Abstract
During neocortical development, the extensive migratory movements of neurons from their place of birth to their final location are essential for the coordinated wiring of synaptic circuits and proper neurological function. Failure or delay in neuronal migration causes severe abnormalities in cortical layering, which consequently results in human lissencephaly (‘smooth brain’), a neuronal migration disorder. The brains of lissencephaly patients have less-convoluted gyri in the cerebral cortex with impaired cortical lamination of neurons. Since microtubule- and actin-associated proteins play important functions in regulating the dynamics of microtubule and actin cytoskeletons during neuronal migration, genetic mutations or deletions of crucial genes involved in cytoskeletal processes lead to lissencephaly in human and neuronal migration defects in mouse. During neuronal migration, microtubule organization and transport are controlled by PAFAH1B1 (LIS1), DCX, YWHAE, and tubulin. Actin stress fibers are modulated by PAFAH1B1 (LIS1), DCX, RELN, and VLDLR/LRP8 (APOER2). There are several important levels of crosstalk between these two cytoskeletal systems to establish accurate cortical patterning in development. The recent understanding of the protein networks that govern neuronal migration by regulating cytoskeletal dynamics, from human and mouse genetics as well as molecular and cellular analyses, provides new insights on neuronal migration disorders and may help us devise novel therapeutic strategies for such brain malformations.
Introduction
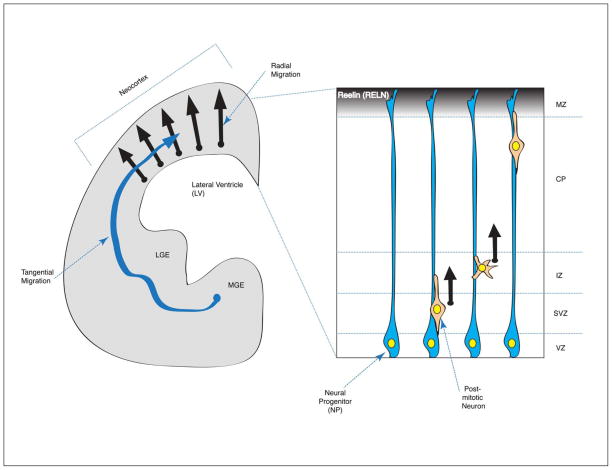
During mammalian central nervous system (CNS) development, the migration of newly born neurons to the appropriate areas within the brain, and the differentiation of post-mitotic neurons are essential for the proper establishment of synaptic circuits and their electrophysiological functions. Failure or delay in neuronal migration in the developing human neocortex results in cortical layer malformations and devastating neurological diseases. Human lissencephaly (‘smooth brain’) is a severe brain malformation disorder found in 1/30,000 births.1,2 Clinically, lissencephaly is characterized by a smooth cerebral surface of the brain without the convolutions known as gyri or sulci. Lissencephaly is always associated with mental retardation and lissencephaly patients suffer from epilepsy and motor function impairment. The most common type of lissencephaly is classical lissencephaly (Type 1), characterized by disorganized or less-defined layering of cortical neuronal lamina rather than the well-defined distinct six neuronal layers formed during normal development. Reduced thickness of the cerebral cortex is also found in the brains of classical lissencephaly patients. This cortical malformation is mainly caused by the misregulation of neuronal migration during early development. During neocortical development, neurons are born from neural progenitors (NPs) in the ventricular zone (VZ) that divide again as they pass through the subventricular zone (SVZ) and intermediate zone (IZ), as intermediate or basal progenitors. After extensive migration toward the pial surface above the marginal zone (MZ), neurons become integrated into defined positions within neuronal layers in the cortical plate (CP) (Figure 1). Therefore, the defects in neuronal migration result in the mispositioning of neurons in the neocortex.
Figure 1.
Schematic representation of the developing mammalian brain. (1) Coronal section of one half of the mammalian developing forebrain. There are two main migratory streams of post-mitotic neurons: the radial migration of excitatory cortical pyramidal neurons from the ventricular zone (VZ) to the cortical plate (CP) (black arrow) and the tangential migration of inhibitory GABAergic interneurons from lateral- and medial- ganglionic eminences (LGE/MGE) into the neocortex (blue arrow). (2) The developing cerebral cortex in mammals is multi-layered with different neuronal cell populations. Near the lateral ventricle (LV) surface, neural progenitors (NPs) reside in the ventricular zone (VZ). This progenitor zone is extended to subventricular and intermediate zones (SVZ and IZ, respectively). Newly born neurons from the division of NPs undergo extensive radial neuronal migration to enter the cortical plate (CP). The marginal zone (MZ) is a most superficial layer to contain Cajal-Retzius cells secreting the RELN (Reelin) glycoprotein.
