Abstract
Meaningful analysis of next-generation sequencing (NGS) data, which are produced extensively by genetics and genomics studies, relies crucially on the accurate calling of SNPs and genotypes. Recently developed statistical methods both improve and quantify the considerable uncertainty associated with genotype calling, and will especially benefit the growing number of studies using low- to medium-coverage data. We review these methods and provide a guide for their use in NGS studies.
Next-generation sequencing (NGS) methods1 provide cheap and reliable large-scale DNA sequencing. They are used extensively for de novo sequencing2, for disease mapping3, for quantifying expression levels through RNA sequencing4–6 and in population genetic studies7–9.
In NGS methods, a whole genome, or targeted regions of the genome, is randomly digested into small fragments (or short reads) that get sequenced and are then either aligned to a reference genome or assembled10. Having aligned the fragments of one or more individuals to a reference genome, ‘SNP calling’ identifies variable sites, whereas ‘genotype calling’ determines the genotype for each individual at each site.
NGS data can suffer from high error rates that are due to multiple factors, including base-calling and alignment errors. Moreover, many NGS studies rely on low- coverage sequencing (<5× per site per individual, on average), for which there is high probability that only one of the two chromosomes of a diploid individual has been sampled at a specified site. Under such circumstances, accurate SNP calling and genotype calling are difficult, and there is often considerable uncertainty associated with the results. It is crucial to quantify and account for this uncertainty, as it influences downstream analyses based on the inferred SNPs and genotypes, such as the identification of rare mutations, the estimation of allele frequencies and association mapping.
One method for reducing uncertainty associated with genotype and SNP calling is to sequence target regions deeply (at >20× coverage). However, the ever-increasing demand for larger samples suggests that medium- (5–20×) or low-coverage sequencing will be the most common and cost-effective study design in many applications of NGS for years to come. For example, the 1000 Genomes Project pilot phase9 relied on approximately 3× coverage to sequence 176 individuals genome-wide. For the identification of low-frequency variants, this design is more cost-efficient than deeper sequencing in fewer individuals. Likewise, in association studies, mapping power is typically maximized by sequencing many individuals at low depth11, rather than sequencing fewer individuals at a high depth.
Alternatively, reducing and quantifying the uncertainty associated with SNP and genotype calling may be accomplished using sophisticated algorithms; therefore, these have recently been the subject of extensive research9,12–15. Most contemporary algorithms use a probabilistic framework. So-called ‘genotype likelihoods’ — which incorporate errors that may have been introduced in base calling, alignment and assembly — are coupled with prior information, such as allele frequencies and patterns of linkage disequilibrium (LD). The result is a SNP and genotype call and an associated measure of uncertainty (which is often described by a ‘quality score’), both of which have a concrete statistical interpretation.
Here we review this research and provide general guidelines for genotype and SNP calling in NGS studies. Converting the raw output of NGS technology into a final set of SNP and genotype data involves a number of steps (summarized in FIG. 1), each of which contributes to the accuracy of the final SNP and genotype calls. We start at the beginning of this process by briefly reviewing recent developments in the methods used for base calling and alignment. We then review and discuss several recent algorithms for SNP and genotype calling and address how the uncertainties in the resulting calls can be accommodated in downstream analyses. Finally, we make some general recommendations for the analysis of NGS data.
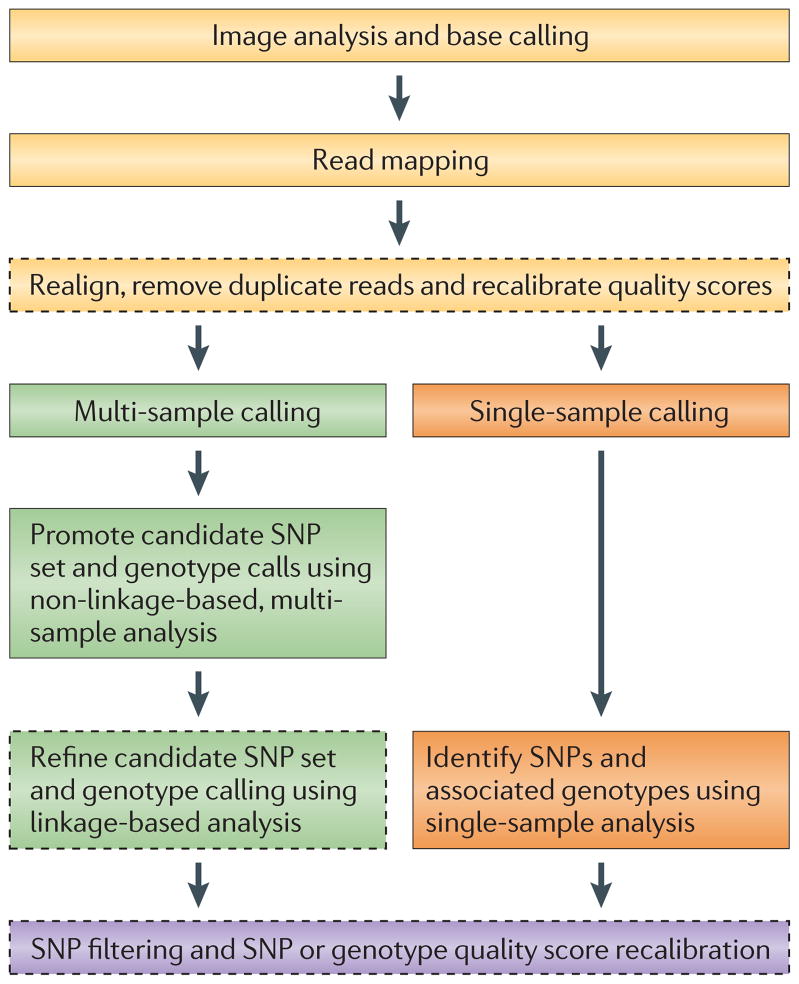
Figure 1. Steps for converting raw next-generation sequencing data into a final set of SNP or genotype calls.
