Abstract
Allergenic proteins must cross-link specific IgE molecules, bound to the surface of mast cells and basophils, to stimulate an immune response. A structural understanding of the allergen-IgE interface is needed to predict cross-reactivities between allergens and to design hypoallergenic proteins. However, there are less than 90 experimentally determined structures available for the approximately 1500 sequences of allergens and isoallergens catalogued in the Structural Database of Allergenic Proteins (SDAP). To provide reliable structural data for the remaining proteins, we previously produced over 500 3D-models using an automated procedure, with strict controls at template choice and model quality evaluation. Here we assessed how well the fold and residue surface exposure of 10 of these models correlated with recently published experimental 3D structures determined by X-ray crystallography or NMR. We also discuss the impact of intrinsically disordered regions on the structural comparison and epitope prediction. Overall, for seven allergens with sequence identities to the original templates higher than 27%, the backbone root-mean square deviations were less than 2Å between the models and the subsequently determined experimental structures for ordered regions. Further, the surface exposure of known IgE epitopes on the models of three major allergens, from peanut (Ara h 1), latex (Hev b 2) and soy (Gly m 4) was very similar to the experimentally determined structures. For three remaining allergens with lower sequence identities to the modeling templates, the 3D folds were correctly identified. However the accuracy of those models is not sufficient for a reliable epitope mapping.
Keywords: template based modeling, allergenic proteins, IgE epitopes, Structural Database of Allergenic Proteins (SDAP)
Introduction
Structural comparisons of allergenic proteins are needed to supply a molecular explanation for clinically observed cross-reactivities between proteins from different organisms1–14, to design hypoallergenic proteins 15–19 and to predict whether new proteins or other biotechnology products are potential allergens20–22. However, there are only 86 experimental 3D structures in the Protein Data Bank (PDB) for allergens23, a small fraction of the 1499 allergen and isoallergens sequences collected in the Structural Database of Allergenic Proteins (SDAP)24. Thus reliable, template-based models of allergens are needed to compare allergen features, and to determine potential cross-reactive IgE binding surfaces25.
In 2008, we used an automated procedure to determine template-based models for 500 of the (at that time) 850 allergenic proteins in SDAP26. Although template-based modeling is a well established and reliable method for predicting the global fold of proteins as documented in recent CASP competitions 27–30, less is known about how reliably one can use 3D models in the context of allergy research. Recently, experimentally determined structures, based on X-ray and NMR data, for several of the proteins we modeled have been published 3,31–39. Here we systematically compared these 10 models, which differed in their length, % ID to the template, and % coverage, to recently determined 3D structures for the same allergen. We evaluated the similarities of the 3D folds, the surface exposure of experimentally established epitopes and the location of glycosylation sites. Our results from this evaluation indicate that the published models in SDAP are useful, not just for determining the overall structure of ordered regions, but also for determining conformational epitopes and other characteristics of allergenic proteins, if the identity of the allergen sequence to the template sequence was higher than 27%.
Materials and Methods
The automated modeling of allergenic proteins previously performed26 is briefly summarized here. We collected fold recognition results from the three fold recognition servers (FUGUE40, mGenThreader41 and 3DPSSM42). The top hits from each server were classified by the SCOP hierarchy and rated as 0 for “reliable”, 1 for “medium” and 2 for “difficult” according to the E-value or the Z-score of the servers. The confidence scores of all templates with the same SCOP fold were added, and a 3D model generated for templates with a confidence score less than 3 and no gap in the alignment greater than 20 residues. In a second round we analyzed the targets which did not pass the SCOP classification filter and used a structural comparison of aligned regions by the program CE43 to find structural similar templates. If the aligned template regions had an RMSD of less than 3Å, they were considered as the same fold and the template with the longest alignment was chosen to generate a 3D model by MPACK44.
To provide objective benchmarks for our automatic modeling procedure 26, we analyzed 3D models for which experimental 3D structures were released to the PDB (http://www.pdb.org/pdb) after November 2008. These 10 models are listed in Table 1 in order of decreasing sequence identity to the template chosen for model generation. Other data in the table include the allergen name (nomenclature of the International Union of Immunological Societies, IUIS), the protein designation (vicilin, lipocalin, etc.), the template used for modeling with the PDB entry and the protein name, sequence identity between target and the template used, the PDB entry of the experimentally determined 3D structure, the backbone root mean square deviation (RMSD) in Å between the model and template, and the coverage of the structural comparison as determined with the tools of DaliLite45 or FATCAT46. The structural superpositions of 3D models (red and yellow in Fig 1) with the experimental structures (green in Fig 1) were prepared with MolMol 47 using the “Fit to first” command. Solvent accessible surface areas (SASA) of individual residues were determined with the GETAREA program48 (http://curie.utmb.edu/getarea.html). All 3D models and links to the NMR or X-ray crystal structures in the PDB are available from the SDAP website (http://fermi.utmb.edu/SDAP/). Our assessment of the quality of the 3D models using the available native structures was also compared with results from three assessment software tools, QMEAN49 (http://swissmodel.expasy.org/qmean/cgi/index.cgi), ProSA50 (https://prosa.services.came.sbg.ac.at/prosa.php) and Verify_3D51 (http://nihserver.mbi.ucla.edu/SAVES/) that provide quality scores based only on the 3D structures of the protein models. They use different scoring functions based on analysis of the statistical properties of the 3D models, such as packing, amino acid specific Ca-Ca distances, environmental propensities of residues, relative to the same general properties of experimental structures.
Table I.
