Abstract
Objective
Cardiac manifestations of neonatal lupus, comprising atrioventricular conduction defects and cardiomyopathy, occur in fetuses exposed to anti-Ro/SSA antibodies, and carry substantial mortality. There is strong evidence of a genetic contribution to the risk. This study was undertaken to evaluate single-nucleotide polymorphisms (SNPs) for associations with cardiac neonatal lupus.
Methods
Children of European ancestry with cardiac neonatal lupus (n = 116) were genotyped using the Illumina 370K SNP platform and merged with 3,351 controls. Odds ratios (ORs) and 95% confidence intervals (95% CIs) for association with cardiac neonatal lupus were determined.
Results
The 17 most significant associations with cardiac neonatal lupus were found in the HLA region. The region near the MICB gene showed the strongest variant (rs3099844; Pdom = 4.52 × 10−10, OR 3.34 [95% CI 2.29–4.89]), followed by a missense variant within C6orf10 (rs7775397; Pdom = 1.35 × 10−9, OR 3.30), which lies between NOTCH4 and BTNL2, and several SNPs near the tumor necrosis factor α gene, including rs2857595 (Padd = 1.96 × 10−9, OR 2.37), rs2230365 (Padd = 1.00 × 10−3, OR 0.46), and rs3128982 (Padd = 6.40 × 10−6, OR 1.86). Outside the HLA region, an association was detected at 21q22, upstream of the transcription regulator ets-related isoform 1 (rs743446; P = 5.45 × 10−6, OR 2.40). HLA notwithstanding, no individual locus previously implicated in autoimmune diseases achieved genome-wide significance.
Conclusion
These results suggest that variation near genes related to inflammatory and apoptotic responses may promote cardiac injury initiated by passively acquired autoantibodies.
Since the first reports of congenital heart block as an autoimmune disease published >30 years ago (1–3), the consistency with which the presence of maternal antibodies directed against components of the Ro/SSA–La/SSB RNP complex has been demonstrated is remarkable. Injury to the fetal heart most often occurs during weeks 18–24 of gestation and is presumed to be dependent on IgG transport by neonatal Fc receptor of maternal IgG autoantibodies (4,5). Maternal health status accompanying the production of the putative autoantibodies is not a risk factor for fetal disease, since many mothers are clinically asymptomatic at the time heart block is detected, and only then are antibodies sought and identified (6). Although advanced conduction abnormalities are the signature phenotype of cardiac neonatal lupus, the spectrum of injury can extend to the myocardium and endocardium (7,8). Third-degree block is not reversible and carries a significant risk of mortality (20–30%, primarily fetal and neonatal) and morbidity (with >60% of surviving children requiring permanent pacing before adulthood) (9).
Fetal exposure to maternal anti-Ro/SSA antibodies is a necessary but not sufficient condition for cardiac neonatal lupus. Three prospective studies of women with the candidate antibodies have estimated the risk of cardiac neonatal lupus to be 2% if the mother has had no previously affected pregnancies (10–12). Accordingly, other factors must be required to promote cardiac injury. Evidence of a fetal genetic factor in the development of cardiac disease is empirically supported by an estimated recurrence rate in subsequent pregnancies that is ~10-fold higher than the risk to the anti-Ro/SSA–positive mother with no affected children (13). The estimated sibling recurrence risk ratio, λs, is ~3,000; that is, siblings of affected infants have a 3,000 times higher risk than the population prevalence of cardiac neonatal lupus per se, which is ~1:15,000 live births (14–16). Since the maternal anti-Ro/SSA antibodies are required for disease expression, conditioning on maternal anti-Ro/SSA antibodies yields a λs of ~10. Although supportive, data on concordance in monozygotic twins are very limited. Of 6 twin sets in which maternal anti-Ro/SSA antibodies were reported to be present and monozygosity stated, 2 (33%) were concordant for cardiac neonatal lupus (9,17–20).
We posit that genetic polymorphisms influence the fetal response to maternal autoantibodies. Herein, we report the first genome-wide association study in children with cardiac neonatal lupus associated with maternal anti-Ro/SSA antibodies. The identification of fetal genes predisposing to cardiac scar might provide insight into the pathogenesis of the disease and delineate risk profiles.
PATIENTS AND METHODS
Patients
All cases included in this study (n = 116) were members of families enrolled in the US-based Research Registry for Neonatal Lupus (RRNL) (9). A fetus, neonate, or child was considered to have cardiac neonatal lupus if the following criteria were met: 1) the presence of a conduction defect (first-degree, second-degree, or third-degree heart block) documented by electrocardiogram (EKG) (in the case of first-degree block, a prolonged PR interval detected solely by in utero echocardiogram was insufficient), echocardiogram, history of pacemaker, or statement in the medical record; and/or presence of cardiac injury which specifically included autopsy evidence of a mononuclear infiltrate in the endocardium, myocardium, and pericardium; and/or endocardial fibroelastosis on echocardiogram always associated with cardiac dysfunction and 2) presence of antibodies to the 52-kd Ro/SSA, 60-kd Ro/SSA, or 48-kd La/SSB RNPs in the maternal serum, as determined by enzyme-linked immunosorbent assay using recombinant proteins or by a commercial laboratory (90% of cases were tested in the research laboratory of JPB and RMC). The 116 cases with cardiac neonatal lupus were compared with 3,351 Caucasian controls genotyped on the Illumina HAP300 SNP chip (~317,000 single-nucleotide polymorphisms [SNPs]) from the Consortium on Systemic Lupus Erythematosus (SLE) Genetics (21).
Genotyping
Genomic DNA samples (750 ng) isolated from RRNL cases (described above) were genotyped on the Illumina BeadStation 500GX following standard manufacturing protocols. Initially, 77% of the samples were genotyped on the Illumina HumanCNV370-Duo_v1 array (370,404 SNPs), and 23% of the samples on the Illumina HumanCNV370-Duo_v3 array (373,398 SNPs). There were 346,110 SNPs common to the HumanCNV370-Duo_v1 and HumanCNV370-Duo_v3 arrays (92.7% overlap). Genotypes were called using the Illumina BeadStudio 3.1 software package using previously generated cluster positions generated by Illumina. Only samples with genotype call rates >93% were included in the genome-wide association analysis. Cluster plots of the strongest associated SNPs were examined for genotype calling quality.
Autoimmune disease loci
A set of 374 SNPs on the Illumina 317K SNP chip that had previously been implicated as being associated with autoimmune diseases were identified from published genome-wide association studies; 207 of these passed our statistical quality control standards. The published genome-wide scans that were reviewed had data on ≥100,000 SNPs and are available from the National Human Genome Research Institute’s Catalog of Published Genome-Wide Association Studies (www.genome.gov/gwastudies) (22). All SNPs considered were reported to have a P for association value of less than 1.0 × 10−5 or were explicitly highlighted by the authors.