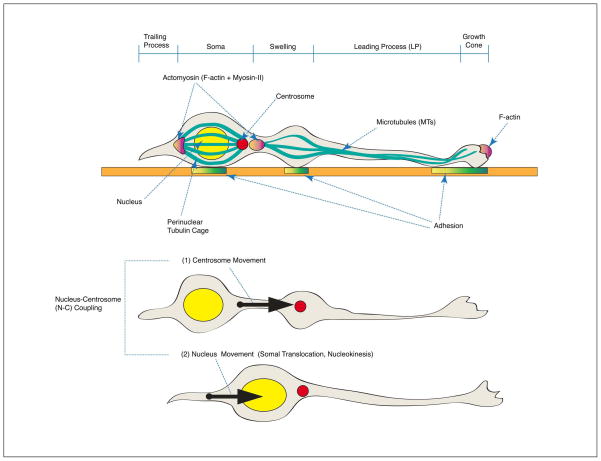
There are two different types of neuronal migration that occur during mammalian forebrain development: (1) radial migration; and (2) tangential migration. During radial migration, the excitatory cortical pyramidal neurons are born from NPs in the VZ and migrate to the CP. In a sequential migration stream, later-born neurons bypass early-born neurons and occupy more superficial layers of the CP, which generates an ‘inside-out’ pattern of cortical layers.3 Radial migration occurs extensively in the cerebral cortex and hippocampus during embryonic brain development. During tangential migration, inhibitory interneurons are generated from the different types of progenitors at the medial or lateral ganglionic eminence (MGE/LGE) and migrate to the neocortex (Figure 1). Radially migrating neurons generally display bipolar morphologies along radial glia with only a few branch points (although they do display transient multipolar morphologies in IZ layer). By contrast, tangentially migrating neurons frequently change the directions of migration and display dynamic morphological transitions. When post-mitotic neurons migrate, cytoskeletal remodeling of microtubules (MTs) and actin is evident in these cells. The volume of the nucleus (also called the soma) compared to those of extended processes is dramatically increased in the migrating neurons. The nucleus is surrounded by MT-enriched arrays in front of (fork-like MTs) and behind (cage-like MTs) the nucleus. In the leading process, the extended bundles of MTs emanate from the microtubule-organizing center (MTOC, also called centrosome) in front of the nucleus. Positioning of the centrosome defines the direction of movements of migrating neurons. The most anterior part of the leading process is the growth cone that senses extrinsic guidance cues and extends to the migration site. This protruded membrane structure is composed of filamentous actin (F-actin) stress fibers that establish new contacts with adhesion substrates (focal adhesion). Migration proceeds during neuronal locomotion in two modes of movements as a two stroke action, with asynchronous movements of the centrosome (C) and later the nucleus (N). The centrosome first moves into a swelling in the leading process and then the nucleus follows (nuclear translocation, also called somal translocation or nucleokinesis) due to a pulling force from MTs and dynein motors located at the centrosome. Centrosome movement forward into the leading process precedes nuclear movement and this coordinated relationship is called nucleus-centrosomal (N-C) coupling.4,5 Eventually, the trailing process is retracted due to actomyosin (F-actin + Myosin)-dependent motor functions, which leads to net movement of the neuron. Myosin-II-dependent motors play a dual role in nuclear movement by generating a forward MT pushing force and a pulling force from behind6 (Figure 2). This repetitive MT-actin remodeling is dynamically regulated during radial migration. Similarly, MGE-derived tangentially migrating cells also undergo MT- and actomyosin-dependent migratory cycles during nucleokinesis.7,8 These dramatic cytoskeletal changes are major common characteristics of neuronal cell migration.
Figure 2.
Cytoskeletal features of migrating neurons during central nervous system (CNS) development. Migrating neurons are polarized from the growth cone, which is the migrating tip of leading process (LP) to the trailing process (TP). The nucleus is surrounded by a perinuclear tubulin cage and the rear side of nucleus is enriched with actomyosin (filamentous actin, F-actin + Myosin-II) that generates pushing forces of nuclear movement (somal translocation, nucleokinesis). Migration occurs in two distinct modes of movements in a two stroke model: (1) Centrosome (C) movement toward the swelling in the LP (2) Nuclear (N) movement in the direction of migration. This N-C coupling consequently provides the pulling force on microtubules (MTs) along the LP, which establishes new contacts to adhesion substrates.