Comparison of 3D models deposited in SDAP to experimentally determined structures
| Homology Modeling | Experimental Structure(s) | DaliLite/FATCAT: Model vs. Experimental | ||||||
|---|---|---|---|---|---|---|---|---|
| Allergen Name | Protein | Template | Sequence Identity (%) | SDAP Model | PDB ID | Structural RMSD (Å) | Coverage (%) | |
| Hev b 2 | rubber (latex), 1,3-glucanase | 1AQ0.pdb | barley, 1,3–1,4-beta-glucanase | 45.9 | 802.pdb | 3EM5.pdb | 1.3 1.2* |
95.5 (300/314) 98.4 (309/314)* |
| Gly m 4 | soybean, stress-induced protein SAM22 | 1FM4.pdb | birch tree, pollen allergen Bet v 1L | 44.9 | 371.pdb | 2K7H.pdb | 2.3 2.7* |
98.1 (153/156) 100.0 (156/156)* |
| Sol i 3 | fire ant, venom allergen III | 1QNX.pdb | yellowjacket, venom allergen 5; Ves v 5 | 42.8 | 256.pdb | 2VZN.pdb | 1.5 1.7* |
92.6 (199/215) 96.3 (207/315)* |
| Ara h 1 | peanut, vicilin | 2CAV.pdb | jack-bean, canavalin | 38.7 | 34.pdb | 3S7E.pdb | 1.5 1.6* |
79.8 (332/416) 83.9 (349/416)* |
| Dau c 1 | carrot, pathogenesis-related protein, PR-10 | 1BV1.pdb | birch tree, pollen; allergen Bet v 1 | 37.9 | 496.pdb | 2WQL.pdb | 1.3 1.3* |
95.4 (146/153) 99.3 (152/153)* |
| Der f 1.0106 | American house dust mite, cysteine protease | 1CS8.pdb | human, procathepsin L | 31.6 | 784.pdb | 3D6S.pdb | 1.9 1.9* |
90.1 (191/212) 99.1 (210/212)* |
| Can f 2 | salivary canine allergen, lipocalin | 1MUP.pdb | house mouse, major urinary protein | 26.8 | 82.pdb | 3L4R.pdb | 1.5 1.6* |
89.8 (141/157) 94.3 (148/157)* |
| Ara h 2 | peanut, conglutin | 1PSY.pdb | castor bean, 2S albumin storage protein; allergen Ric c 1 | 22.4 | 283.pdb | 3OB4.pdb | 3.1 2.2* |
41.0 (55/134) 63.4 (85/134)* |
| Bla g 4 | German cockroach, calycin | 1I4U.pdb | lobster crustacyanin, subunit C1 | 15.4 | 63.pdb | 3EBK.pdb | 3.0 3.0* |
35.2 (64/182) 77.0 (141/182)* |
| Aed a 2 | mosquito, salivary, odorant binding protein, protein D7 | 1OOH.pdb | Drosophila melanogaster, odorant binding protein LUSH | 9.8 | 4.pdb | 3DXL.pdb | 3.2 2.5* |
53.7 (66/123) 94.0 (116/123)* |
| 11.3 | 356.pdb | 3.2 2.9* |
65.3 (81/124) 94.0 (116/124)* |
|||||
Flexible structural superposition with FATCAT.
Figure 1.
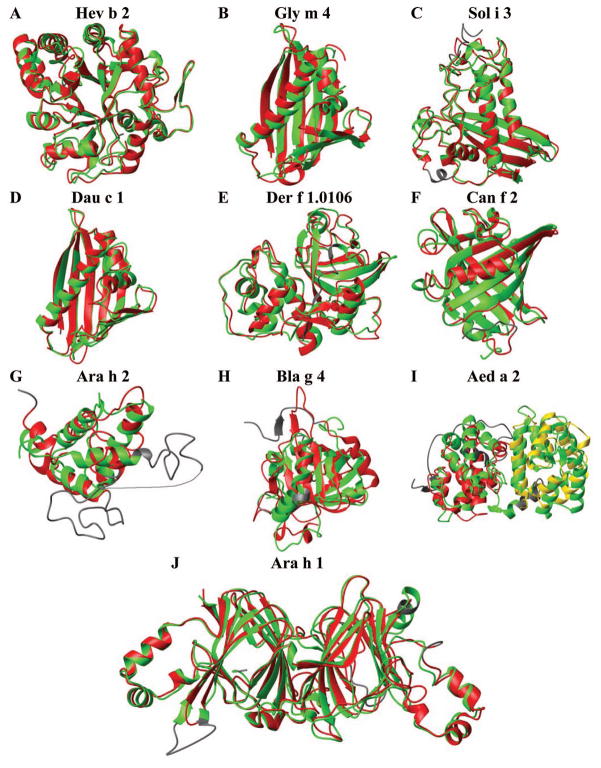
Structural overlays of aligned regions of the SDAP allergen model structures (red, yellow) with the corresponding experimental structures (green) from Table 1. A) 1,3-glucanase from latex rubber (Hev b 2) B) Stress-induced protein SAM22 from soybean (Gly m 4) C) Venom allergen III from fire ant (Sol i 3) D) Pathogenesis-related protein PR-10 from carrot (Dau c 1) E) Cysteine protease from American dust mite (Der f 1.0106) F) Canine salivary lipocalin (Can f 2) G) Conglutin from peanut (Ara h 2) H) Calycin from German cockroach (Bla g 4) I) Salivary odorant binding protein D7 from mosquito (Aed a 2) J) Vicilin from peanut (Ara h 1). The regions depicted in grey could not be aligned due to missing residues in either the model or the experimental structure. For Ara h 2, a disordered loop between helix 1 and 2, which was differently oriented in the Ric c 1 NMR structure used as template, is also depicted in grey since significant residues (shown with a thin line) are missing from the experimental structure.