Pre-processing steps (shown in yellow) transform the raw data from next-generation sequencing technology into a set of aligned reads that have a measure of confidence, or quality score, associated with the bases of each read. The per-base quality scores produced by base-calling algorithms may need to be recalibrated to accurately reflect the true error rates. Depending on the number of samples and the depth of coverage, either a multi-sample calling procedure (green) or a single-sample calling procedure (orange) may then be applied to obtain SNP or genotype calls and associated quality scores. Note that the multi-sample procedure may include a linkage-based analysis, which can substantially improve the accuracy of SNP or genotype calls. Finally, post-processing (purple) uses both known data and simple heuristics to filter the set of SNPs and/or improve the associated quality scores. Optional, although recommended, steps are shown in dashed lines.
Base calling and alignment
The main principle underlying NGS technologies is sequencing-by-synthesis. In brief, tens-to-hundreds of millions of clusters of small ssDNA templates are ‘read’ simultaneously by sequentially building up complementary bases. The synthesis process is captured in a series of fluorescence images, and base-calling algorithms infer the actual nucleotide information from the obtained fluorescence-intensity data for each cluster of DNA templates. They then assign a measure of uncertainty (or quality score) to each base call. The resulting short-read data are then assembled into a genome. When a reference genome is available, the primary approach used to assemble a newly sequenced genome is alignment (also known as ‘read mapping’), in which the basic task is to align each short read onto an available reference genome. Here we review the key aspects of base calling and alignment.
Base calling and quality scores
Base-calling procedures vary according to the sequencing platform used, all of which are prone to a different type of error. For the 454 platform, base calling involves inferring the length of each homopolymer from the observed fluorescence intensity. The main challenge stems from the fact that the variance of signal intensity for a specific homopolymer length is large, resulting in high error rates in insertion and deletion (indel) calls. For the Illumina platform, indel errors are rare, but the overall miscall error rate is typically around 1%. Here, the main complication arises from the synthesis process becoming desynchronized between different copies of DNA templates in the same cluster. Base calling becomes less accurate in later cycles as the extent of asynchrony is exacerbated with each sequencing cycle. The SOLiD platform uses a two-base encoding scheme in which each fluorescent dye colour represents four dinucleotide combinations. Each base of the DNA template is examined twice in this system and a length m nucleotide sequence is represented as a length m – 1 colour sequence. A major complication in ‘colour calling’ arises from biases in fluorescence intensities that appear in later machine cycles.
In addition to identifying nucleotides, base-calling algorithms produce per-base quality scores by using noise estimates from image analysis. Some sequencing platforms adopt quality values that are defined specifically for the platforms, but those quality values can be easily transformed into the standard Phred16 quality score, given by
| (1) |
Note that a Phred score of 20 corresponds to a 1% error rate in base calling.
The typical error rate of NGS data ranges from a few tenths of a per cent to several per cent, depending on the platform. Reducing the error rate of base calls and improving the accuracy of the per-base quality score have important implications for assembly, polymorphism detection and downstream population-genomic analyses. As such, several base-calling algorithms have been developed to optimize data acquisition for the more widely used NGS platforms: examples include Pyrobayes17 for the 454 platform; Rsolid18 for the SOLiD platform; and Ibis19 and BayesCall20,21 for the Illumina platform. These algorithms provide ~5–30% improvement in error rates over the base-calling methods developed by the manufacturers of the NGS platforms, and it has been shown that improved base-call accuracy can lead to a significant reduction in false-positive SNP calls and facilitate assembly when the coverage is low to moderate. However, some of the new methods tend to be either too computationally intensive to be of broad practical use or have not been tested thoroughly. Although more-accurate image analysis and base-calling algorithms for NGS platforms continue to be developed, the default software packages currently accompanying NGS platforms are the ones that are most widely adopted by users.
Alignment and assembly
The accuracy of the alignment has a crucial role in variant detection. Incorrectly aligned reads may lead to errors in SNP and genotype calling, so it is important for alignment algorithms to be able to cope with sequencing errors, as well as with potentially real differences (both point mutations and indels) between the reference genome and the sequenced genome that are due to polymorphisms. Furthermore, it is important for aligners to produce well-calibrated alignment (or mapping) quality values, as variant calls and their posterior probabilities depend on those scores.
The amount of sequence identity required between each read and the reference sequence is determined by a trade-off between accuracy and read depth. The optimal choice of the tolerable number of mismatches may differ between different organisms. For example, as populations of Drosophila melanogaster are more variable than human populations, using mapping criteria that are optimized for analyses of human sequences may lead to a severe loss of sequencing depth in D. melanogaster. This, in turn, may lead to a potential for biases in the downstream analyses, as regions that harbour many natural polymorphisms will be underrepresented. Likewise, using alignment criteria that are appropriate for fruitflies in humans would lead to a large amount of incorrectly aligned reads.
Most alignment algorithms for NGS data are based on either ‘hashing’ or an effective data compression algorithm called the ‘Burrows–Wheeler transform’ (BWT)22. BWT-based aligners (for example, Bowtie23, SOAP2 (REF. 15) and BWA24) are fast, memory-efficient and particularly useful for aligning repetitive reads; however, they tend to be less sensitive than the state-of-the-art hash-based algorithms (for example, MAQ12, Novoalign and Stampy25). The Novoalign and Stampy aligners currently produce the most accurate overall results, while also being practical in terms of running time (see REF. 25 for a detailed comparison of the performance of various aligners).