A diverse collection of cytoskeletal proteins is involved in the tight control of neuronal migration during neocortical development. Extensive studies on the genes mutated in human neuronal migration defects have uncovered critical roles of their protein products in the regulation of cytoskeleton dynamics during neuronal migration and development. In this review, we will focus on some of the genetic studies in humans and mouse model system that identified those genes. We will also highlight recent approaches focusing on cellular mechanisms of action of these key proteins working in neuronal migration in the context of the microtubule and actin cytoskeletons.
PAFAH1B1 (LIS1)
Human and mouse genetics
Human PAFAH1B1 (platelet-activating factor acetylhydrolase isoform 1b regulatory subunit 1, formerly known as LIS1, Lissencephaly-1) was the first identified causal gene of neuronal migration disorders in any organism. Heterozygous de novo mutation or deletion of human PAFAH1B1 gene on chromosome 17p13.3 is responsible for 40% of patients with isolated lissencephaly sequence (ILS).1,2,9,10 Thus far, more than 70 distinct intragenic heterozygous mutations have been identified from human genetic studies.11 Severe brain malformations are the main characteristics of these patients, including simplified gyration in the cerebral cortex (agyria, pachygyria), disrupted cortical lamination, enlarged ventricles and neuronal heterotopias, leading to short lifespan of ILS patients.9
PAFAH1B1 (LIS1) protein is evolutionarily conserved from yeast to mammals. In order to study the in vivo function of mouse Pafah1b1 (Lis1), our group generated Pafah1b1 knockout (ko, null) and hypomorphic conditional knockout (hc) alleles by gene targeting. Pafah1b1 heterozygous (Pafah1b1ko/+) mice display mild neuronal migration defects in the cortex and hippocampus.12 Further reduction of PAFAH1B1 (LIS1) protein levels by producing compound heterozygous (Pafah1b1hc/ko) mice results in enhanced anatomical abnormalities in cortical and hippocampal structures. Pafah1b1 compound heterozygous (Pafah1b1hc/ko) mice display severe thinning of the cerebral cortex and broadly diffuse CP laminar organization. The hippocampal structures in these mice were also markedly disorganized due to neuronal migration defects during hippocampal development.13 These PAFAH1B1 dosage-dependent phenotypes in mouse animal models support a haploinsufficiency model for lissencephaly resulting from heterozygous deletion or mutation of PAFAH1B1 in humans. Epileptic phenotypes and deficits in learning and motor function were also observed in these Pafah1b1 mouse models, which have been seen in most human ILS patients.14,15
PAFAH1B1 (LIS1), a member of NUD family proteins and a subunit of PAFAH
PAFAH1B1 (LIS1) is a 45kDa protein with a N-terminal, homodimerization and coiled-coil domain. The C-terminus of PAFAH1B1 contains seven WD-40 (tryptophan-aspartic acid-40) repeats that are required for dynein/MT binding. The PAFAH1B1 homolog nudF was identified as one of the nuclear distribution nud mutants in the bread mold Aspergillus nidulans. The nud mutants exhibit defects in nuclear movement into fungal hyphae during sporulation. Several nud mutants were genetically identified in Aspergillus nidulans and one of the nud mutants was cytoplasmic dynein, a MT minus end-directed motor. Cytoplasmic dynein is a MT motor that plays a key role in generating the pulling power of retrograde cargo transport along MTs. In addition, dynactin, a dynein-regulatory accessory protein, was also identified as a nud mutant, confirming the participation of dynein in nuclear movements. Several NUD protein-dynein interactions are evolutionarily conserved, supporting the critical functions of PAFAH1B1-NUD proteins in cytoplasmic dynein regulatory pathways that control nuclear movement and nucleus positioning.
Among mammalian NUD family proteins, NUDE (nudE nuclear distribution gene E homolog) and NUDC (nudC nuclear distribution gene C homolog) were identified as PAFAH1B1 (mammalian nudF homolog protein)-binding partners. Mammalian nudE homologues are now termed NDE1 (mammalian NudE, mNudE) and NDEL1 (mNudE-like) (see Box 1, NDE1 and NDEL1). NUDC directly binds to PAFAH1B1 and dynein/dynactin complex and plays a chaperonin-like role for PAFAH1B1 stabilization.16,17 A recent study demonstrated that Nudc siRNA-mediated knockdown (KD) in rat brain also results in neuronal migration defects.18
Box 1.