Results and Discussion
Comparison of the backbone folds
The ten models used in this comparison are a good sampling, as they include proteins with many different sizes and a wide range of sequence identities from 9.8% to 45.9% between target and template used for modeling (Table 1). Backbone folds were correctly predicted with high precision for all 7 allergens where the sequence identities between templates and targets was >27%. The RMSD values between the models and the experimental structures were in most cases below 2Å, with a coverage for the structural alignment between models and experimental structures of 80–98% determined by DaliLite45 or FATCAT46. For the three allergens, Ara h 2, Bla g 4 and Aed a 2, the global folds were correctly identified by our modeling procedure despite low sequence identities. All 10 chosen templates were structurally similar to the experimental structures with significant P-values as determined by the flexible structure comparison method FATCAT46. Two of the best models were for proteins larger than 300 residues, for the major latex allergen Hev b 252, a 1,3-glucanase 53, and the peanut vicilin, Ara h 1. The Hev b 2 model, based on the X-ray crystal structure of 1,3–1,4-beta-glucanase from barley54, captured the (α/β)8 TIM barrel structure in the X-ray crystal structure (PDB code 3EM5) 31 with remarkable accuracy (Fig. 1A). The RMSD was only 1.3Å for a structural alignment covering 95% of the protein, including the irregular loop structures. Similar results (RMSD value of 1.5AǺ with 89% coverage) were obtained for the canine lipocalin allergen Can f 2 model, where the sequence was only 27% identical to the template (Fig 1 F).
Despite low sequence identities to their templates, the global folds of the 3D models of the cockroach allergen Bla g 4 and the mosquito salivary protein antigen Aed a 2 had a sizable fraction of structural overlap. About a third of the backbone fold of the Bla g 4 model was structurally equivalent to the crystal structure 3EBK38 with an RMSD value of 3AǺ (Fig. 1H) with rigid superposition by DaliLite and more than 2/3 (77%) of the global fold was structurally similar in a flexible superposition using FATCAT46. The structurally similar regions in that flexible superposition include the 8-strand β-barrel characteristic of the lipocalin fold and part of an α-helix between K154-K162. Thus, even though we used templates with low sequence identities, the only ones available at that time, the model did correctly predict the 3D fold of Bla g 4. The rigid superposition of the model and the X-ray crystal structure by MolMol is shown in Fig. 1 H, where three 3 β-strands of Bla g 4 (V98-T101, Y106-G111 and I121-R126 in the 3EBK labeling) were found as structurally equivalent.
The fold prediction was also surprisingly good for models determined for the two domains of the mosquito Aed a 2. As is also the case for the two cupin domains of Ara h 155, Aed a 2 has two similar domains. Although only 15% of the residues in the two domains are identical, alignment of the two sequences suggested they had a common fold. The same template, the Drosophila protein LUSH (1OOH)56, was thus used for modeling both domains, although its sequence was only 9% and 11% identical to either of the Aed a 2 domains. Structural alignments of the models of the N and C-terminal domains to the corresponding domains of Aed a 2 in the X-ray crystal structure (3DXL39) gave an RMSD of 3.2AǺ with a coverage of 53.7% and 65.3%, respectively (Figure 1, I). Flexible superposition with FATCAT recognized more than 90% of the folds as structurally similar, with RMSD values of less than 3Ǻ.
The successful template choices depended on the internal quality control of our automated modeling procedure, which required that the 3 fold recognition servers consistently agree on a template. The alignments for modeling were also selected based on a scoring function that included a gapping penalty, and overall length of the alignment of the template with the target sequence, and low conformational energy26.
Mapping IgE epitopes on the models
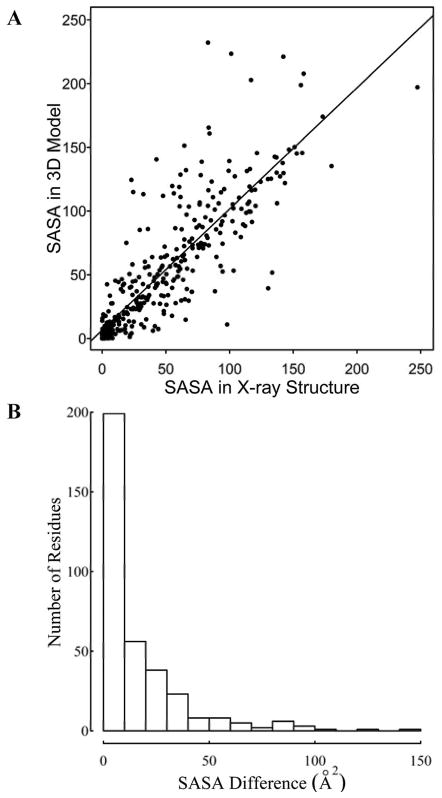
In many applications of 3D modeling for allergenic proteins, the goal is to determine the solvent accessibility of side chains on the allergens, and possibly predict the structure of (or at least the residues involved in) conformational epitopes22. Most data on IgE epitopes comes from dot spot assays, which measure the binding of IgE in allergic patient sera to linear peptides synthesized on membranes7,22 or microarrays 57. IgE epitope information is available for a few of the allergens in this study, including those from peanut Ara h 158,59 and Ara h 260, latex, Hev b 261, and soybean, Gly m 462. The Ara h 1 model was based on the jack-bean canavalin crystal structure 2CAV63; the overall sequence identity to this template was below 40%. The model correlated very well with the subsequently determined X-ray crystal structure of Ara h 133 (3S7E); the RMSD was 1.5Ǻ with 80% coverage (Fig. 1J). Further, the orientation and solvent accessible surface area (SASA) of the linear IgE epitopes (i.e., the peptides that bound to IgE in dot spot assays) were nearly identical in the model and the crystal structure (Fig. 2), and the conformations of the eight individual epitopes were also quite similar, with RMSD values ranging from 0.3Å to 0.9Å. Indeed, as Fig. 3 shows graphically, the solvent exposures of more than 90% of the residues in the model were within 10 Ǻ2 of those in the crystal structure. There was a significant difference (>50 Ǻ2) in solvent exposure of only 27 residues (Fig. 3B) most of them for surface exposed residues (see Supplementary Material S1). The single glycosylation site (residues 521–524, NASS; arrow in Fig. 2) was also located at similar surface exposed areas in both the model and crystal structure, as was its distance to the side chains of the IgE epitopes that surround it.
Figure 2.