In general, alignment is more difficult for regions with higher levels of diversity between the reference genome and the sequenced genome. This difficulty can be ameliorated by the use of longer reads and paired-end reads (see REF. 25 for further quantitative details). However, assembling highly diverse regions such as the major histocompatibility complex (MHC) remains a challenge. Using de novo assembly algorithms — which are based on graph-theoretic ideas26–30 that try to exploit overlap information to stitch together the reads into contiguous sequences — may provide a viable solution to this challenge. Combining such methods with alignment to study genetic variation in complex regions is likely to be an active area of research in the forthcoming years.
Recalibration of per-base quality scores
The raw Phred-scaled quality scores produced by base-calling algorithms may not accurately reflect the true base-calling error rates14,15,31. In such a case, the raw quality scores need to be recalibrated so that a Phred score of Q more-accurately corresponds to an error rate of 10− Q/10, as implied by equation 1. Obtaining well-calibrated quality scores is important, as SNP and genotype calling at a specific position in the genome depends on both the base calls and the per-base quality scores of the reads overlapping the position.
In SOAPsnp14,15, per-base quality scores are recalibrated by comparing a sequenced genome to the reference genome at sites with no known SNPs. A related alignment-based recalibration algorithm has been implemented in the GATK software32,33, which takes into account several covariates such as machine cycle and dinucleotide context. For all supposedly non- polymorphic sites, the bases that align to those sites are put into different categories classified by the following features: the raw quality score (produced by base calling), the position of the base in the read, the dinucleotide context and the read group. For each category, the algorithm estimates the empirical quality score by using the number of mismatches with respect to the reference genome. Recalibrated quality scores are then estimated by adding to the raw quality scores the residual differences between empirical quality scores and the mismatch rates implied by the raw quality scores, which are conditioned on various subsets of the features. This recalibration algorithm, which is adopted in the 1000 Genomes Project9, can be applied to various sequencing platforms. As described above, the algorithm uses a set of supposedly non-polymorphic sites. If a comprehensive database of known SNPs is not available for the species under consideration, then one can first identify candidate polymorphic sites that are highly likely to be real and use the remaining sites in the recalibration procedure. In such a case, another round of SNP calling should be performed with recalibrated quality scores.
Genotype and SNP calling
The process of converting base calls and quality scores into a set of genotypes for each individual in a sample is often divided into two steps: genotype calling and SNP calling. SNP calling aims to determine in which positions there are polymorphisms or in which positions at least one of the bases differs from a reference sequence; the latter is also sometimes referred to as ‘variant calling’. Genotype calling is the process of determining the genotype for each individual and is typically only done for positions in which a SNP or a ‘variant’ has already been called. We use the word ‘calling’ here to signify the estimation of one unique SNP or genotype. However, we note that some analyses can proceed without determining the exact identity of each genotype, but instead allow uncertainty regarding genotypes to be incorporated directly into the analyses.
Genotype and SNP calling can proceed, as in early studies, by counting alleles at each site and using simple cutoff rules for when to call a SNP or genotype. More-recent methods incorporate uncertainty in a probabilistic framework. In the probabilistic framework, it is also possible to further incorporate additional information regarding allele frequencies and/or patterns of LD. We review these different approaches below, starting with the simple methods based on counting alleles.
Early methods for calling genotypes
Early NGS studies based both SNP and genotype calling on separate analyses of data from each individual sampled. Typically, analyses would first involve a filtering step in which only high-confidence bases would be kept. The most common cutoff used would be a Phred-type quality score of Q20 (QPhred = 20). Genotype calling would then proceed for each individual by counting the number of times each allele is observed and using fixed cutoffs. SNP calling would then be performed based on the inferred genotypes. For example34,35, one would first use a Q20 filter and then call a heterozygous genotype if the proportion of the non-reference allele is between 20% and 80%; otherwise, a homozygous genotype would be called. This is a fairly standard procedure and works well when the sequencing depth is high (>20×), so that the probability of a heterozygous individual falling outside the 20–80% range is small. Related methods for genotype calling form the basis for the commercially available software in Roche’s GSMapper, the CLC Genomic Workbench and the DNSTAR Lasergene software. These methods can be improved by using more empirically determined cutoffs (as described in REF. 36).
Probabilistic methods
For moderate or low sequencing depths, genotype calling based on fixed cutoffs will typically lead to under-calling of heterozygous genotypes and the use of a simple filtering based on quality score leads to a loss of information regarding individual read qualities. An additional disadvantage of this type of genotype calling is that it typically does not provide measures of uncertainty in the genotype inference. For this reason, several probabilistic methods have been developed that use the quality score to provide a posterior probability for each genotype12–15.
In brief, it is assumed that one can compute a genotype likelihood, p(X|G), for a genotype G. The symbol X represents, in this generic notation, all of the read data for a particular individual at a particular site. In conjunction with a genotype prior, p(G), Bayes’ formula is used to calculate p(G|X), which is the posterior probability of genotype G. The genotype with the highest posterior probability is generally chosen, and this probability, or perhaps the ratio between the highest and the second highest probabilities, is used as a measure of confidence. The advantages of the probabilistic methods are that they provide measures of statistical uncertainty when calling genotypes, they lead to higher accuracy of genotype calling, and they provide a natural framework for incorporating information regarding allele frequencies and patterns of LD.
Calculating genotype likelihoods
The genotype likelihood can be calculated using the quality scores for each read. Let Xi be the data in read i for a particular individual and a particular site with genotype G. The probability p(Xi|G) is then given by a simple rescaling of the quality score of Xi, and the genotype likelihood, p(X|G), can be calculated directly from the data by taking the product of p(Xi|G) over all i. There is an implicit assumption here of independence among reads, which may be violated in the presence of alignment errors or PCR artefacts. It has been suggested12 that a weighting scheme should be used that takes correlated errors into account. The genotype likelihoods can also be improved by recalibrating the per-base quality scores using empirical data, as discussed in the section regarding base calling and alignment. When genotype calling is preceded by SNP calling, the information from the SNP-calling step can be incorporated into the genotype-calling algorithm, leading to genotype likelihoods that are calculated by conditioning on the site containing a polymorphism.