Statistical analysis
Admixture and SNP statistical quality control
To account for the potential confounding influences of population substructure, a principal components analysis (PCA) was computed using all SNPs with <5% missing data, no differential missingness between cases and controls (P < 0.05), no significant departures from Hardy-Weinberg equilibrium expectations (P < 0.0001 for cases and P < 0.01 for controls), and minor allele frequency >0.05. Due to the complexity of performing a PCA on >300K SNPs in ~3,500 individuals, recent advanced numerical algebraic techniques were used to reduce computation time (23,24). Velicer’s algorithm (25) and the Tracy-Widom test (26,27) were used to identify the number of principal components. The association analyses (described below) were computed adjusting for the first 3 principal components from this analysis. At the outset of our study, genome-wide analysis was performed on 120 cardiac neonatal lupus cases. Four individuals were removed due to relatedness (1 of each of 4 pairs of siblings).
Association analysis
Testing for association was completed using the program SNPGWA (www.phs.wfubmc.edu). For each SNP, missing data proportions for cases and controls, minor allele frequency, and exact tests for departures from Hardy-Weinberg equilibrium were calculated. The following 5 tests of genotypic association were computed: 2df overall test for 2 × 3 tables, dominant model, additive model (Cochran-Armitage trend test), recessive model, and lack-of-fit to an additive model. The genetic model and odds ratios (ORs) are defined relative to the minor allele. At least 10 individuals who were homozygous for the minor allele were required before computing the recessive model. The primary inference for this study was based on the additive genetic model, unless the lack-of-fit to an additive model was statistically significant (P < 0.05). When the lack-of-fit test was significant, then the minimum P value from the dominant, additive, or recessive models was used. ORs, 95% confidence intervals (95% CIs), and P values were calculated for all models.
RESULTS
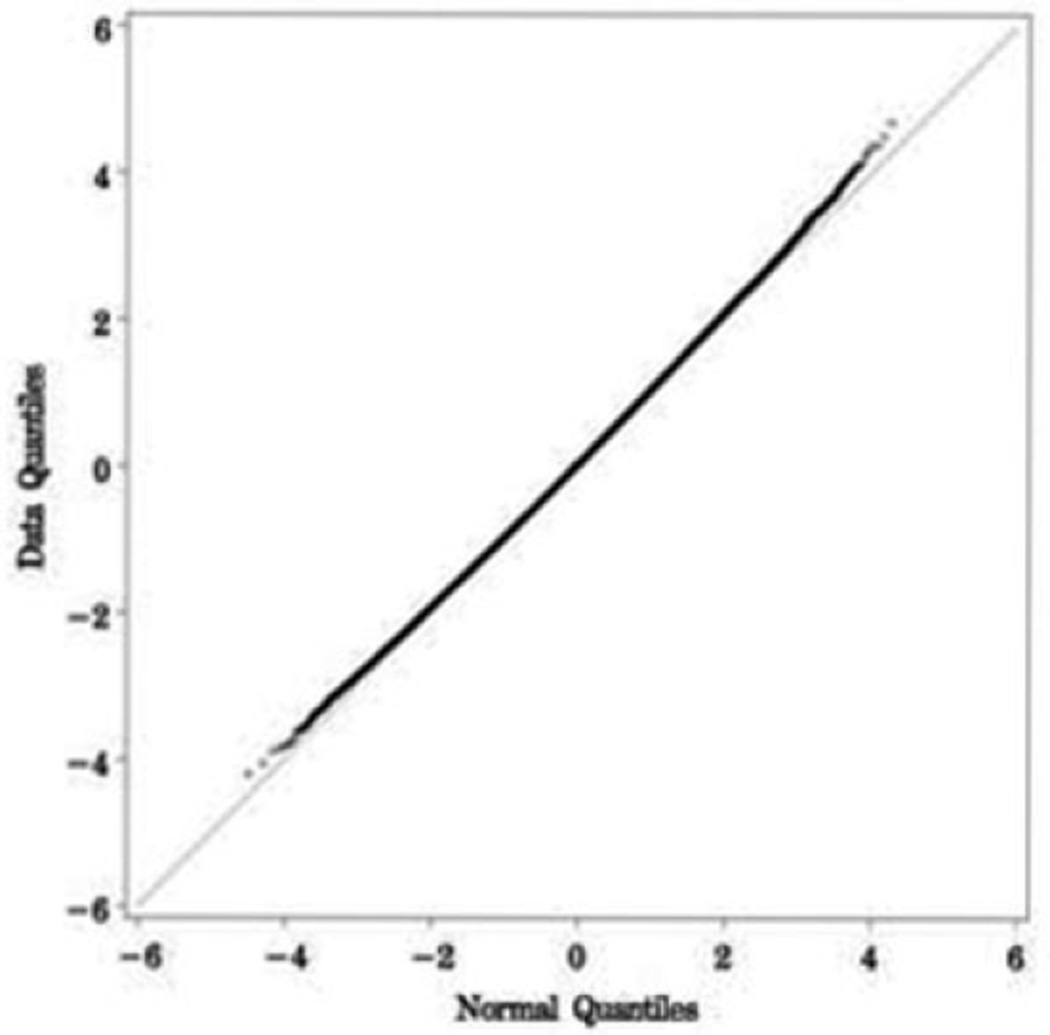
The 116 cardiac neonatal lupus cases and 3,351 out-of-study controls were all of self-reported Caucasian ancestry (Table 1), which was consistent with the PCA. The case group was 51.7% female, and the control group was 65.7% female. Advanced heart block was present in 96% of the cases. PCA suggested adjusting for 3 principal components. Adjusting for these 3 principal components accounted for 9.8% of the genetic variation. To examine the inflation of the test statistics due to potential sources of bias (e.g., population substructure), the chi-square value from the additive genetic model adjusting for these 3 principal components via logistic regression was compared with its theoretical mean of 1. An inflation factor of 1.026 was observed, suggesting little evidence of inflation in the test statistic. Similarly, the Q–Q plot provided no evidence of systematic bias (Figure 1).
Table 1.
Demographic characteristics of the 116 patients from the Research Registry for Neonatal Lupus*
| Caucasian | 116 (100) |
| Female† | 60 (51.7) |
| Second-degree or third-degree heart block | 107 (92) |
| Second-degree or third-degree heart block and cardiomyopathy | 2 (1.7) |
| Second-degree heart block | 2 (1.7) |
| First-degree heart block | 2 (1.7) |
| Isolated cardiomyopathy | 1 (0.8) |
| First-degree heart block and cardiomyopathy | 2 (1.7) |
Values are the number (%) of patients.
The control group was 65.7% female.
Figure 1.
Q–Q plot of the genome-wide association study of cardiac neonatal lupus. The number and magnitude of observed associations (data quantile) were compared with expectations under no association (normal quantile). Each point represents 1 single-nucleotide polymorphism (of 346,110).