NDE1, NDEL1: evolutionarily-conserved PAFAH1B1 (LIS1)-binding partners
NDE1 (nuclear distribution gene E homolog 1) and NDEL1 (NDE1-like), mammalian homologues of NudE proteins, directly interact with PAFAH1B1 through its conserved coiled-coil domain.22,44 PAFAH1B1-dynein-NDE1 forms a complex that generates a persistent force of transport.45 In recent human genetic studies, NDE1 frame-shift mutations with protein decay have been reported and these mutations cause micro-lissencephaly (brain size reduction with cortical gyral simplification). NDE1-mutated patients have abnormal cortical gyral pattern and partial cortical layering defects in the brain. Similar mutation-carrying NDE1 proteins in vitro cannot localize to the kinetochores or centrosomes where normal NDE1 protein is recruited.46,47
Nde1 homozygous knockout (KO) mice (Nde1−/−) display microcephaly (small brain) with a fairly well-preserved CP. Detailed examination of the Nde1 KO mouse cortex revealed that migrating neurons exhibited moderately retarded migration with thin superficial CP layers.48 By contrast, Ndel1 heterozygous (Ndel1ko/+) mice display no obvious phenotype but further reduction of NDEL1 protein level in compound heterozygous (Ndel1hc/ko) mice leads to cortical patterning abnormalities such as irregularly diffuse CP neuronal layering. In addition, Ndel1 mutants (Ndel1hc/ko) display significant splitting of the hippocampal pyramidal cell layer, suggesting neuronal migration defects in radial migration during hippocampal development.49 Nde1 and Pafah1b1 double mutants (Nde1−/−; Pafah1b1ko/+) display synergistic effects on neuronal migration49, while Ndel1 and Pafah1b1 double mutants (Nde1hc/ko; Pafah1b1ko/+) display synergistic effects on brain size and organization.50 Thus, the phenotypes of loss-of-function of NDE1 and NDEL1 are somewhat different, suggesting that NDE1 and NDEL1 may play distinct roles in PAFAH1B1 pathway during brain development.
MT regulation
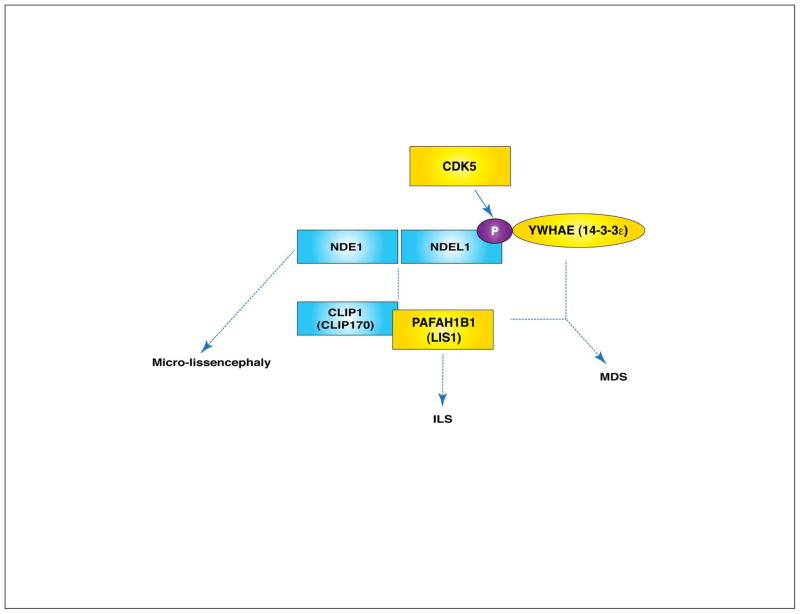
We recently demonstrated that Ndel1 and Pafah1b1 have distinct dosage-dependent effects on neuronal migration, neurite outgrowth and N-C coupling.51,52 However, cellular functions of NDEL1 in migrating neurons seem to be mediated by MT cytoskeleton regulation that is somewhat similar to PAFAH1B1/dynein-dependent MT reorganization. In post-mitotic migrating neurons, CDK5-mediated phosphorylation of NDEL1 enhances PAFAH1B1/dynein-regulated MT dynamics and promotes the binding to YWHAE. Importantly, both NDE1 and NDEL1 proteins are primarily localized to the centrosomes in mammalian cells, which suggests these NUD family proteins are important for MT reorganization by mediating central functions of PAFAH1B1 at the centrosomes. Together, NDE1 and NDEL1 have key roles in cortical neuronal migration by integrating signals from centrosome/MT-associated proteins (Figure 4).
Interestingly, the PAFAH1B1 protein was first discovered to be a non-catalytic subunit of PAFAH (platelet-activating factor acetylhydrolase). The fully-functional PAFAH complex is composed of two PAFAH catalytic subunits (PAFAH1B2 and PAFAH1B3, formerly known as PAFAH alpha 2 and PAFAH alpha 1, respectively) and homodimers of regulatory subunits, PAFAH1B1. This PAFAH complex inactivates the intracellular messenger PAF (platelet-activating factor) by removing the acetyl moiety19 (Figure 3). The PAF receptor-encoding gene PTAFR is expressed in developing brain and PAF receptor-deficient mice (Ptafr−/−) show cerebellum disorganization. Importantly, the speed of neuronal migration is significantly slower in double mutants for Pafah1b1 and the PAF receptor (Ptafr−/−; Pafah1b1ko/+) than Pafah1b1 heterozygous (Pafah1b1ko/+), suggesting the crucial role of PAF signaling pathway in neuronal migration.20
Figure 3.