Martin et al.37 also suggested estimating error rates directly from the read data for each site independently, instead of using quality scores. The advantage of such an approach is that genotype and SNP calling do not depend on the accuracy of the calculated quality scores. However, a disadvantage is that the considerable information regarding errors gained through the base-calling and alignment process is lost. These approaches are very recent, and no research has been done to systematically compare the advantages and disadvantages of directly estimating error rates from the data. Ideally, error rates should be estimated from the data while incorporating information obtained during base calling and alignment.
Methods for calculating genotype likelihoods will probably be a topic of much future research.
Assigning priors using single samples
In addition to computing the genotype likelihood, a prior probability for each genotype must be assumed in order to produce posterior probabilities for the genotypes. Suppose that a single individual is sequenced. The prior-genotype probability may be chosen to assign equal probability to all genotypes, or it can be based on external information — for example, from the reference sequence, SNP databases or an available population sample. In the analysis of human data in SOAPsnp14,15, a prior is chosen by the use of dbSNP38. For example, if a G/T polymorphism is reported in dbSNP, the prior probabilities are set to be 0.454 for each of the genotypes GG and TT, 0.0909 for GT and less than 10−4 for all other genotypes14,15. A similar approach is used in MAQ12. Notice that there is a strong weight against heterozygotes in order to avoid mistaking sequencing errors for real polymorphisms.
Assigning priors using multiple samples
Priors can be improved by jointly analysing multiple individuals. This can be done by considering allele frequencies, or genotype frequencies, estimated from larger data sets — for example, using maximum likelihood14,37. If allele frequencies are known, genotype probabilities can then be calculated using the Hardy–Weinberg equilibrium (HWE) assumption or other assumptions that relate allele frequencies to genotype frequencies. Uncertainty in the estimate of the allele frequency can be incorporated by also assigning a prior to the allele frequency itself, instead of estimating the allele frequency. This prior can be derived either from the data or from population genetic theory.
In general, the use of information from multiple individuals when calling genotypes for a single individual should be of great help. For example, imagine that the genotype likelihoods for two genotypes — for example, genotypes AT and AA — are equally large. Based on this information alone, we should be equally likely to choose AT or AA when performing genotype calling. However, if we were provided with the information (from a large sample) that the frequency of the A allele in the study population was small — for example, around 1% — we would be unlikely to choose the genotype AA. This is because the prior probability of observing AA is 10−4, whereas the prior probability of observing AT is approximately 2 × 10−2, assuming HWE.
Incorporating LD information
The approaches discussed so far assume that genotype calling is done independently for each site. However, much can be gained from taking advantage of the pattern of LD at nearby sites. Several different population genetic methods have been developed for imputation of missing data in SNP data sets39–45. In brief, these methods use the pattern at linked sites to infer genotypes. As an example, consider a population in which the only haplotypes observed are ATA and CGC at three sites. If an individual is sampled with genotypes A or C in the first site, A or C in the third site, but an unknown genotype in the second site, we might think that the unknown genotype in the second site is actually T or G. A straightforward adaptation of these algorithms enables them to be used with NGS data. The use of LD patterns is a cornerstone of the 1000 Genomes Project, and it leads to a significant improvement in genotype-calling accuracy9.
A comparison of genotype-call accuracies
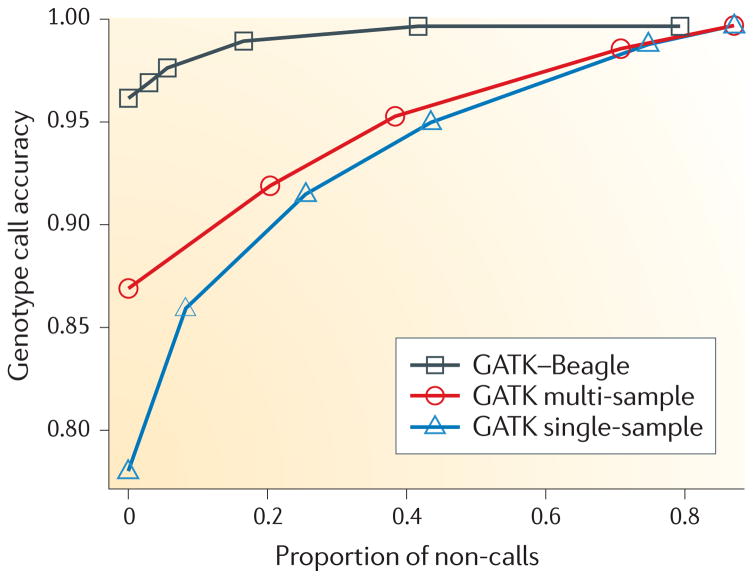
FIGURE 2 compares the accuracy of the genotype calls resulting from three comparable methods for calling genotypes: independently for each individual, jointly for all individuals without using an LD-based analysis and jointly for all samples using LD-based analysis. For high call rates, the use of multiple individuals leads to a substantial increase in the accuracy of genotype calling over using single samples from approximately 80% to 87%. The use of LD information provides an even greater improvement in accuracy: approximately 96%. To obtain a similar level of accuracy without the use of LD information would require that approximately 40% of the genotypes were non-calls; that is, they are left as missing data. Clearly, the use of LD patterns can substantially improve genotype calling when multiple samples have been sequenced. Even greater benefits can be derived when a high-quality reference data set, such as HapMap or SeattleSNPs, is available. The gain in accuracy is mostly obtained for SNPs of moderate- or high-allele frequencies. SNP and genotype calling for rare mutations, which would not be represented in any reference panel, may not improve much by the use of LD information.