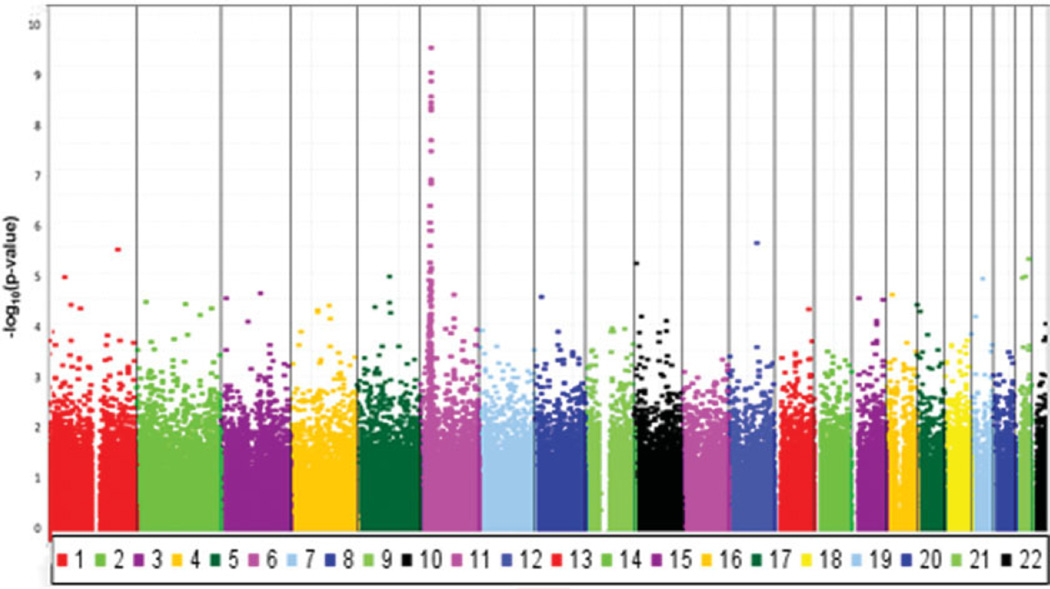
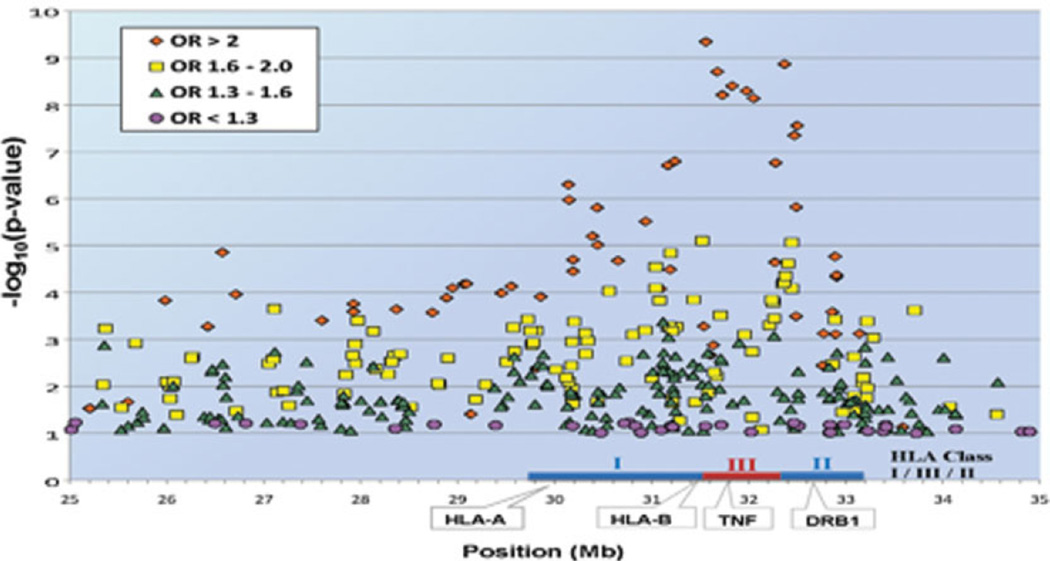
Combined association results for the entire genome are shown in Figure 2. The 17 SNPs with the most significant associations resided in the HLA region at 6p21.3. As indicated by the farthest outlier in the Q–Q plot (Figure 1), the strongest association across the entire genome was found at rs3099844 (Pdom = 4.52 × 10−10, OR 3.34 [95% CI 2.29–4.89]), which lies in a region of copy number variation (http://projects.tcag.ca/variation). Interestingly, rs3099844 is near the class III major histocompatibility complex (MHC) region and 94 kb from the tumor necrosis factor α (TNFα) gene, which contains a polymorphism rs1800629 (SNP not genotyped) that is associated with cardiac neonatal lupus (Figure 3). (See Supplementary Table 1, available on the Arthritis & Rheumatism Web site at http://www3.interscience.wiley.com/journal/76509746/home, for a list of the most significant associations with cardiac neonatal lupus in the HLA region.)
Figure 2.
Combined association results for the entire genome. Each point represents 1 single-nucleotide polymorphism (of 370,000) versus the P value.
Figure 3.
Graphic analysis of the HLA–A and HLA–B single-nucleotide polymorphisms with P < 0.1. OR = odds ratio.
The rs3099844 polymorphism is also near several flanking variants in the NFKBIL1–LTA–TNF–LTB–AIF1 region, including a signal that is located between the NCR3 and AIF1 genes (rs2857595; Padd = 1.96 × 10−9, OR 2.37 [95% CI 1.79–3.14]). In addition, within this region, there is evidence suggestive of association (rs2230365; P = 1.00 × 10−3, OR 0.46 [95% CI 0.29–0.74] and rs3128982; P = 6.40 × 10−6, OR 1.86 [95% CI 1.41–2.43]). Outside of the NFKBIL1–LTA–TNF–LTB–AIF1 region, there is an intronic variant in HLA class III in the MSH5 gene, which is the next strongest association in this region (rs3131379; Padd = 3.95 × 10−9, OR 2.59 [95% CI 1.89–3.56]).
Within HLA class I, associated regions included PSORS1C1 (rs3130544; P = 1.94 × 10−7, OR 2.77 [95% CI 1.89–4.07]) and missense mutations within transcription factor 19 (TCF19) (rs7750641; Pdom = 1.58 × 10−7, OR 2.79 [95% CI 1.90–4.09]). The most centromeric side of the class I region contains a variant in proximity to the RING-finger protein 39 (RNF39) (rs9261290; Pdom = 1.58 × 10−7, OR 2.65 [95% CI 1.79–3.93]). Within HLA class II, there was a significant association between cardiac neonatal lupus and a missense variant within C6orf10 (rs7775397; Pdom = 1.35 × 10−9, OR 3.30 [95% CI 2.24–4.86]), which lies between NOTCH4 and butyrophilin-like protein 2 (BTNL2) in the class III–class II boundary.