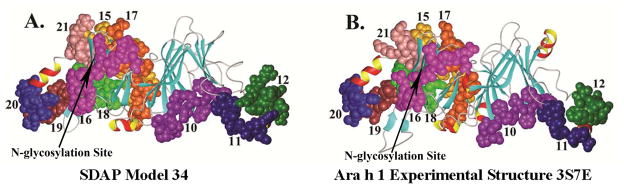
Comparison of the structural regions of Ara h 1 that correspond to the linear peptides that bind IgE antibodies. Epitopes 10–21, listed in the Ara h 1 entry page of SDAP (http://fermi.utmb.edu/SDAP/), were mapped on the 3D model (A) and X-ray crystal structure (B) of Ara h 1.
Figure 3.
Comparison of the solvent accessible surface areas (SASA) per residue in the Ara h 1 model from SDAP and the crystal structure (3S7E,33). (A) Correlation plot (B) Histogram for the absolute differences of the SASA of residues in the 3D model and the crystal structure.
Similarly high structural equivalence was found for the locations of the individual epitopes of the latex allergen Hev b 2 (Fig. S2) and the soybean allergen Gly m 4 (Fig. S3). The structural deviations of the individual epitope backbone segments were near 1Å with an average over all RMSD values of the segments of 0.7Å for Hev b 2 and slightly higher, 1.0Å, for Gly m 4. The highest deviating segments were found for 3 epitopes of Gly m 4 (1.4Å, 1.7Å and 2.7Å) in loop regions. For the more challenging model of Ara h 2 we found larger variations (Fig. S4). Whereas the surface positions of epitopes 3, 4, 8 and 10 were similar in the model and crystal structure, epitopes 6 and 7 showed large deviations. Note, however, that only 4/8 residues of Epitope 7 could be compared, as epitope 7 is part of a flexible loop (R59 to H84) that is missing in the X-ray crystal structure (thin line in Fig. 1G).
Weaknesses and Strengths of the 3D models by quality assessment tools
The above results show that, before mapping epitopes on a SDAP model, the user should take into account three factors included in all the structure files: the % identity of the allergen sequence to the template used and the overall coverage of the sequences. In addition, the user should determine the degree of disordered structure in the template. The user can also use several different, publicly assessable tools to further determine the quality of the models, based only on the coordinates. In supplementary material(Table S1), we show the results for assessing the quality of our 10 models using three current software tools, including QMEAN 49, ProSA 50 and Verify_3D 51. The quality scores of all three assessment tools for the seven high quality models of Hev b 2, Gly m 4, Sol i 3, Ara h 1, Dau c 1, Der f 1 and Can f 2 are in a range that one would expect for native structures, and are thus consistent with the analysis using the experimentally known 3D structures. Also all servers rank the qualities of the three models for Ara h 2, Bla g 4 and Aed a 2 consistently lower. However, there are some differences in the degree of quality assessment of those models. Whereas Verify_3D flags all three models of lower quality with “Fail”, the OMEAN Z-scores and the ProSA Z-scores for the models of Ara h 2 and Aed a 2 are in a borderline region when compared to the scores of native structures. Thus in practice those 3D models might be useful for the structural classification of allergens in super families, but not for reliable epitope mapping or the design of hypoallergenic proteins. As those tools provide quality scores for the 3D allergen models, they can be applied by end-users to the remaining predicted models in SDAP to give them further confidence for use of the models or reasons for further analysis by local quality plots.
Impact of structural flexibility on 3D models and experimental structures
Flexible regions were the major reason for differences between the models and the experimental structures. Intrinsically disordered regions are found prominently in the protein universe64. Although the overwhelming majority (80%) of allergens have a structured 3D fold, a sizable fraction contain locally disordered regions65. Peanut Ara h 2 is a good illustration of how disordered regions present problems for both experimentalists and modelers. Despite its importance as an allergen60,66, and its small size, the protein eluded crystallization efforts by several groups. An X-ray crystal structure was only recently determined, using a fusion protein with an N-terminal deleted form of Ara h 2 with the maltose binding protein (MBP) 37 (PDB structure 3OB4). The MBP fusion partner might impact the structure of the N-terminal region of the protein. Our model of Ara h 2 was based on an NMR structure for 2S albumin Ric c 1 from castor bean (PDB code 1PSY; 67). Although the Ric c 1 template was only 22% identical, the Ara h 2 model’s four helices overlay well with the corresponding ones in the crystal structure (Fig. 1G). The relatively high backbone RMSD (3.1 AǺ) reflects mainly differences in the conformation of the kinked helix 1. The NMR ensemble showed considerable variation especially in the loop region connecting helices 1 and 2 (gray line in Fig. 1G), which was also predicted by DisProt68 to be disordered. The crystal structure lacks also structural information for 15 of the residues (R59 to H84) in that loop (thin line in Fig. 1G). Two IgE epitopes (epitope 6 and 7 in the SDAP epitope table for Ara h 2) have been located in that area of the protein60, illustrating that IgE epitopes can overlap with flexible loop regions as documented by a recent statistical study 65. Thus determining the actual structure of these two epitopes will probably require a co-crystal structure of Ara h 2 with relevant monoclonal antibodies.
We also saw some structural differences due to flexibility even in allergen structures where the identity to the template was much higher. Soybean Gly m 469,70, a member of the pathogen related protein family PR-10, was modeled on an X-ray crystal structure of the birch pollen allergen Bet v 1 15. Although the overall fold is correct (RMSD 2.3Å), the model deviates from the NMR solution structures of Gly m 43 in flexible areas, where the structures in the bundle also differ from each other.