Figure 2. A comparison of three genotype callers.
A subset of the data (chromosome 20, bases 20,000,000–25,000,000) for the 62 CEU individuals in both the HapMap Public Release no. 28 and the 1000 Genomes Pilot Project was genotype-called using the following methods: GATK Unified Genotyper32,33 applied to each individual independently (blue); GATK Unified Genotyper applied to all individuals collectively (red); and GATK Unified Genotyper applied to all individuals collectively, followed by Beagle42 using linkage disequilibrium (LD) information for genotype calling (black). For each of several quality thresholds, genotype calls with quality greater than the threshold were compared to HapMap data. Every such threshold thus entails both a proportion of called HapMap data and accuracy, relative to HapMap. For high call rates, genotyping the individuals collectively and using the LD-based method Beagle provided marked improvements.
SNP calling
So far, we have only discussed genotype calling. This issue is slightly different from the issue of how to call SNPs. In early NGS papers, in which only a single genome was analysed, SNP calling and genotype calling were more or less identical, as an inferred heterozygous genotype or a homozygous non-reference genotype would imply the presence of a SNP. For larger data sets containing many individuals, a SNP would be called if any individual was heterozygous or homozygous for a non-reference allele. However, this might not be an optimal way of proceeding, because the expected false-positive rate will increase linearly with the sample size. Furthermore, the information from multiple individuals is not best combined using called genotypes. Ideally, the joint posterior probability would be used to ascertain the probability that all genotypes are homozygous for the reference type, resulting in both a SNP call and an associated confidence. Alternatively, SNP calling can proceed, for example, by using a likelihood ratio test of the hypothesis of the population allele frequency being zero, using the methods for calculating the likelihood function described in REFS 11,37.
A list of programs for genotype and SNP calling is given in TABLE 1.
Table 1. A list of available non-commercial NGS genotype-calling software.
| Software | Available from | Calling method | Prerequisites | Comments | Refs |
|---|---|---|---|---|---|
| SOAP2 | http://soap.genomics.org.cn/index.html | Single-sample | High-quality variant database (for example, dbSNP) | Package for NGS data analysis, which includes a single individual genotype caller (SOAPsnp) | 15 |
| realSFS | http://128.32.118.212/thorfinn/realSFS/ | Single-sample | Aligned reads | Software for SNP and genotype calling using single individuals and allele frequencies. Site frequency spectrum (SFS) estimation | - |
| Samtools | http://samtools.sourceforge.net/ | Multi-sample | Aligned reads | Package for manipulation of NGS alignments, which includes a computation of genotype likelihoods (samtools) and SNP and genotype calling (bcftools) | 53 |
| GATK | http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit | Multi-sample | Aligned reads | Package for aligned NGS data analysis, which includes a SNP and genotype caller (Unifed Genotyper), SNP filtering (Variant Filtration) and SNP quality recalibration (Variant Recalibrator) | 32,33 |
| Beagle | http://faculty.washington.edu/browning/beagle/beagle.html | Multi-sample LD | Candidate SNPs, genotype likelihoods | Software for imputation, phasing and association that includes a mode for genotype calling | 42 |
| IMPUTE2 | http://mathgen.stats.ox.ac.uk/impute/impute_v2.html | Multi-sample LD | Candidate SNPs, genotype likelihoods | Software for imputation and phasing, including a mode for genotype calling. Requires fine-scale linkage map | 44 |
| QCall | ftp://ftp.sanger.ac.uk/pub/rd/QCALL | Multi-sample LD | ‘Feasible’ genealogies at a dense set of loci, genotype likelihoods | Software for SNP and genotype calling, including a method for generating candidate SNPs without LD information (NLDA) and a method for incorporating LD information (LDA). The ‘feasible’ genealogies can be generated using Margarita (http://www.sanger.ac.uk/resources/software/margarita) | 54 |
| MaCH | http://genome.sph.umich.edu/wiki/Thunder | Multi-sample LD | Genotype likelihoods | Software for SNP and genotype calling, including a method (GPT_Freq) for generating candidate SNPs without LD information and a method (thunder_glf_freq) for incorporating LD information | - |
A more complete list is available from http://seqanswers.com/wiki/Software/list. LD, linkage disequilibrium; NGS, next-generation sequencing.
Filtering
If the posterior probabilities in each site are calculated accurately, then all information regarding errors is taken into account and there is no reason to add any additional filtering or perform any additional manipulation of the data. However, for many real data this is not the case, and genotype and SNP calls can be greatly improved by using a number of filtering steps. For example, the 1000 Genomes Project9 eliminated entire sequencing batches that showed too high a discrepancy with known genotypes from the HapMap data. This type of filtering will only be available in projects that aim to resequence individuals who have already been subject to genome-wide SNP genotyping. Projects based on organisms other than humans, or NGS on humans for which genotyping data are not already available, should therefore expect to observe higher error rates than those observed in the 1000 Genomes data.
Other types of filtering based on deviations from the HWE — generally low-quality scores, systematic differences in quality scores for major and minor alleles, aberrant LD patterns, extreme read depths, strand bias, and so on — can also help to improve the accuracy of genotype and SNP calling. The appropriate filters depend on the sequencing protocol and the upstream analyses. For example, a site with strand bias (where a disproportional number of plus and minus strands are observed) could be an indication of a problematic site that is more error prone and should be filtered out. However, if the sequencing has been performed on captured sequences, such as those used for exome capturing, then the bias might not be an indication of a problematic site but rather an artefact of the capture array.