Outside the HLA region, an interesting association was observed between rs743446 (21q22) and cardiac neonatal lupus, near ets-related isoform 1 (ERG) (Prec = 5.45 × 10−6, OR 2.4 [95% CI 1.64–3.49]; minor allele frequency 0.64 in cases and 0.49 in controls) (Table 2). Importantly, flanking SNPs were noted. Although they were not within the top 200 statistical associations, the multiple SNPs in proximity to rs743446 were within an extensive linkage disequilibrium block providing weak evidence of association (P < 0.005) (see Supplementary Table 2, available on the Arthritis & Rheumatism Web site at http://www3.interscience.wiley.com/journal/76509746/home).
Table 2.
Strongest statistical associations with cardiac neonatal lupus in non-HLA regions*
| Marker | Chr. | Position (kb) |
Nearest genes | MAF in cases |
MAF in controls |
OR (95% CI) | Model | P |
|---|---|---|---|---|---|---|---|---|
| rs743446 | 21 | 38978 | 54 kb from NCRNA00114; 22 kb from ERG | 0.64 | 0.49 | 2.40 (1.64–3.49) | Recessive | 5.45 × 10−6 |
| rs2403106 | 12 | 82532 | 480 kb from TMTC2 | 0.15 | 0.07 | 2.48 (1.70–3.61) | Additive | 2.62 × 10−6 |
| rs1890645 | 1 | 193798 | None within 500 kb | 0.38 | 0.30 | 2.98 (1.88–4.73) | Recessive | 3.52 × 10−6 |
| rs1391511 | 10 | 4709 | 149 kb from AKR1E2; 15 kb from LOC338588 | 0.50 | 0.36 | 1.84 (1.41–2.40) | Additive | 6.62 × 10−6 |
Chr. = chromosome; MAF = minor allele frequency; OR = odds ratio; 95% CI = 95% confidence interval.
Associations outside the HLA region were also noted at rs2403106 (12q21; Padd = 2.62 × 10−6, OR 2.48 [95% CI 1.70–3.61]) and rs1391511 (10p15; Padd = 6.6 × 10−6, OR 1.84 [95% CI 1.41–2.4]). Neither of these polymorphisms was in proximity to any known genes or structural variation. Also, rs1890645, in a region of a known copy number polymorphism, was associated with cardiac neonatal lupus (1q31; Prec = 3.52 × 10−6, OR 2.98 [95% CI 1.88–4.73]).
Given that this sample is the world’s largest for any genetic study of cardiac neonatal lupus and the only genome-wide association study, it is important to highlight some suggestive associations that others might use to attempt more focused association studies (see Supplementary Table 1, available on the Arthritis & Rheumatism Web site at http://www3.interscience.wiley.com/journal/76509746/home). In the context of the functional category of cell adhesion, SNP rs2432143 is within an intron of ITGA1 at 5q11 (Pdom = 4.5 × 10−5, OR 2.31 [95% CI 1.54–3.45]). Within 1p34.2, rs9960 (Padd = 1.2 × 10−5, OR 1.94 [1.44–2.62]) is in the region containing the erythroblast membrane–associated protein (ERMAP), a gene that also plays a role in cell adhesion. In proximity to the CD200 cell surface glycoprotein receptor 2 (CD200R1L) resides rs6438101 (3q13.2; Prec = 2.5 × 10−5, OR 2.29 [95% CI 1.56–3.36]). In a group related to membrane potential, at 1q43, rs1072319 and rs1578180 are 2 SNPs at an intron of the cholinergic receptor, muscarinic 3 (CHRM3), with suggestive associations at Prec = 7.4 × 10−4 (OR 3.2 [1.63–6.4]) and Prec = 7.8 × 10−4 (OR 3.22 [1.63–6.37]), respectively.
SNP rs6767890, a candidate involving lipid metabolism, was associated with cardiac neonatal lupus (3p24; Pdom = 1.2 × 10−5, OR 2.25 [95% CI 1.54–2.30]) and is within the raft-linking protein (RFTN1). RFTN1 is expressed in B cells, may regulate B cell antigen receptor–mediated signaling, and may be important in the formation and maintenance of lipid rafts. With regard to a function of signal transduction, an example is rs1913342, which is located at locus 1q42, within intron 1 of CDC42BPA (Prec = 6.55 × 10−4, OR 4.12 [95% CI 1.81–9.23]). Also within this group, SNP rs3746314 (19q12; Pdom = 1.3 × 10−5, OR 2.37 [95% CI 1.61–3.48]) is near the Pleckstrin homology domain-containing family F member 1 (PLEKHF1) gene, an apoptosis-inducing protein gene that is expressed in the heart and placenta. In a category of candidates with unknown function, the SNP rs29911 (5q15; Padd = 1.2 × 10−5, OR 2.17 [95% CI 1.53–3.07]) is in the region containing KIAA0825 and is located in a known copy number region. At 21q22.3, a variant is in proximity to WD repeat domain 4 protein (WDR4) (rs2839586; Prec = 2.0 × 10−4, OR 4.63 [95% CI 2.04–10.53]) and rs2839597 (11 kb downstream from rs2839586, in the promoter region of WDR4) (Prec = 9.5 × 10−4, OR 3.94 [95% CI 2.08–9.68]).
To test whether polymorphisms implicated in other autoimmune diseases might influence the risk for cardiac neonatal lupus, 204 SNPs reported in genome-wide association studies of autoimmune diseases that passed our quality control filters were screened for evidence of association (Table 3). (For additional data, see Supplementary Table 3, available on the Arthritis & Rheumatism Web site at http://www3.interscience.wiley.com/journal/76509746/home.) Not surprisingly given the overall sample size, the data did not support a significant association of any previously described autoimmune-associated polymorphisms with cardiac neonatal lupus. The highest association was displayed by 3 SNPs in the IRF5 region, which is one of the genes with a most robustly established association with SLE (21) (rs10239340; Padd = 2.40 × 10−3, OR 0.64 [95% CI 0.48–0.85]). It is interesting to note that if the autoimmune disease loci do not correlate with cardiac neonatal lupus, then the expected inflation factor for these loci should coincide with that of the rest of the genome (i.e., 1.01). However, the observed inflation factor for the autoimmune-associated SNPs was significantly larger (i.e., 1.22; P < 0.03), suggesting collectively an enrichment of association with these loci. Whether this pattern reflects maternal enrichment of these risk variants and/or cardiac neonatal lupus loci requires further study. As noted above and consistent with many autoimmune diseases, the HLA region contained several strong associations (Figure 3 and Supplementary Table 1, available on the Arthritis & Rheumatism Web site at http://www3.interscience.wiley.com/journal/76509746/home).
Table 3.