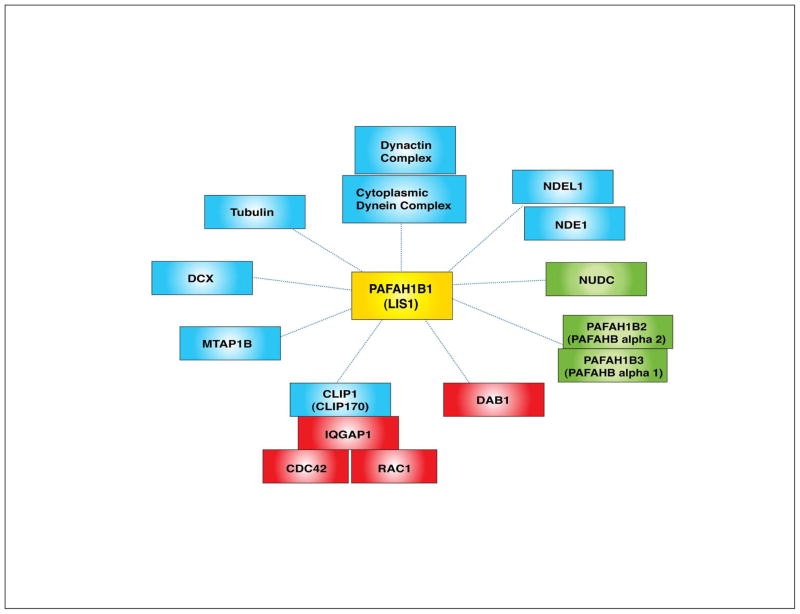
PAFAH1B1 (platelet-activating factor acetylhydrolase 1 regulatory subnuit 1, formerly known as LIS1)-binding partners. The binding of PAFAH1B1 (LIS1) to the cytoplasmic dynein/dynactin complex is evolutionarily conserved from fungi to mammals. Nuclear distribution proteins such as NDE1, NDEL1 and NUDC interact directly with PAFAH1B1. PAFAH1B1 itself is a microtubule-associated protein (MAP) and is localized at tubulin/MT-rich subcellular compartments in cells like centrosomes. Other MAPs, such as DCX and MTAP1B, are also PAFAH1B1-binding partners. PAFAH1B1 was also identified as a non-catalytic subunit of PAFAH and PAFAH1B1 associates with PAFAH catalytic subunits such as PAFAH1B2 (PAFAH alpha 2) and PAFAH1B3 (PAFAH alpha 1). When PAFAH1B1 binds to CLIP1 (CLIP170), a MT plus end protein, it forms a complex with IQGAP1 and CDC42/RAC1. Through this interaction, PAFAH1B1 participates in F-actin dynamics in growth cones during neuronal migration. Interestingly, PAFAH1B1 also binds to DAB1, an actin-regulatory protein acting in RELN signaling pathway. PAFAH1B1 has dual roles in distinct regulatory pathways of MT and actin cytoskeletons. (Blue proteins: MT regulators, red proteins: actin regulators, green proteins: Unclear detailed function in MT/actin regulation)
MT regulation
PAFAH1B1 (LIS1) directly binds to tubulin and stabilizes MTs by regulating MT dynamics.21,22 Most importantly, PAFAH1B1 has been implicated in cytoplasmic dynein/dynactin-mediated MT cytoskeletal changes.10 Dynein is a MT minus end-directed motor protein that regulates MT dynamics and generates MT pulling forces in the cells. This dynein/MT modulation by PAFAH1B1 (LIS1) is evolutionarily conserved from Aspergillus to mammals. The Aspergillus PAFAH1B1 homolog (nudF) mutants exhibit less MT dynamics with reduced rates of MT polymerization at MT plus ends.23 Mammalian PAFAH1B1 directly binds to several subunits of cytoplasmic dynein and dynactin.22,24,25 Pafah1b1-deficient cells display an increase in perinuclear localization of MT arrays and a decrease in MT plus end distribution at the cell periphery. These phenotypes seen in PAFAH1B1 deficiency are consistent with a loss of cytoplasmic dynein/dynactin complex at plasma membranes in these cells.22,25,26 In addition to this, PAFAH1B1 protein is enriched at MT-concentrated intracellular compartments such as the centrosome in mammalian cells.24,25 A decrease in PAFAH1B1 protein levels causes defects in N-C coupling with reduced distance between nucleus and centrosome during neuronal cell migration.4,27 The pulling force of nuclear movement during nucleokinesis is generated by the PAFAH1B1/dynein complex at the centrosome. Therefore, defects in N-C coupling of Pafah1b1-deficient neurons result in slower migration speeds of these cells. PAFAH1B1 and dynein KD cells display similar migration defects in rat cortical cultures, suggesting that PAFAH1B1 and dynein converge on the same MT regulatory pathways in cortical development.28 PAFAH1B1 itself is an atypical MAP (microtubule-associated protein), and interacts with other MAPs such as DCX (doublecortin)29 and MTAP1B (microtubule-associated protein 1b)30. DCX binding to MTs elevates the rate of MT polymerization and stabilizes MT arrays. Similarly, MTAP1B binding on MTs enhances the growth of dynamic MTs.29,30 PAFAH1B1 also binds to CLIP1 (formerly known as CLIP170), a MT plus end binding protein that helps loading of vesicle cargoes along MT arrays to the cell periphery31,32 (Figure 3).