Incorporating genotype uncertainty in analyses of NGS data
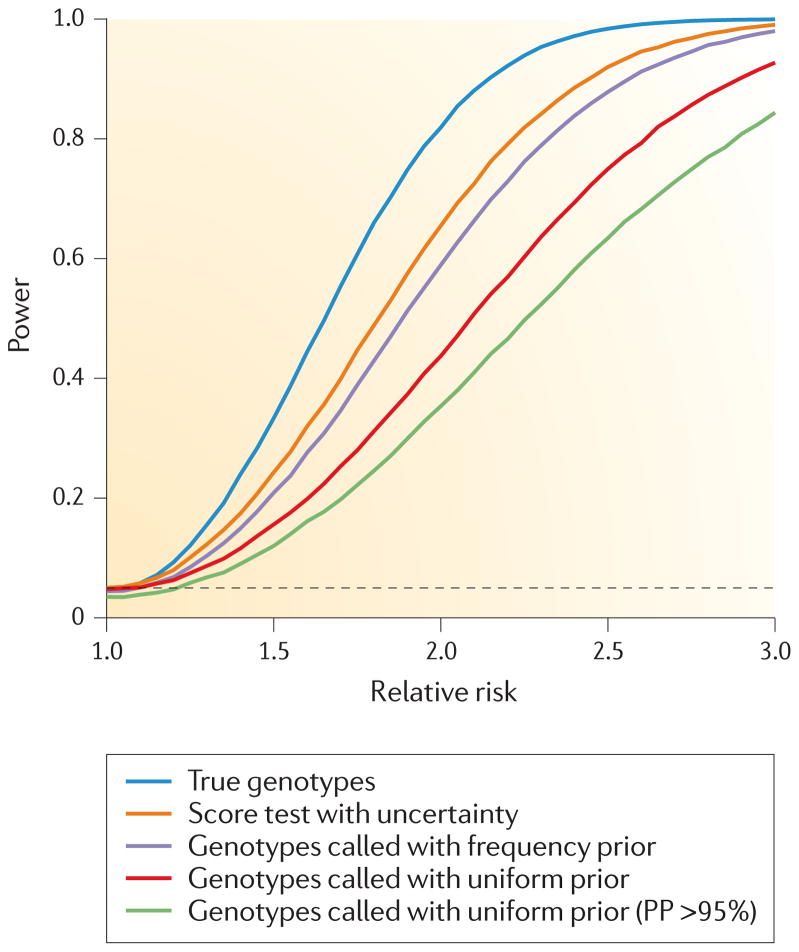
The choice of NGS genotype-calling strategies is ultimately related to the subsequent use of the data. Different applications might call for different genotype-calling strategies. As genotype calling for low- or moderate-coverage data entails some uncertainty, in many applications it may be particularly important to take this uncertainty into account. One of the most important applications of NGS is association-mapping studies. In the presence of genotype-calling uncertainty, standard methods for obtaining p-values using allelic tests are not valid because of potential over-calling of heterozygotes or homozygotes11. However, if the error structure is the same in cases and controls, tests that are robust to violations from the HWE will not suffer from an excess of false positives. Nonetheless, they may suffer reduced power, as even a low level of genotyping errors can lead to a surprisingly strong decrease in power46 (FIG. 3). The decrease in power cannot be circumvented by increased filtering that is based on genotype quality score, as such filtering will typically only lead to a further reduction in power (FIG. 3).
Figure 3. The power of association mapping for next-generation sequencing data.
Simulations of the power to detect association (p-value <0.05; dashed line) using various approaches for genotype calling at a 5% significance level. For each effect size, 50,000 simulations were performed for 1,000 cases and 1,000 controls assuming a population minor allele frequency (MAF) of 1% and a disease prevalence of 10%. The individual depth was simulated assuming a Poisson distribution with mean coverage of 4× and the sequence reads were sampled from the true genotypes, assuming an error rate of 1%. Genotype probabilities were calculated either by assuming a uniform genotype prior (red and light green) or by using the inferred MAF and Hardy–Weinberg equilibrium (purple and orange). Genotypes were called based on either the highest genotype probability (red and purple) or only for genotypes with a posterior-genotype probability (PP) >95% (light green). The called genotypes were tested using logistic regression, whereas the score statistic used the probability of the genotype, and therefore effectively integrates over the uncertainty in the genotype calls.
However, the use of genotype posteriors allows the construction of valid tests that combine the probabilities from all individuals and effectively sums over all possible genotypes11,37. For sequencing data, such methods have been described for allelic tests15,37, and methods used for haplotype data, such as score statistics47 and Bayesian models48, are attractive approaches. These methods and others are reviewed in the context of haplotype imputation in REF. 45. Such methods lead to valid statistical tests in association mapping and can provide increased mapping power (FIG. 3). For LD-based methods, this means performing the so-called multiple imputation: obtaining samples of multiple possible inferred data sets and weighting each by their relative probability. Most LD-based methods for genotype calling are developed for this purpose and can readily be applied to provide multiple samples.
