expert Opin Pharmacother. 2001;2(5):813–818. doi: 10.1517/14656566.2.5.813. [DOI] [PubMed] [Google Scholar]
- 31.Rupp ME, Hamer KE. Effect of subinhibitory concentrations of vancomycin, cefazolin, ofloxacin, L-ofloxacin and D-ofloxacin on adherence to intravascular catheters and biofilm formation by Staphylococcus epidermidis. J Antimicrob Chemother. 1998;41(2):155–161. doi: 10.1093/jac/41.2.155. [DOI] [PubMed] [Google Scholar]
- 32.Hatt JK, Rather PN. Role of bacterial biofilms in urinary tract infections. Curr Top Microbiol Immunol. 2008;322:163–192. doi: 10.1007/978-3-540-75418-3_8. [DOI] [PubMed] [Google Scholar]
- 33.Frei E, Hodgkiss-Harlow K, Rossi PJ, Edmiston CE, Jr, Bandyk DF. Microbial pathogenesis of bacterial biofilms: a causative factor of vascular surgical site infection. Vasc Endovascular Surg. 2011;45(8):688–696. doi: 10.1177/1538574411419528. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez L, Breidenstein EB, Song D, Hancock RE. Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2012;56(2):1128–1132. doi: 10.1128/AAC.05336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breidenstein EB, de la Fuente-Nunez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Anuntagool N, Wuthiekanun V, White NJ, et al. Lipopolysaccharide heterogeneity among Burkholderia pseudomallei from different geographic and clinical origins. Am J Trop Med Hyg. 2006;74(3):348–352. [PubMed] [Google Scholar]
- 37.Coulon C, Vinogradov E, Filloux A, Sadovskaya I. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS One. 2010;5(12):e14220. doi: 10.1371/journal.pone.0014220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawasdidoln C, Taweechaisupapong S, Sermswan RW, Tattawasart U, Tungpradabkul S, Wongratanacheewin S. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One. 2010;5(2):e9196. doi: 10.1371/journal.pone.0009196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Minimum inhibitory concentration values of Staphylococcus aureus planktonic and biofilm cells
| Antibiotics | Planktonic cells | Biofilm cells |
|---|---|---|
| Ciprofloxacin | 0.5 ± 0.1 μg/mL | 128 ± 25 μg/mL |
| Amoxicillin | 4 ± 0.9 μg/mL | 512 ± 110 μg/mL |
| Cotrimoxazole | 4 ± 0.0 μg/mL | 256 ± 60 μg/mL |
| Cefaclor | 8 ± 1.8 μg/mL | >1024 μg/mL |
Table S2.
Minimum inhibitory concentration values of Pseudomonas aeruginosa planktonic and biofilm cells
| Antibiotics | Planktonic cells | Biofilm cells |
|---|---|---|
| Ciprofloxacin | 0.125 ± 0.02 μg/mL | 64 ± 13 μg/mL |
| Ceftazidime | 2 ± 0.4 μg/mL | 256 ± 80 μg/mL |
| Cotrimoxazole | 256 ± 60 μg/mL | 512 ± 100 μg/mL |
| Amikacin | 2 ± 0.0 μg/mL | 0.02 ± 0.004 μg/mL |