Screening of SNPs previously found to be associated with autoimmune diseases for associations with cardiac neonatal lupus*
| Marker | Chr. | Position (kb) |
Locus | MAF in cases |
MAF in controls |
OR (95% CI) | P | Associated diseases |
|---|---|---|---|---|---|---|---|---|
| rs7517847 | 1 | 67454 | IL23R | 0.42 | 0.42 | 0.56 (0.31–1.01) | 5.47 × 10−2† | CD, IBD |
| rs1801274 | 1 | 159746 | FCGR2A | 0.57 | 0.49 | 1.36 (1.04–1.78) | 2.69 × 10−2‡ | UC |
| rs2022013 | 1 | 181620 | NMNAT2 | 0.34 | 0.42 | 0.72 (0.55–0.95) | 2.19 × 10−2‡ | SLE |
| rs13017599 | 2 | 61018 | REL | 0.27 | 0.34 | 0.71 (0.53–0.96) | 2.35 × 10−2‡ | RA |
| rs3197999 | 3 | 49697 | MST1 | 0.23 | 0.30 | 0.63 (0.43–0.93) | 1. × 10−2§ | CD |
| rs6439924 | 3 | 141652 | CLSTN2 | 0.23 | 0.18 | 1.46 (1.07–1.99) | 1.78 × 10−2‡ | CD |
| rs6897932 | 5 | 35910 | IL7R | 0.21 | 0.25 | 0.67 (0.46–0.99) | 4.42 × 10−2§ | MS, type 1 DM |
| rs4613763 | 5 | 40428 | PTGER4 | 0.17 | 0.11 | 1.59 (1.11–2.27) | 1.07 × 10−2‡ | CD |
| rs20541 | 5 | 132024 | IL13 | 0.15 | 0.19 | 0.67 (0.44–1.03) | 6.81 × 10−2§ | Psoriasis |
| rs729302 | 7 | 128356 | IRF5-TNPO3 | 0.24 | 0.31 | 0.59 (0.41–0.87) | 6.95 × 10−3§ | SLE |
| rs10239340 | 7 | 128456 | IRF5-TNPO3 | 0.28 | 0.37 | 0.64 (0.48–0.85) | 2.40 × 10−3‡ | SLE |
| rs12537284 | 7 | 128505 | IRF5-TNPO3 | 0.18 | 0.13 | 1.45 (1.03–2.05) | 3.24 × 10−2‡ | SLE |
| rs7111341 | 11 | 2170 | INS | 0.31 | 0.25 | 1.32 (1.00–1.75) | 4.76 × 10−2‡ | Type 1 DM |
| rs4788084 | 16 | 28447 | IL27 | 0.34 | 0.39 | 0.76 (0.58–1.01) | 6.07 × 10−2‡ | Type 1 DM |
| rs8049439 | 16 | 28745 | IL27 | 0.31 | 0.38 | 0.73 (0.55–0.97) | 3.02 × 10−2‡ | IBD |
SNPs = single-nucleotide polymorphisms; CD = Crohn’s disease; IBD = inflammatory bowel disease; UC = ulcerative colitis; SLE = systemic lupus erythematosus; RA = rheumatoid arthritis; MS = multiple sclerosis; type 1 DM = type 1 diabetes mellitus (see Table 2 for other definitions).
Recessive model.
Additive model.
Dominant model.
DISCUSSION
Through the application of genome-wide association methods and subsequent replication studies, it is anticipated that validated markers will emerge that can be used as novel diagnostic and prognostic tools for risk stratification in counseling mothers with anti-Ro/SSA antibodies. In this first genome-wide association study of 116 Caucasian children with cardiac neonatal lupus, the most strongly associated variants (MICB region at class I, NFKBIL1–LTA–TNF–LTB–AIF1 at class III, and C6orf10 at class II) were found in the MHC region, a locus with extensive linkage disequilibrium. Outside the HLA locus, an association was identified at locus 21q22, 22 kb upstream of ERG. With the exception of the HLA region, no locus previously implicated in autoimmune diseases was found to have genome-wide significance in children with cardiac neonatal lupus.
In recent genome-wide association studies of SLE patients, the most significant association was found in the HLA region at 6p21.3. In this 7.05-Mb region, 93 SNPs had a P value of less than 10−6, a result which represents the long-range linkage disequilibrium related to the extended HLA–A1;B8;DR3 haplotype (28). The association of this extended haplotype with the generation of antibodies to Ro/SSA and La/SSB in the context of Sjögren’s syndrome and SLE has been consistently demonstrated (29,30). The class III TNFα polymorphism at position −308 (TNFα, rs1800629, TNFα –308A polymorphism-TNF2 allele, proinflammatory), which is also part of the extended haplotype, is associated with a number of autoimmune diseases, including Sjögren’s syndrome (31), SLE (32), subacute cutaneous lupus erythematosus (33), rheumatoid arthritis (34), and ulcerative colitis (35). In earlier limited studies of families enrolled in the RRNL (36), the TNF2 allele was significantly overrepresented in the mothers as well as the affected and unaffected siblings compared with healthy controls. The present study corroborated these findings in children with cardiac neonatal lupus. Specifically, 2 SNPs, which are in proximity to rs1800629, reached genome wide-significance (rs3099844 [Pdom = 4.5 × 10−10] and rs2857595 [Padd = 1.96 × 10−9]). Notably, there is strong linkage disequilibrium between rs3099844 and rs1800629 (r2 = 0.64) and between rs2857595 and rs1800629 (r2 = 0.86). What is yet to be established is whether the HLA candidates reflect the inheritance of maternal associations expected in women with anti-Ro/SSA and anti-La/SSB antibodies. Association analysis in these families using the transmission disequilibrium test (TDT) will help to elucidate this relationship.
The genetic factors may represent a dual hit by differentially influencing disease in this maternal/fetal dyad. On the maternal side the genes promote the necessary autoantibodies, and on the fetal side the genes promote tissue inflammation in a permissive in utero environment, e.g., hypoxia. The functional biology is supported by an in vitro model of cardiac neonatal lupus in which TNFα was secreted by macrophages cocultured with anti-Ro/SSA–bound human fetal cardiocytes (37).
The identification of associations with HLA class I genes in the present study is consistent with the results of a prior study of 40 children with cardiac neonatal lupus that revealed an enrichment of HLA-Cw7 (38). HLA-Cw7 is inclusive of the PSORS1 region (which is associated with psoriasis [39]). In the present study, associations in this region were found for rs3130544 and rs7750641. The functional effects of these candidate genes within PSORS1 may relate to stimulation of receptors for class I MHC and dysfunction of natural killer (NK) or T cell self tolerance. A combination of class I MHC and ligand (on NK or T cells) may augment susceptibility to autoimmune injury during pregnancy.