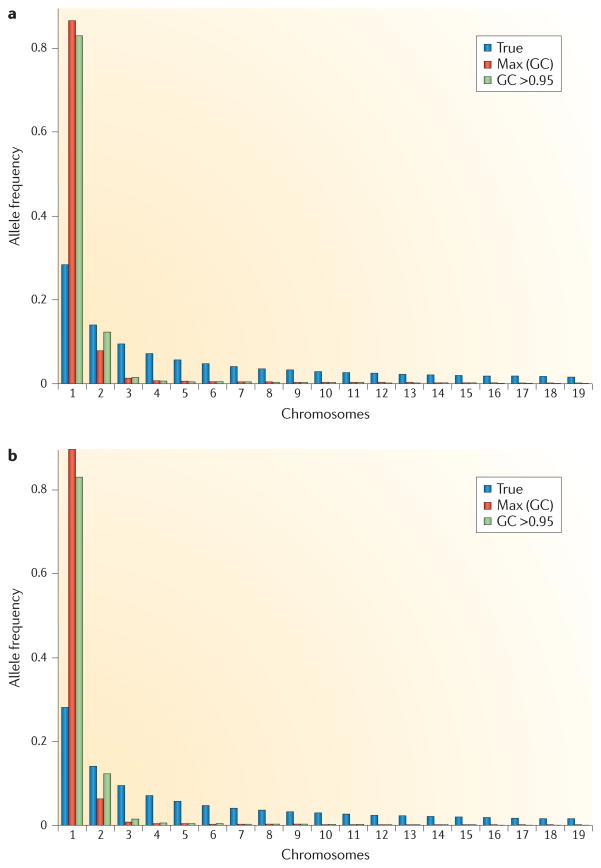
Uncertainty in genotype calls will also be an important consideration in population-genetic studies. In such studies, many inferences are based on allele frequencies, and ignoring genotype call uncertainty can lead to biased estimates49–51. The distribution of allele frequencies will be biased (FIG. 4), leading to biases in most of the common statistical methods applied in population genetics. An approach taken to address this problem in REF. 52 involved calculating the posterior probability of the allele frequency for each site. Population-genetic approaches for estimating variability, detecting selection and quantifying population substructure can then proceed by summing over these posterior probabilities.
Figure 4. The site frequency spectrum in next-generation sequencing data.
Fifty megabases of sequence were simulated for 50 individuals assuming a mean (Poisson-distributed) sequencing depth of 4× per individual, a per-site error rate of 0.003 and that 2% of all sites were variable. Because of the presence of missing data for one method, the data for all methods were subsampled down to a sample size of 20 chromosomes, using only called genotypes. Panels a and b show the site frequency spectrum (SFS) for all sites and for sites with a probability of harbouring a SNP of >95%, respectively. Both panels show the true SFS (True), the SFS using genotype calls (GC) obtained by always choosing the genotype with the highest posterior probability (Max (GC)), and when only calling genotypes with a posterior probability of >95% (GC >0.95). Notice that genotype-based inferences tend to overestimate the amount of singletons. The excess of singletons can be reduced or eliminated by using priors, or filtering processes, that are biased against singletons. However, such procedures will typically tend to introduce other biases.
Recommendations
The analysis of NGS data is a fast-evolving field, and new statistical methods for analysing the data are constantly being developed. As such, recommendations for how to analyse NGS data may change from month to month. For example, a number of new tools have been developed as part of the 1000 Genomes Project9, but many of them have still not been published or subjected to peer review.
At present, we make the following recommendations regarding genotype and SNP calling. First, base calling and calculation of quality scores should be carried out using methods that have been thoroughly tested and benchmarked. We then recommend a recalibration of per-base quality scores as in GATK or SOAPsnp. For aligning short reads to a reference genome, we recommend using a sensitive aligner such as Novoalign or Stampy; the latter can run in a hybrid mode that uses the efficient aligner BWA. Second, SNP calling should proceed by using methods that can incorporate data from all individuals in the sample simultaneously. SNP calling can be done using likelihood ratio tests or Bayesian procedures, in which the prior distributions for the allele frequencies are estimated from the data. Third, genotype calling should also proceed by combining data from multiple individuals in a Bayesian framework. Fourth, when possible, LD-based methods should be used to improve the accuracy of genotype and SNP calls.
Several additional steps can be taken to improve genotype calls, such as local realignments, combining results from multiple SNP- and genotype-calling algorithms and post hoc filtering based on quality scores9. Finally, we also urge the incorporation of uncertainty in the subsequent statistical procedures for analysing the data. In particular, association-mapping studies based on low- or medium-coverage data should use tests of association based on summing over all possible genotypes and weighing them by their respective probabilities (as described in REF. 11).
Conclusions
Genotype calling and SNP calling for NGS data have matured from simple methods based on counting alleles to sophisticated methods that provide probabilistic measures of uncertainty, and they can incorporate information from many individuals and linked sites. The probabilistic methods rely on accurate calculations of genotype likelihoods that incorporate information regarding alignment or assembly uncertainty and base-calling uncertainty. Therefore, more research is warranted into the accuracy of genotype-likelihood calculations and into the methods for improving genotype-likelihood calculations. More research is also needed in a number of other areas, including improved development of LD-based methods and in methods for incorporating genotype probabilities into downstream analyses. NGS will be central in genomic and medical genetic studies for years to come, and it is worthwhile now to focus attention on forming a solid foundation for future research in these areas.
Acknowledgments
This work was supported in part by NIH grants NIGMS R01-HG003229–05 and R01-HG003229–0551 to R.N., an NSF CAREER grant DBI-0846015 to Y.S.S. and an NIH National Research Service Award Trainee appointment on T32-HG00047 to J.S.P.