Actin regulation
Interestingly, PAFAH1B1-CLIP1 at MT plus ends is part of another large complex with CDC42 and RAC1, members of the small GTPase family proteins that function as actin regulatory proteins. An effector of these small GTPases, IQGAP1, participates in this interaction and further stabilizes this complex.33 PAFAH1B1 promotes F-actin polymerization at distal ends of leading processes by modulating CDC42/RAC1-dependent actin stabilization. PAFAH1B1 has critical roles in regulating these small GTPases by activating CDC42/RAC1 and antagonistically inactivating RHOA (encoded by RhoA gene). Pafah1b1 deficiency in heterozygous cells (Pafah1b1ko/+) leads to the misregulation of actin cytoskeleton at the growth cones in the leading processes of migrating neurons. Pafah1b1-deficient neurons have reduced F-actin in the leading edge and less numbers of filopodia, which results in slower migration of these cells.33,34 This actin cytoskeleton remodeling by PAFAH1B1 also contributes to proper neuronal migration. Intriguingly, PAFAH1B1 binds to DAB1 (disabled homolog 1), a scaffold adaptor protein downstream of RELN (see RELN section). By interacting with phosphorylated-DAB1 (P-DAB1) in a phosphorylation-dependent manner, PAFAH1B1 controls F-actin polymerization in the growth cones of the migrating neurons35 (Figure 3). DAB1-mediated crosstalk between PAFAH1B1 and RELN pathways has pivotal roles linking cytoskeletal dynamics of actin to those of MTs.
Deletion of human chromosome 17p13.3
Human and mouse genetics
Large deletions of human chromosome 17p13.3 cause Miller-Dieker syndrome (MDS), a severe form of lissencephaly (often complete agyria) accompanied by craniofacial dysmorphisms.2 In contrast to ILS patients, where 40% display PAFAH1B1 deletions, 100% MDS patients have larger contiguous gene deletions consisting of both PAFAH1B1 and YWHAE. More than 20 genes are located in this deleted region. Among these genes, the human YWHAE gene is located near the telomeric tip of chromosome 17p about 1Mb from the PAFAH1B1 gene. YWHAE gene encodes the 14-3-3 epsilon (tyrosine 3-monooxygenase 5-monooxygenase activation protein, epsilon peptide) protein, a member of 14-3-3 protein family that is an evolutionarily conserved signaling molecule.36,37 The location of mouse Ywhae gene is also conserved on mouse chromosome 11 where mouse Pafah1b1 gene is located, and these are conserved synteny regions on human chromosome 17p13.3 region and mouse chromosome 11.
Mouse Ywhae null homozygous mutants display hippocampal and cerebral disorganization. Double compound heterozygous mice with both Pafah1b1 and Ywhae mutations (Pafah1b1ko/+; Ywhae−/−) display more severe migration defects than those of single mutants, consistent with the more severe brain phenotypes seen in MDS patients.38 This synergistic effects of reduced YWHAE protein levels on the severity of neuronal migration in Pafah1b1-deficient mice suggest that YWHAE may modulate components of the PAFAH1B1 pathway to control the motility of neurons.