Despite the problems noted above for flexible regions, which may not be completely resolved by any technique, experimental structures are still the most direct and reliable procedure to map conformational epitopes or to graft conformational epitopes between different allergens9,71,72. Thus it is very encouraging that many structures have been published recently for newly identified allergens, or those for which we were unable to find a suitable modeling template in our 2008 study. These include NMR structures of the major mugwort pollen allergen Art v 1 73 and X-ray crystal structures of the mold allergen Alt a 174, pollen allergen Che a 313, dust mite allergen Der p 7 75, and antibody complexes of the house dust mite allergens Der p 1 and Der f 176 and the cockroach allergen Bla g 218. At least one of these, the fungal allergen Alt a 1, showed a novel fold, a unique β-barrel dimer structure74. However, based on the number of allergens for which no structure exists, the analysis of IgE epitopes will continue to rely on 3D models for some time to come 4,5,10,11,77–83. As the accuracy of prediction methods for template based modeling 27,30,84–86 and intrinsically disordered regions87,88 is continually improving, as is the number of potential templates in the PDB for the protein families of most allergens89,90, we can anticipate that future models will provide even better descriptions of epitope surfaces.
Conclusion
Our comprehensive 3D modeling effort for the allergens in SDAP was a major effort towards determining the 3D structures of all allergens. This analysis shows that the automatic, template based modeling procedure, probably thanks to the quality controls implemented, yielded reliable models if the identity of the allergen sequence to the template sequence was higher than 27%. In those cases the 3D models could be considered as high-quality models with backbone deviations of typically between 1Å and 2 AǺ in structured regions. However, not all 3D models have the same reliability. Current quality assessment tools can help end-users from over-interpreting the structural information. The user should be especially wary of regions that do not align to the template, or those based on templates with a large content of disordered or flexible loops. Proteins with large disordered regions will require improved tools to determine accurate models. The examples of three major allergens show that it is possible to use 3D models to determine the IgE epitopes and glycosylation sites with great accuracy if a reliable template can be found. This structural information will allow a better comparison of allergens from different organisms for potential cross-reactivity and to design hypoallergenic proteins than just using sequence information alone.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Health (R56 AI 064913) and the U.S. Environmental Protection Agency STAR Research Assistance Agreement (No. RD 834823). Use of the computational resources of the Sealy Center for Structural Biology and Molecular Biophysics at UTMB is gratefully acknowledged.
References
- 1.Chapman MD, Pomes A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. J Allergy Clin Immunol. 2007;119(2):414–420. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Schein CH, Ivanciuc O, Braun W. Bioinformatics approaches to classifying allergens and predicting cross-reactivity. Immunol Allergy Clin N Am. 2007;27(1):1–27. doi: 10.1016/j.iac.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkner H, Neudecker P, Mittag D, Ballmer-Weber BK, Schweimer K, Vieths S, Rosch P. Cross-reactivity of pollen and food allergens: soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins. Biosci Rep. 2009;29(3):183–192. doi: 10.1042/BSR20080117. [DOI] [PubMed] [Google Scholar]
- 4.Robotham JM, Hoffman GG, Teuber SS, Beyer K, Sampson HA, Sathe SK, Roux KH. Linear IgE-epitope mapping and comparative structural homology modeling of hazelnut and English walnut 11S globulins. Mol Immunol. 2009;46(15):2975–2984. doi: 10.1016/j.molimm.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 5.Cabanos C, Tandang-Silvas MR, Odijk V, Brostedt P, Tanaka A, Utsumi S, Maruyama N. Expression, purification, cross-reactivity and homology modeling of peanut profilin. Protein Expr Purif. 2010;73(1):36–45. doi: 10.1016/j.pep.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Mattsson L, Lundgren T, Olsson P, Sundberg M, Lidholm J. Molecular and immunological characterization of Can f 4: a dog dander allergen cross-reactive with a 23 kDa odorant-binding protein in cow dander. Clin Exp Allergy. 2010 doi: 10.1111/j.1365-2222.2010.03533.x. [DOI] [PubMed] [Google Scholar]
- 7.Maleki SJ, Teuber SS, Cheng H, Chen D, Comstock SS, Ruan S, Schein CH. Computationally predicted IgE epitopes of walnut allergens contribute to cross-reactivity with peanuts. Allergy. 2011;66(12):1522–1529. doi: 10.1111/j.1398-9995.2011.02692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schein CH, Ivanciuc O, Midoro-Horiuti T, Goldblum RM, Braun W. An Allergen Portrait Gallery: Representative Structures and an Overview of IgE Binding Surfaces. Bioinform Biol Insights. 2010;4:113–125. doi: 10.4137/BBI.S5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm J, Ferreras M, Ipsen H, Wurtzen PA, Gajhede M, Larsen JN, Lund K, Spangfort MD. Epitope grafting, re-creating a conformational Bet v 1 antibody epitope on the surface of the homologous apple allergen Mal d 1. J Biol Chem. 2011;286(20):17569–17578. doi: 10.1074/jbc.M110.194878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecker J, Diethers A, Etzold S, Seismann H, Michel Y, Plum M, Bredehorst R, Blank S, Braren I, Spillner E. Generation and epitope analysis of human monoclonal antibody isotypes with specificity for the Timothy grass major allergen Phl p 5a. Mol Immunol. 2011;48(9–10):1236–1244. doi: 10.1016/j.molimm.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Rouge P, Culerrier R, Granier C, Rance F, Barre A. Characterization of IgE-binding epitopes of peanut (Arachis hypogaea) PNA lectin allergen cross-reacting with other structurally related legume lectins. Mol Immunol. 2010;47(14):2359–2366. doi: 10.1016/j.molimm.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Tordesillas L, Pacios LF, Palacin A, Quirce S, Armentia A, Barber D, Salcedo G, Diaz-Perales A. Molecular basis of allergen cross-reactivity: non-specific lipid transfer proteins from wheat flour and peach fruit as models. Mol Immunol. 2009;47(2–3):534–540. doi: 10.1016/j.molimm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 13.Verdino P, Barderas R, Villalba M, Westritschnig K, Valenta R, Rodriguez R, Keller W. Three-dimensional structure of the cross-reactive pollen allergen Che a 3: visualizing cross-reactivity on the molecular surfaces of weed, grass, and tree pollen allergens. J Immunol. 2008;180(4):2313–2321. doi: 10.4049/jimmunol.180.4.2313. [DOI] [PubMed] [Google Scholar]