Glossary
- Likelihoods
Functions expressing the probability of observing the data — for example, next-generation sequencing data — given a parameter, such as a genotype or an allele frequency
- Posterior probabilities
In this context, these are the probabilities of a particular genotype given observed data: they are calculated by incorporating the information from the next-generation sequencing data as well as some prior information
- Hashing
A procedure of creating a data structure that helps to accelerate alignment. It stores information about which reads or where in the reference genome a particular substring or subsequence occurs. Some hash-based aligners hash the reads, while others hash the reference genome
- Paired-end reads
Sequencing of both the forward and reverse template of a DNA molecule, which is possible by inserting a primer sequence between the two ends of the read. The use of this technique greatly helps to increase assembly and alignment accuracy
- CEU individuals
One of the 11 populations in HapMap phase three. It consists of Utah residents with Northern and Western European ancestry from the Centre d’Etude du Polymorphisme Humain (CEPH) collection
- Bayes’ formula
A mathematical expression showing that a posterior probability can be found as the prior probability multiplied by the likelihood divided by a constant
- Correlated errors
Errors that do not occur independently of each other. An error that is observed in one position might increase the chance of observing another error in a neighbouring position
- Prior probability
In the context of this Review, the probability of a genotype calculated without incorporating information from the next-generation sequencing data. Prior probabilities can be obtained from a set of reference data
- Maximum likelihood
The statistical principle of estimating a parameter by finding the value of the parameters that maximizes the likelihood function
- Imputation
The substitution of some value for a missing data point. In this context, it is the use of a set of reference haplotypes to infer a genotype for an individual, when data are missing or incomplete
- Likelihood ratio test
A method for testing statistical hypotheses based on comparing the maximum likelihood under two different models. In this context, the allele frequency in one model equals zero, whereas the frequency in the second model might be larger than zero
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Rasmus Nieslen’s homepage: http://cteg.berkeley.edu/nielsen.htmlYun S. Song’s homepage: http://www.eecs.berkeley.edu/~yss1000 Genomes Project: http://www.1000genomes.orgNature Reviews Genetics series on Study Designs: http://www.nature.com/nrg/series/studydesigns/index.htmlALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Metzker M. Sequencing technologies — the next generation. Nature Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. This article provides an excellent Review of NGS technologies and their applications. [DOI] [PubMed] [Google Scholar]
- 2.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nature Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature Biotech. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nature Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 9.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. This 1000 Genomes paper provides an application of many of the state-of-the-art methods for analysis of NGS data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nature Methods. 2009;6:S6–S12. doi: 10.1038/nmeth.1376. [DOI] [PubMed] [Google Scholar]
- 11.Kim SY, et al. Design of association studies with pooled or un-pooled next-generation sequencing data. Genet Epidemiol. 2010;34:479–491. doi: 10.1002/gepi.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Ruan J, Durbin RM. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. This paper describes MAQ, a forerunner of efficient, hash-based alignment algorithms for short reads. MAQ also produces genotype calls. The concept of read-mapping quality is introduced in this paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JB, et al. Multiplex padlock targeted sequencing reveal human hypermutable CpG variations. Genome Res. 2009;19:1606–1615. doi: 10.1101/gr.092213.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 17.Quinlan AR, et al. Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nature Methods. 2008;5:179–181. doi: 10.1038/nmeth.1172. [DOI] [PubMed] [Google Scholar]
- 18.Wu H, Irizarry RA, Bravo HC. Intensity normalization improves color calling in SOLiD sequencing. Nature Methods. 2010;7:336–337. doi: 10.1038/nmeth0510-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao WC, Stevens K, Song YS. BayesCall: a model-based basecalling algorithm for high-throughput short-read sequencing. Genome Res. 2009;19:1884–1895. doi: 10.1101/gr.095299.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao WC, Song Y. S naiveBayesCall: an efficient model-based base-calling algorithm for high-throughput sequencing. Lect Notes Comp Sci. 2010;6044:233–247. doi: 10.1089/cmb.2010.0247. [DOI] [PubMed] [Google Scholar]
- 22.Burrows M, Wheeler D. HP Labs Technical Reports. Digital Equipment Corporation; 1994. A block-sorting lossless data compression algorithm. Technical Report 124. [online], http://www.hpl.hp.com/techreports/Compaq-DEC/SRC-RR-124.html. [Google Scholar]
- 23.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2010 Oct 27; doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundquist A, Ronaghi M, Tang H, Pevzner P, Batzoglou S. Whole-genome sequencing and assembly with high-throughput, short-read technologies. PLoS ONE. 2007;2:e484. doi: 10.1371/journal.pone.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler J, et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaisson MJP, Brinza D, Pevzner P. A De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Res. 2009;19:336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 2011 Apr 10; doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harismendy O, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, et al. The diploid sequence of an Asian individual. Nature. 2009;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedges D, et al. Exome sequencing of a multigenerational human pedigree. PLoS ONE. 2009;4:e8232. doi: 10.1371/journal.pone.0008232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin ER, et al. SeqEM: an adaptive genotype-calling approach for next- generation sequencing studies. Bioinformatics. 2010;26:2803–2810. doi: 10.1093/bioinformatics/btq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai JY, et al. Imputation methods to improve inference in SNP association studies. Genet Epidemiol. 2006;30:690–702. doi: 10.1002/gepi.20180. [DOI] [PubMed] [Google Scholar]
- 40.Minichiello MJ, Durbin R. Mapping trait loci by use of inferred ancestral recombination graphs. Am J Hum Genet. 2006;79:910–922. doi: 10.1086/508901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchini J, Howie B, Myers S, McVean G, Donnely P. A new multipoint method for genome-wide association studies via imputation of genotypes. Nature Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 44.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nature Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. This Review provides a comprehensive overview of available statistical methods for imputing genotypes and discusses various uses of imputation. [DOI] [PubMed] [Google Scholar]
- 46.Huang L, et al. The relationship between imputation error and statistical power in genetic association studies in diverse populations. Am J Hum Genet. 2009;85:692–698. doi: 10.1016/j.ajhg.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Servin B, Stephens M. Imputation-based analysis of association studies: candidate genes and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hellmann I, et al. Population genetic analysis of shotgun assemblies of genomic sequences from multiple individuals. Genome Res. 2008;18:1020–1029. doi: 10.1101/gr.074187.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson PLF, Slatkin M. Accounting for bias from sequencing error in population genetic estimates. Mol Biol Evol. 2008;25:199–206. doi: 10.1093/molbev/msm239. [DOI] [PubMed] [Google Scholar]
- 51.Johnson PLF, Slatkin M. Inference of population genetic parameters in metagenomics. A clean look at messy data. Genome Res. 2006;16:1320–1327. doi: 10.1101/gr.5431206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le SQ, Durbin R. SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples. Genome Res. 2010 Oct 27; doi: 10.1101/gr.113084.110. [DOI] [PMC free article] [PubMed] [Google Scholar]