YWHAE mediated MT regulation
YWHAE protein binds to CDK5-phosphorylated-NDEL1 (P-NDEL1) directly. CDK5 (cyclin-dependent kinase 5) plays a key role in the regulation of neuronal migration in mice (see Box 2. CDK5). YWHAE binding to NDEL1 protects P-NDEL1 from PPP2 (protein phosphatase 2, formerly known as PP2A)-dependent dephosphorylation and enhances P-NDEL1 stability.38 These CDK5-mediated phosphorylation sites of NDEL1 are involved in dynein and PAFAH1B1 interaction. Hence, the P-NDEL1/YWHAE complex binds tightly to cytoplasmic dynein and PAFAH1B1 along MTs. Loss of the YWHAE protein results in mislocalization of NDEL1 and PAFAH1B1 offloaded from MT plus end tips38, suggesting that YWHAE also converges on PAFAH1B1/NDEL1/dynein-regulated MT remodeling pathway during neuronal migration.
Box 2.
CDK5: a key kinase to regulate neuronal migration in mice
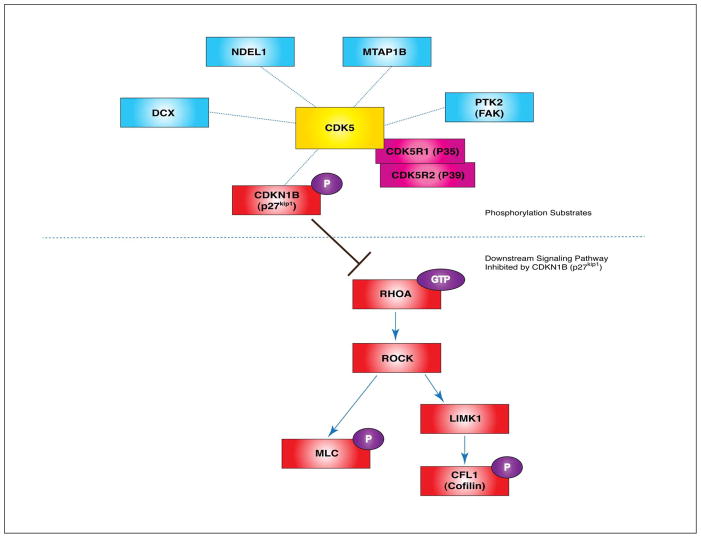
CDK5 (cyclin-dependent kinase 5) is a multifunctional serine/threonine kinase involved in neuronal migration by regulating phosphorylation events of several substrates. Upon binding of neuronal activators CDK5R1 (CDK5 regulatory subunit 1, formerly known as p35) and CDK5R2 (CDK5 regulatory subunit 2, formerly known as p39), CDK5 becomes enzymatically active in the developing brain. Cdk5 KO mice and Cdk5r1/Cdk5r2 DKO mice display inverted cortical layering, suggesting that the CDK5-CDK5R1/CDK5R2 pathway is essential for normal cortical development and neuronal migration.79
MT regulation
CDK5 phosphorylates NDEL1, a PAFAH1B1-binding MT/dynein regulator and P-NDEL1 increases peripheral MT polymerization.22,44 DCX is also a substrate of CDK5 phosphorylation. P-DCX reduces the affinity of DCX binding to MT arrays, which results in DCX localization at perinuclear MTs.27 MTAP1B is phosphorylated by CDK5 and P-MTAP1B regulates MT-enriched axon elongation in migrating neurons.80 Finally, CDK5-mediated PTK2 (protein tyrosine kinase 2, formerly known as FAK) phosphorylation is required for MT organization, nuclear movement and neuronal migration.81
Actin regulation
Another important substrate of CDK5 in neuronal migration is CDKN1B (cdk inhibitor 1B, formerly known as p27kip1), an actin-regulatory protein. P-CDKN1B (phosphorylated form of CDKN1B) is stabilized and suppresses RHOA-GTPase activity.82 RHOA has critical roles in actin cytoskeleton regulation by two major pathways: RHOA-ROCK(Rho kinase)-LIMK1-CFL1 phosphorylation and RhoA-ROCK-MLC (myosin light chain) phosphorylation. Therefore, CDK5-dependent CDKN1B phosphorylation is required for the inhibition of RHOA activity to reorganize the actomyosin network during neuronal migration (Figure 6).
VLDLR/LRP8 (APOER2)
Human VLDLR mutations cause severe lissencephaly with cerebellar malformation.83 Homozygous mutant patients display mental retardation and gyral simplification in the cortex. As noted above, VLDLR and LRP8 are the members of the lipoprotein receptor family proteins at the plasma membrane and act as co-receptors for RELN.84,85 Compound DKO mice of VLDLR/LRP8 display perturbation of neuronal migration that phenocopies the Reeler mutant.86 VLDLR/LRP8 play critical roles in actin-MT regulation by interacting with DAB1 through their cytosolic domain and VLDLR/LRP8-DAB1 interaction triggers cytoskeletal remodeling during neuronal polarization and migration (Figure 5) (see also RELN section).