- 14.Dall’Antonia F, Gieras A, Devanaboyina SC, Valenta R, Keller W. Prediction of IgE-binding epitopes by means of allergen surface comparison and correlation to cross-reactivity. J Allergy Clin Immunol. 2011;128(4):872–879. e878. doi: 10.1016/j.jaci.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Markovic-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H. Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J Mol Biol. 2003;325(1):123–133. doi: 10.1016/s0022-2836(02)01197-x. [DOI] [PubMed] [Google Scholar]
- 16.Campana R, Vrtala S, Maderegger B, Jertschin P, Stegfellner G, Swoboda I, Focke-Tejkl M, Blatt K, Gieras A, Zafred D, Neubauer A, Valent P, Keller W, Spitzauer S, Valenta R. Hypoallergenic derivatives of the major birch pollen allergen Bet v 1 obtained by rational sequence reassembly. J Allergy Clin Immunol. 2010;126(5):1024–1031. 1031 e1021–1028. doi: 10.1016/j.jaci.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Thalhamer T, Dobias H, Stepanoska T, Proll M, Stutz H, Dissertori O, Lackner P, Ferreira F, Wallner M, Thalhamer J, Hartl A. Designing hypoallergenic derivatives for allergy treatment by means of in silico mutation and screening. J Allergy Clin Immunol. 2010;125(4):926–934. e910. doi: 10.1016/j.jaci.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 18.Glesner J, Wunschmann S, Li M, Gustchina A, Wlodawer A, Himly M, Chapman MD, Pomes A. Mechanisms of allergen-antibody interaction of cockroach allergen Bla g 2 with monoclonal antibodies that inhibit IgE antibody binding. PLoS One. 2011;6(7):e22223. doi: 10.1371/journal.pone.0022223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marazuela EG, Hajek R, Villalba M, Barber D, Breiteneder H, Rodriguez R, Batanero E. A non-allergenic Ole e 1-like protein from birch pollen as a tool to design hypoallergenic vaccine candidates. Mol Immunol. 2012;50(1–2):83–90. doi: 10.1016/j.molimm.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Lehrer SB, McClain S. Utility of animal models for predicting human allergenicity. Regul Toxicol Pharmacol. 2009;54(3 Suppl):S46–51. doi: 10.1016/j.yrtph.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Selgrade MK, Bowman CC, Ladics GS, Privalle L, Laessig SA. Safety assessment of biotechnology products for potential risk of food allergy: implications of new research. Toxicol Sci. 2009;110(1):31–39. doi: 10.1093/toxsci/kfp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanciuc O, Schein CH, Garcia T, Oezguen N, Negi SS, Braun W. Structural analysis of linear and conformational epitopes of allergens. Regul Toxicol Pharmacol. 2009;54(3 Suppl):S11–19. doi: 10.1016/j.yrtph.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, Young J, Yukich B, Zardecki C, Berman HM, Bourne PE. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39(Database issue):D392–401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanciuc O, Schein CH, Braun W. SDAP: Database and computational tools for allergenic proteins. Nucleic Acids Res. 2003;31(1):359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gadermaier E. In shape--the art of mapping conformational epitopes. Int Arch Allergy Immunol. 2012;157(4):321–322. doi: 10.1159/000332948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, Braun W. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol Immunol. 2008;45(14):3740–3747. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79 (Suppl 10):37–58. doi: 10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
- 28.Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77 (Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 29.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Skolnick J. Template-based protein structure modeling using TASSER(VMT) Proteins. 2011 doi: 10.1002/prot.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuentes-Silva D, Mendoza-Hernandez G, Stojanoff V, Palomares LA, Zenteno E, Torres-Larios A, Rodriguez-Romero A. Crystallization and identification of the glycosylated moieties of two isoforms of the main allergen Hev b 2 and preliminary X-ray analysis of two polymorphs of isoform II. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63(Pt 9):787–791. doi: 10.1107/S1744309107039838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padavattan S, Schmidt M, Hoffman DR, Markovic-Housley Z. Crystal Structure of the Major Allergen from Fire Ant Venom, Sol i 3. J Mol Biol. 2008;383(1):178–185. doi: 10.1016/j.jmb.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Chruszcz M, Maleki SJ, Majorek KA, Demas M, Bublin M, Solberg R, Hurlburt BK, Ruan S, Mattisohn CP, Breiteneder H, Minor W. Structural and immunologic characterization of Ara h 1, a major peanut allergen. J Biol Chem. 2011;286(45):39318–39327. doi: 10.1074/jbc.M111.270132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markovic-Housley Z, Basle A, Padavattan S, Maderegger B, Schirmer T, Hoffmann-Sommergruber K. Structure of the major carrot allergen Dau c 1. Acta Crystallographica Section D-Biological Crystallography. 2009;65:1206–1212. doi: 10.1107/S0907444909034854. [DOI] [PubMed] [Google Scholar]