Cardiac neonatal lupus was also associated with non-HLA regions, including loci at 21q22, 12q21, 1q31, and 10p15. A cluster of associated SNPs at 21q22 is in proximity to ERG-ETS2/WDR4. ERG is a transcription factor that serves as a “brake” to both apoptosis and inflammation, components previously described in the cascade to injury and replacement of the atrioventricular node by fibrosis (36,40,41). Yi and coworkers demonstrated that ERG protects fibroblasts against apoptosis induced by serum deprivation (42). Recently, Yuan and coworkers demonstrated that ERG plays a role in repressing the expression of interleukin-8 (IL-8) (43), a mediator of inflammatory cell accumulation produced by numerous cell types, including macrophages and fibroblasts. In addition, it has been demonstrated that ERG plays a role in augmenting the expression of transforming growth factor β (TGFβ) receptor type II (44). Thus, a polymorphism associated with low expression or diminished function of the encoded protein may represent the absence of a needed protective factor for fetuses exposed to maternal anti-Ro/SSA antibodies.
In psoriasis, risk alleles also reside at non-HLA genes such as chromosomes 1 and 5 (45), which include IL23R and IL13, respectively. Although they did not reach genome-wide significance, SNPs associated with each of these genes were associated with cardiac neonatal lupus. IL-13 is a prototypic Th2 cytokine that is also strongly profibrotic. Mice deficient in IL-13 are protected against a fluorescein isothiocyanate–induced model of lung fibrosis, and IL-13 can stimulate fibroblast collagen production independently of TGFβ (46). However, it has recently been demonstrated that signaling through IL-13Rα2, initially thought to be a decoy receptor for IL-13, results in the production of TGFβ (47). Moreover, in vivo gene silencing of IL-13Rα2 using small interfering RNA attenuates bleomycin-induced lung fibrosis, with decreases in TGFβ1 secretion and collagen formation (48).
In this passively acquired autoimmune disease, with the exception of HLA, there were no significant genome-wide associations with identified polymorphisms that have been implicated in other autoimmune diseases, such as SLE, in which there are genetic contributions relating to dysfunction of the acquired immune system. Overall, however, there was an inflation factor of 1.22 for these autoimmune SNPs in cardiac neonatal lupus, compared with an inflation factor of 1.01 for the entire genome. Specifically, the enrichment of a variant of IRF5 is intriguing, and the variation at 7q32 has attracted a substantial amount of attention (21). However, it is acknowledged that maternal inheritance may account for some of these genes for which the associations are just below the level of significance. There are several possible and non–mutually exclusive interpretations. First, the associations may simply reflect the maternal enrichment of the genome for autoimmunity risk alleles, given the autoantibody state of the mother. Second, there may be an enrichment in autoimmunity-predisposing risk alleles in children with cardiac neonatal lupus independent of the maternal state. Third, there may be no significant enrichment, consistent with the relative immaturity of the human fetal immune system, which relies almost exclusively on adaptive maternal immunity, and the genetic contribution to cardiac injury may be restricted to the innate immune system and tissue reactivity.
These data require careful consideration and recognition of their inferential limits. As discussed above, maternal inheritance may be a limitation. Specifically, at 6p21, it is difficult to distinguish whether inheritance reflects transmission of genes related to maternal autoimmunity or whether these genes represent enrichment of genes that are biologically important. TDT will be informative. The limited power of the study is another limitation. However, the strength of the study is in the precise clinical phenotyping of the cases, with 96% having advanced atrioventricular block, the most characteristic cardiac manifestation associated with maternal anti-Ro/SSA antibodies. Given the rarity of the disease, the study included 4 cases of first-degree block, persisting after birth in 3 as shown by EKG, suggesting sustained injury, and 1 case of isolated cardiomyopathy. Arguably, combining cases expressing full progression of disease with those having less advanced disease may also represent a limitation.
Given the current interest in identifying a biomarker to predict more serious lifelong injury, isolated in utero prolongation of the PR interval has become an important focus of attention (10,49,50). As such, these cases could provide potential candidates to increase the power of the study of a rare disease. However, given the uncertainty as to the pathologic significance of transient first-degree block, we did not include these children in the analysis due to concerns of underestimating the genetic influence. Thus, unlike SLE, in which the manifestations are heterogeneous with regard to organ system, severity, and temporal course, cardiac neonatal lupus represents a more homogeneous phenotype.
This study represents the first large-scale investigation of genes associated with cardiac neonatal lupus. Identification of risk alleles is an incremental step toward the discovery of a fetal genetic component that contributes to the development of lifelong cardiac damage in newborns exposed to maternal anti-Ro/SSA antibodies. These analyses support the potential of this first cohort to provide clues that are immediate and of high impact in the study of the genetics of cardiac neonatal lupus, with candidates identified in several pathways relevant to the pathogenesis of disease, antigen presentation, apoptosis, and inflammation. The absence of a significant association with genes previously identified as being associated with autoimmune diseases emphasizes the passive nature of this unique disease.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the International Consortium on Systemic Lupus Erythematosus Genetics (SLEGEN) and Drs. Kathy L. Moser, Lindsey A. Criswell, Marta E. Alarcon-Riquelme, Chaim O. Jacob, Robert P. Kimberly, Betty P. Tsao, and Timothy J. Vyse for the use of the SLEGEN control data.
Supported by NIH grants AR-42455 and N01-AR-42271 and by American Heart Association grant-in-aid 0655938T. Ms Marion and Drs. Ramos and Langefeld’s work was supported by the Wake Forest University Health Sciences Center for Public Health Genomics. Drs. Kaufman and Harley are recipients of US Department of Veterans Affairs Merit Awards. Dr. Harley’s work was also supported by NIH grants P01-AI-083194-01, P20-RR-020143-06, U19-AI-082714-01, P01-AR-049084-07, N01-AR-62277, R01-DE-018209-01A1, R37-AI-024717-21, and R01-AR-042460-15. The International Consortium on Systemic Lupus Erythematosus Genetics was funded by the Alliance for Lupus Research.
Footnotes
The International Consortium on Systemic Lupus Erythematosus Genetics comprises lupus researchers who are dedicated to genetic discoveries in systemic lupus erythematosus. Its members include John B. Harley, MD, Marta E. Alarcon-Riquelme, MD, Lindsey A. Criswell, MD, MPH, Chaim O. Jacob, MD, PhD, Robert P. Kimberly, MD, Kathy L. Moser, PhD, Betty P. Tsao, PhD, Timothy J. Vyse, MA, MRCP, PhD, and Carl D. Langefeld, PhD.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Clancy had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Clancy, Langefeld.
Acquisition of data. Kaufman, Adler, Harley, Buyon.
Analysis and interpretation of data. Marion, Ramos, Langefeld.