- 35.Chruszcz M, Chapman MD, Vailes LD, Stura EA, Saint-Remy JM, Minor W, Pomes A. Crystal structures of mite allergens Der f 1 and Der p 1 reveal differences in surface-exposed residues that may influence antibody binding. J Mol Biol. 2009;386(2):520–530. doi: 10.1016/j.jmb.2008.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madhurantakam C, Nilsson OB, Uchtenhagen H, Konradsen J, Saarne T, Hogbom E, Sandalova T, Gronlund H, Achour A. Crystal structure of the dog lipocalin allergen Can f 2: implications for cross-reactivity to the cat allergen Fel d 4. J Mol Biol. 2010;401(1):68–83. doi: 10.1016/j.jmb.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 37.Mueller GA, Gosavi RA, Pomés A, Wünschmann S, Moon AF, London RE, Pedersen LC. Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011;66(7):878–885. doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan YW, Chan SL, Ong TC, Yit le Y, Tiong YS, Chew FT, Sivaraman J, Mok YK. Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes. J Biol Chem. 2009;284(5):3148–3157. doi: 10.1074/jbc.M807209200. [DOI] [PubMed] [Google Scholar]
- 39.Calvo E, Mans BJ, Ribeiro JM, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc Natl Acad Sci U S A. 2009;106(10):3728–3733. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310(1):243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 41.McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19(7):874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- 42.Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299(2):499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 43.Shindyalov IN, Bourne PE. A database and tools for 3-D protein structure comparison and alignment using the Combinatorial Extension (CE) algorithm. Nucleic Acids Res. 2001;29(1):228–229. doi: 10.1093/nar/29.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanciuc O, Oezguen N, Mathura VS, Schein CH, Xu Y, Braun W. Using property based sequence motifs and 3D modeling to determine structure and functional regions of proteins. Curr Med Chem. 2004;11(5):583–593. doi: 10.2174/0929867043455819. [DOI] [PubMed] [Google Scholar]
- 45.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16(6):566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 46.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 47.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 48.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19(3):319–333. [Google Scholar]
- 49.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27(3):343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 52.Kurup VP, Sussman GL, Yeang HY, Elms N, Breiteneder H, Arif SA, Kelly KJ, Bansal NK, Fink JN. Specific IgE response to purified and recombinant allergens in latex allergy. Clin Mol Allergy. 2005;3:11. doi: 10.1186/1476-7961-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chye ML, Cheung KY. beta-1,3-Glucanase is highly-expressed in laticifers of Hevea brasiliensis. Plant Mol Biol. 1995;29(2):397–402. doi: 10.1007/BF00043663. [DOI] [PubMed] [Google Scholar]
- 54.Muller JJ, Thomsen KK, Heinemann U. Crystal structure of barley 1,3-1,4-beta-glucanase at 2. 0-A resolution and comparison with Bacillus 1,3-1,4-beta-glucanase. J Biol Chem. 1998;273(6):3438–3446. doi: 10.1074/jbc.273.6.3438. [DOI] [PubMed] [Google Scholar]
- 55.Schein CH, Ivanciuc O, Braun W. Common physical-chemical properties correlate with similar structure of the IgE epitopes of peanut allergens. J Agric Food Chem. 2005;53(22):8752–8759. doi: 10.1021/jf051148a. [DOI] [PubMed] [Google Scholar]
- 56.Kruse SW, Zhao R, Smith DP, Jones DN. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat Struct Biol. 2003;10(9):694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cerecedo I, Zamora J, Shreffler WG, Lin J, Bardina L, Dieguez MC, Wang J, Muriel A, de la Hoz B, Sampson HA. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay. J Allergy Clin Immunol. 2008;122(3):589–594. doi: 10.1016/j.jaci.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 58.Burks AW, Cockrell G, Stanley JS, Helm RM, Bannon GA. Recombinant peanut allergen Ara h I expression and IgE binding in patients with peanut hypersensitivity. J Clin Invest. 1995;96(4):1715–1721. doi: 10.1172/JCI118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolarich D, Altmann F. N-Glycan analysis by matrix-assisted laser desorption/ionization mass spectrometry of electrophoretically separated nonmammalian proteins: application to peanut allergen Ara h 1 and olive pollen allergen Ole e 1. Anal Biochem. 2000;285(1):64–75. doi: 10.1006/abio.2000.4737. [DOI] [PubMed] [Google Scholar]
- 60.Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, Helm RM, West CM, Bannon GA. Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Arch Biochem Biophys. 1997;342(2):244–253. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- 61.Barre A, Culerrier R, Granier C, Selman L, Peumans WJ, Van Damme EJ, Bienvenu F, Bienvenu J, Rouge P. Mapping of IgE-binding epitopes on the major latex allergen Hev b 2 and the cross-reacting 1,3beta-glucanase fruit allergens as a molecular basis for the latex-fruit syndrome. Mol Immunol. 2009;46(8–9):1595–1604. doi: 10.1016/j.molimm.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Mittag D, Batori V, Neudecker P, Wiche R, Friis EP, Ballmer-Weber BK, Vieths S, Roggen EL. A novel approach for investigation of specific and cross-reactive IgE epitopes on Bet v 1 and homologous food allergens in individual patients. Mol Immunol. 2006;43(3):268–278. doi: 10.1016/j.molimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 63.Ko TP, Day J, McPherson A. The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2. 0 A resolution. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 4):411–420. doi: 10.1107/s0907444900002237. [DOI] [PubMed] [Google Scholar]
- 64.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9 (Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue B, Soeria-Atmadja D, Gustafsson MG, Hammerling U, Dunker AK, Uversky VN. Abundance and functional roles of intrinsic disorder in allergenic proteins and allergen representative peptides. Proteins. 2011;79(9):2595–2606. doi: 10.1002/prot.23077. [DOI] [PubMed] [Google Scholar]
- 66.Maleki SJ, Viquez O, Jacks T, Dodo H, Champagne ET, Chung SY, Landry SJ. The major peanut allergen, Ara h 2, functions as a trypsin inhibitor, and roasting enhances this function. J Allergy Clin Immunol. 2003;112(1):190–195. doi: 10.1067/mai.2003.1551. [DOI] [PubMed] [Google Scholar]
- 67.Pantoja-Uceda D, Bruix M, Gimenez-Gallego G, Rico M, Santoro J. Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis. Biochemistry. 2003;42(47):13839–13847. doi: 10.1021/bi0352217. [DOI] [PubMed] [Google Scholar]