REFERENCES
- 1.Chameides L, Truex RC, Vetter V, Rashkind WJ, Galioto FM, Jr, Noonan JA. Association of maternal systemic lupus erythematosus with congenital complete heart block. N Engl J Med. 1977;297:1204–1207. doi: 10.1056/NEJM197712012972203. [DOI] [PubMed] [Google Scholar]
- 2.Scott JS, Maddison PJ, Taylor PV, Esscher E, Scott O, Skinner RP. Connective-tissue disease, antibodies to ribonucleoprotein, and congenital heart block. N Engl J Med. 1983;309:209–212. doi: 10.1056/NEJM198307283090403. [DOI] [PubMed] [Google Scholar]
- 3.Reed BR, Lee LA, Harmon C, Wolfe R, Wiggins J, Peebles C, et al. Autoantibodies to SS-A/Ro in infants with congenital heart block. J Pediatr. 1983;103:889–891. doi: 10.1016/s0022-3476(83)80707-0. [DOI] [PubMed] [Google Scholar]
- 4.Buyon JP, Clancy RM, Friedman DM. Autoimmune associated congenital heart block: integration of clinical and research clues in the management of the maternal/foetal dyad at risk. J Intern Med. 2009;265:653–662. doi: 10.1111/j.1365-2796.2009.02100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buyon J, Roubey R, Swersky S, Pompeo L, Parke A, Baxi L, et al. Complete congenital heart block: risk of occurrence and therapeutic approach to prevention. J Rheumatol. 1988;15:1104–1108. [PubMed] [Google Scholar]
- 6.Rivera TL, Izmirly PM, Birnbaum BK, Byrne P, Brauth JB, Katholi M, et al. Disease progression in mothers of children enrolled in the Research Registry for Neonatal Lupus. Ann Rheum Dis. 2009;68:828–835. doi: 10.1136/ard.2008.088054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nield LE, Silverman ED, Smallhorn JF, Taylor GP, Mullen JB, Benson LN, et al. Endocardial fibroelastosis associated with maternal anti-Ro and anti-La antibodies in the absence of atrioventricular block. J Am Coll Cardiol. 2002;40:796–802. doi: 10.1016/s0735-1097(02)02004-1. [DOI] [PubMed] [Google Scholar]
- 8.Moak JP, Barron KS, Hougen TJ, Wiles HB, Balaji S, Sreeram N, et al. Congenital heart block: development of late-onset cardiomyopathy, a previously underappreciated sequela. J Am Coll Cardiol. 2001;37:238–242. doi: 10.1016/s0735-1097(00)01048-2. [DOI] [PubMed] [Google Scholar]
- 9.Buyon JP, Hiebert R, Copel J, Craft J, Friedman D, Katholi M, et al. Autoimmune-associated congenital heart block: demographics, mortality, morbidity and recurrence rates obtained from a national neonatal lupus registry. J Am Coll Cardiol. 1998;31:1658–1666. doi: 10.1016/s0735-1097(98)00161-2. [DOI] [PubMed] [Google Scholar]
- 10.Friedman DM, Kim MY, Copel JA, Davis C, Phoon CK, Glickstein JS, et al. Utility of cardiac monitoring in fetuses at risk for congenital heart block: the PR Interval and Dexamethasone Evaluation (PRIDE) prospective study. Circulation. 2008;117:485–493. doi: 10.1161/CIRCULATIONAHA.107.707661. [DOI] [PubMed] [Google Scholar]
- 11.Cimaz R, Spence DL, Hornberger L, Silverman ED. Incidence and spectrum of neonatal lupus erythematosus: a prospective study of infants born to mothers with anti-Ro autoantibodies. J Pediatr. 2003;142:678–683. doi: 10.1067/mpd.2003.233. [DOI] [PubMed] [Google Scholar]
- 12.Brucato A, Frassi M, Franceschini F, Cimaz R, Faden D, Pisoni MP, et al. Risk of congenital complete heart block in newborns of mothers with anti-Ro/SSA antibodies detected by counterimmunoelectrophoresis: a prospective study of 100 women. Arthritis Rheum. 2001;44:1832–1835. doi: 10.1002/1529-0131(200108)44:8<1832::AID-ART320>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Llanos C, Izmirly PM, Katholi M, Clancy RM, Friedman DM, Kim MY, et al. Recurrence rates of cardiac manifestations associated with neonatal lupus and maternal/fetal risk factors. Arthritis Rheum. 2009;60:3091–3097. doi: 10.1002/art.24768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritzler MJ, Pauls JD, Kinsella TD, Bowen TJ. Antinuclear, anticytoplasmic, and anti-Sjögren’s syndrome antigen A (SS-A/Ro) antibodies in female blood donors. Clin Immunol Immunopathol. 1985;36:120–128. doi: 10.1016/0090-1229(85)90045-5. [DOI] [PubMed] [Google Scholar]
- 15.Harmon CE, Lee LA, Huff JC, Norris DA, Weston WL. The frequency of autoantibodies to the SS-A/Ro antigen in pregnancy sera [abstract] Arthritis Rheum. 1984;27(Suppl 4):S20. [Google Scholar]
- 16.Siren MK, Julkunen H, Kaaja R. The increasing incidence of isolated congenital heart block in Finland. J Rheumatol. 1998;25:1862–1864. [PubMed] [Google Scholar]
- 17.Gawkrodger DJ, Beveridge GW. Neonatal lupus erythematosus in four successive siblings born to a mother with discoid lupus erythematosus. Br J Dermatol. 1984;111:683–687. doi: 10.1111/j.1365-2133.1984.tb14151.x. [DOI] [PubMed] [Google Scholar]
- 18.Cooley HM, Keech CL, Melny BJ, Menahem S, Morahan G, Kay TW. Monozygotic twins discordant for congenital complete heart block. Arthritis Rheum. 1997;40:381–384. doi: 10.1002/art.1780400223. [DOI] [PubMed] [Google Scholar]
- 19.Siren MK, Julkunen H, Kaaja R, Ekblad H, Koskimies S. Role of HLA in congenital heart block: susceptibility alleles in children. Lupus. 1999;8:60–67. doi: 10.1191/096120399678847407. [DOI] [PubMed] [Google Scholar]
- 20.Barquero Genoves IM, Figueras Aloy J, Guerola Serret M, Jimenez Gonzalez R. A complete congenital heart block in twins. An Esp Pediatr. 1996;44:164–166. In Spanish. [PubMed] [Google Scholar]
- 21.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. Washington: NIH/National Human Genome Research Institute; A catalog of published genome-wide association studies. URL: http://www.genome.gov/gwastudies/. [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Narayanaswamy C, Raghavarao D. Principal component analysis of large dispersion matrices. Appl Stat. 1991;40:309–316. [Google Scholar]
- 25.Velicer WF. Determining the number of components from the matrix of partial correlations. Psychometrika. 1976;41:321–327. [Google Scholar]
- 26.Tracy C, Widom H. Level-spacing distributions and the Airy kernel. Commun Math Phys. 1994;159:151–174. [Google Scholar]
- 27.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham RR, Ortmann W, Rodine P, Espe K, Langefeld C, Lange E, et al. Specific combinations of HLA-DR2 and DR3 class II haplotypes contribute graded risk for disease susceptibility and autoantibodies in human SLE. Eur J Hum Genet. 2007;15:823–830. doi: 10.1038/sj.ejhg.5201827. [DOI] [PubMed] [Google Scholar]
- 29.Harley JB, Reichlin M, Arnett FC, Alexander EL, Bias WB, Provost TT. Gene interaction at HLA-DQ enhances autoantibody production in primary Sjögren’s syndrome. Science. 1986;232:1145–1147. doi: 10.1126/science.3458307. [DOI] [PubMed] [Google Scholar]
- 30.Scofield RH, Frank MB, Neas BR, Horowitz RM, Hardgrave KL, Fujisaku A, et al. Cooperative association of T cell β receptor and HLA-DQ alleles in production of anti-Ro in systemic lupus erythematosus. Clin Immunol Immunopathol. 1994;72:335–341. doi: 10.1006/clin.1994.1150. [DOI] [PubMed] [Google Scholar]
- 31.Guggenbuhl P, Veillard E, Quelvenec E, Jego P, Semana G, Jean S, et al. Analysis of TNFα microsatellites in 35 patients with primary Sjögren’s syndrome. Joint Bone Spine. 2000;67:290–295. [PubMed] [Google Scholar]
- 32.Rood MJ, van Krugten MV, Zanelli E, van der Linden MW, Keijsers V, Schreuder GM, et al. TNF-308A and HLA-DR3 alleles contribute independently to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2000;43:129–134. doi: 10.1002/1529-0131(200001)43:1<129::AID-ANR16>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 33.Werth VP, Zhang W, Dortzbach K, Sullivan K. Association of a promoter polymorphism of tumor necrosis factor-α with subacute cutaneous lupus erythematosus and distinct photoregulation of transcription. J Invest Dermatol. 2000;115:726–730. doi: 10.1046/j.1523-1747.2000.00118.x. [DOI] [PubMed] [Google Scholar]
- 34.Martinez A, Fernandez-Arquero M, Pascual-Salcedo D, Conejero L, Alves H, Balsa A, et al. Primary association of tumor necrosis factor–region genetic markers with susceptibility to rheumatoid arthritis. Arthritis Rheum. 2000;43:1366–1370. doi: 10.1002/1529-0131(200006)43:6<1366::AID-ANR21>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 35.Sashio H, Tamura K, Ito R, Yamamoto Y, Bamba H, Kosaka T, et al. Polymorphisms of the TNF gene and the TNF receptor superfamily member 1B gene are associated with susceptibility to ulcerative colitis and Crohn’s disease, respectively. Immunogenetics. 2002;53:1020–1027. doi: 10.1007/s00251-001-0423-7. [DOI] [PubMed] [Google Scholar]
- 36.Clancy RM, Backer CB, Yin X, Kapur RP, Molad Y, Buyon JP. Cytokine polymorphisms and histologic expression in autopsy studies: contribution of TNF-α and TGF-β1 to the pathogenesis of autoimmune-associated congenital heart block. J Immunol. 2003;171:3253–3261. doi: 10.4049/jimmunol.171.6.3253. [DOI] [PubMed] [Google Scholar]
- 37.Miranda-Carus ME, Askanase AD, Clancy RM, Di Donato F, Chou TM, Libera MR, et al. Anti-SSA/Ro and anti-SSB/La autoantibodies bind the surface of apoptotic fetal cardiocytes and promote secretion of TNF-α by macrophages. J Immunol. 2000;165:5345–5351. doi: 10.4049/jimmunol.165.9.5345. [DOI] [PubMed] [Google Scholar]
- 38.Clancy R, Yin X, Askanese A, Miranda-Carus E, Nelson JL, Sestak A, et al. HLA-DBR1 relationships in neonatal lupus families [abstract] Arthritis Rheum. 2003;48(Suppl 9):S410–S411. [Google Scholar]
- 39.Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, Jenisch S, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clancy RM, Kapur RP, Molad Y, Askanase AD, Buyon JP. Immunohistologic evidence supports apoptosis, IgG deposition, and novel macrophage/fibroblast crosstalk in the pathologic cascade leading to congenital heart block. Arthritis Rheum. 2004;50:173–182. doi: 10.1002/art.11430. [DOI] [PubMed] [Google Scholar]
- 41.Clancy RM, Neufing PJ, Zheng P, O’Mahony M, Nimmerjahn F, Gordon TP, et al. Impaired clearance of apoptotic cardiocytes is linked to anti-SSA/Ro and-SSB/La antibodies in the pathogenesis of congenital heart block. J Clin Invest. 2006;116:2413–2422. doi: 10.1172/JCI27803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi H, Fujimura Y, Ouchida M, Prasad DD, Rao VN, Reddy ES. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene. 1997;14:1259–1268. doi: 10.1038/sj.onc.1201099. [DOI] [PubMed] [Google Scholar]
- 43.Yuan L, Nikolova-Krstevski V, Zhan Y, Kondo M, Bhasin M, Varghese L, et al. Antiinflammatory effects of the ETS factor ERG in endothelial cells are mediated through transcriptional repression of the interleukin-8 gene. Circ Res. 2009;104:1049–1057. doi: 10.1161/CIRCRESAHA.108.190751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Im YH, Kim HT, Lee C, Poulin D, Welford S, Sorensen PH, et al. EWS-FLI1, EWS-ERG, and EWS-ETV1 oncoproteins of Ewing tumor family all suppress transcription of transforming growth factor β type II receptor gene. Cancer Res. 2000;60:1536–1540. [PubMed] [Google Scholar]
- 45.Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolodsick JE, Toews GB, Jakubzick C, Hogaboam C, Moore TA, McKenzie A, et al. Protection from fluorescein isothiocyanate-induced fibrosis in IL-13-deficient, but not IL-4-deficient, mice results from impaired collagen synthesis by fibroblasts. J Immunol. 2004;172:4068–4076. doi: 10.4049/jimmunol.172.7.4068. [DOI] [PubMed] [Google Scholar]
- 47.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13α2 receptor is involved in induction of TGF-β1 production and fibrosis. Nat Med. 2006;12:99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 48.Scotton CJ, Chambers RC. Molecular targets in pulmonary fibrosis: the myofibroblast in focus. Chest. 2007;132:1311–1321. doi: 10.1378/chest.06-2568. [DOI] [PubMed] [Google Scholar]
- 49.Sonesson SE, Salomonsson S, Jacobsson LA, Bremme K, Wahren-Herlenius M. Signs of first-degree heart block occur in one-third of fetuses of pregnant women with anti–SSA/Ro 52-kd antibodies. Arthritis Rheum. 2004;50:1253–1261. doi: 10.1002/art.20126. [DOI] [PubMed] [Google Scholar]
- 50.Rein AJ, Mevorach D, Perles Z, Gavri S, Nadjari M, Nir A, et al. Early diagnosis and treatment of atrioventricular block in the fetus exposed to maternal anti-SSA/Ro-SSB/La antibodies: a prospective, observational, fetal kinetocardiogram-based study. Circulation. 2009;119:1867–1872. doi: 10.1161/CIRCULATIONAHA.108.773143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.