- 68.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35(Database issue):D786–793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleine-Tebbe J, Herold DA, Vieths S. Soy allergy due to cross reactions to major birch pollen allergen Bet v 1. Allergologie. 2008;31(8):303–313. [Google Scholar]
- 70.Kleine-Tebbe J, Vogel L, Crowell DN, Haustein UF, Vieths S. Severe oral allergy syndrome and anaphylactic reactions caused by a Bet v 1-related PR-10 protein in soybean, SAM22. J Allergy Clin Immunol. 2002;110(5):797–804. doi: 10.1067/mai.2002.128946. [DOI] [PubMed] [Google Scholar]
- 71.Hochwallner H, Schulmeister U, Swoboda I, Focke-Tejkl M, Civaj V, Balic N, Nystrand M, Harlin A, Thalhamer J, Scheiblhofer S, Keller W, Pavkov T, Zafred D, Niggemann B, Quirce S, Mari A, Pauli G, Ebner C, Papadopoulos NG, Herz U, van Tol EA, Valenta R, Spitzauer S. Visualization of clustered IgE epitopes on alpha-lactalbumin. J Allergy Clin Immunol. 2010;125(6):1279–1285. e1279. doi: 10.1016/j.jaci.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 72.Dai YC, Chuang WJ, Chua KY, Shieh CC, Wang JY. Epitope mapping and structural analysis of the anti-Der p 1 monoclonal antibody: insight into therapeutic potential. J Mol Med (Berl) 2011;89(7):701–712. doi: 10.1007/s00109-011-0744-4. [DOI] [PubMed] [Google Scholar]
- 73.Razzera G, Gadermaier G, de Paula V, Almeida MS, Egger M, Jahn-Schmid B, Almeida FC, Ferreira F, Valente AP. Mapping the interactions between a major pollen allergen and human IgE antibodies. Structure. 2010;18(8):1011–1021. doi: 10.1016/j.str.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 74.Chruszcz M, Chapman MD, Osinski T, Solberg R, Demas M, Porebski PJ, Majorek KA, Pomes A, Minor W. Alternaria alternata allergen Alt a 1: A unique beta-barrel protein dimer found exclusively in fungi. J Allergy Clin Immunol. 2012;130(1):241–247. e249. doi: 10.1016/j.jaci.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mueller GA, Edwards LL, Aloor JJ, Fessler MB, Glesner J, Pomes A, Chapman MD, London RE, Pedersen LC. The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins. J Allergy Clin Immunol. 2010;125(4):909–917. e904. doi: 10.1016/j.jaci.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chruszcz M, Pomes A, Glesner J, Vailes LD, Osinski T, Porebski PJ, Majorek KA, Heymann PW, Platts-Mills TA, Minor W, Chapman MD. Molecular determinants for antibody binding on group 1 house dust mite allergens. J Biol Chem. 2012;287(10):7388–7398. doi: 10.1074/jbc.M111.311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lopez-Torrejon G, Diaz-Perales A, Rodriguez J, Sanchez-Monge R, Crespo JF, Salcedo G, Pacios LF. An experimental and modeling-based approach to locate IgE epitopes of plant profilin allergens. J Allergy Clin Immunol. 2007;119(6):1481–1488. doi: 10.1016/j.jaci.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 78.Chen JC, Chiu LL, Lee KL, Huang WN, Chuang JG, Liao HK, Chow LP. Identification of critical amino acids in an immunodominant IgE epitope of Pen c 13, a major allergen from Penicillium citrinum. PLoS One. 2012;7(4):e34627. doi: 10.1371/journal.pone.0034627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xia L, Willison LN, Porter L, Robotham JM, Teuber SS, Sathe SK, Roux KH. Mapping of a conformational epitope on the cashew allergen Ana o 2: a discontinuous large subunit epitope dependent upon homologous or heterologous small subunit association. Mol Immunol. 2010;47(9):1808–1816. doi: 10.1016/j.molimm.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 80.Hantusch B, Krieger S, Untersmayr E, Scholl I, Knittelfelder R, Flicker S, Spitzauer S, Valenta R, Boltz-Nitulescu G, Scheiner O, Jensen-Jarolim E. Mapping of conformational IgE epitopes on Phl p 5a by using mimotopes from a phage display library. J Allergy Clin Immunol. 2004;114(6):1294–1300. doi: 10.1016/j.jaci.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 81.Barre A, Sordet C, Culerrier R, Rance F, Didier A, Rouge P. Vicilin allergens of peanut and tree nuts (walnut, hazelnut and cashew nut) share structurally related IgE-binding epitopes. Mol Immunol. 2008;45(5):1231–1240. doi: 10.1016/j.molimm.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 82.Shen HD, Tam MF, Huang CH, Chou H, Tai HY, Chen YS, Sheu SY, Thomas WR. Homology modeling and monoclonal antibody binding of the Der f 7 dust mite allergen. Immunol Cell Biol. 2011;89(2):225–230. doi: 10.1038/icb.2010.77. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Q, Willison LN, Tripathi P, Sathe SK, Roux KH, Emmett MR, Blakney GT, Zhang HM, Marshall AG. Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2011;83(18):7129–7136. doi: 10.1021/ac201501z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu D, Zhang J, Roy A, Zhang Y. Automated protein structure modeling in CASP9 by I-TASSER pipeline combined with QUARK-based ab initio folding and FG-MD-based structure refinement. Proteins. 2011;79 (Suppl 10):147–160. doi: 10.1002/prot.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moult J, Fidelis K, Kryshtafovych A, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)--round IX. Proteins. 2011;79 (Suppl 10):1–5. doi: 10.1002/prot.23200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kryshtafovych A, Fidelis K, Moult J. CASP9 results compared to those of previous CASP experiments. Proteins. 2011;79 (Suppl 10):196–207. doi: 10.1002/prot.23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deng X, Eickholt J, Cheng J. A comprehensive overview of computational protein disorder prediction methods. Mol Biosyst. 2012;8(1):114–121. doi: 10.1039/c1mb05207a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roche DB, Buenavista MT, Tetchner SJ, McGuffin LJ. The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res. 2011;39(Web Server issue):W171–176. doi: 10.1093/nar/gkr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J Allergy Clin Immunol. 2008;121(4):847–852. e847. doi: 10.1016/j.jaci.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 90.Ivanciuc O, Garcia T, Torres M, Schein CH, Braun W. Characteristic motifs for families of allergenic proteins. Molecular Immunology. 2009;46(4):559–568. doi: 10.1016/j.molimm.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.