CRK mediated Actin regulation
CRK (v-crk sarcoma virus CT10 oncogene homolog) is one of the genes located within the MDS critical region at the telomeric end of human chromosome 17p39, adjacent to YWHAE, and additional loss of CRK may contribute to the more severe phenotype of MDS. CRK family proteins function as adaptor molecules to reorganize the actin cytoskeleton by regulating adhesion signals.40 Intriguingly, CRK and CRKL (CRK-like) interact with P-DAB1 that is implicated in RELN-mediated actin regulatory pathway.41 Crk null mice display craniofacial defects during early development42 and Crk/Crkl KD prevents RELN-induced dendrite extension in hippocampal neurons43, which are consistent with the severe craniofacial and neurological phenotypes seen in MDS patients.
DCX
Human and mouse genetics
DCX (doublecortin) is the most common genetic cause of X-linked lissencephaly and the human gene is located in Xq22.3-q23. Clinically, heterozygous DCX mutations in females (DCX+/−) result in subcortical band heterotopia (SBH, misplaced neurons in the white matter rather than the CP), while hemizygous mutation in males (DCXy/−) results in ILS. Female phenotypes with DCX mutation are variable due to random X-inactivation, resulting in genetic mosaicism in these patients.53,54 One distinct feature in DCX-associated ILS compared to that of PAFAH1B1-associated ILS is that PAFAH1B1 has a more severe posterior (P) cortical abnormality, while DCX has a more severe anterior (A) phenotype (DCX P>A vs. PAFAH1B1 P<A).
Mouse Dcx genetic male mutants (Dcxy/−) display mild histological defects in the cortex and more notable disorganization of the hippocampus55, probably due to compensatory mechanisms from doublecortin-like kinase (Dclk). In support of this, Dcx/Dclk double knockout (DKO) mice display severe defects in neuronal migration with abnormal lamination in the CP with the accumulation of multipolar neurons in the IZ.56,57 More careful analysis of neuronal migration by live imaging of brain slices from Dcx genetic male mouse mutants (Dcxy/−)58 or acute Dcx KD cells in rat brain slices59 uncovered significant migration defects in Dcx-deficient migrating neurons.
MT regulation
DCX is a MAP expressed in NPs and differentiating neurons. DCX protein has two evolutionarily conserved tandem repeat domains that are required for MT binding and stabilization. DCX stabilizes MTs and enhances MT polymerization as well as the bundling capacity of MTs. Dcx mutant and Dcx KD immature neurons exhibit weakened N-C coupling and ultimately delayed centrosomal and nuclear movement.27,60 DCX is also a phosphoprotein that serves as a substrate of several kinases, including: MARK1 (microtubule affinity-regulating kinase 1) and PRKA (formerly known as PKA, protein kinase A) at residue Ser47 61, and CDK5 mainly at residue Ser297 62. MARK1- and PRKA-mediated phosphorylation of DCX reduces its MT binding activity. P-Ser47-DCX (phosphorylated form of DCX at Ser47) is required for proper localization of DCX protein at the leading processes of migrating neurons. Ser297 of DCX is the in vivo phosphorylation target of CDK5 and this phophorylation event enhances the MT binding ability of DCX. DCX may directly interact with PAFAH1B129, and this interaction may contribute to MT stabilization near perinuclear regions in migrating neurons to provide power for nuclear translocation (nucleokinesis). Thus, DCX increases MT stabilization and MT nucleation by increasing tubulin polymerization, and is one of the key MT regulators in corticogenesis during early development.
Actin regulation
DCX is also phosphorylated by MAPK8 (mitogen-activated protein kinase 8, formerly known as JNK) at residue Thr321, Thr331, and Ser334. Phosphorylation of these sites of DCX are responsible for DCX recruitment to the growth cones of leading processes, the F-actin-rich zone. MAPK8-mediated DCX phosphorylation is essential for neurite outgrowth of migrating neurons.63 Through binding to MAPK8IP1 (MAPK8-interacting protein 1, formerly known as JIP-1), DCX may control actin dynamics by regulating the RELN pathway, since JIP-1 directly binds to LRP8 (APOER2), a RELN receptor. DCX interaction with PPP1R9B (protein phosphatase 1 regulatory subunit 9B, formerly known as Spinophilin or Neurabin-II) may also provide a mechanism linking actin and DCX. DCX itself is co-sedimented with F-actin in vitro and adding PPP1R9B elevates DCX-F-actin binding affinity.64 PPP1R9B is an actin-binding protein functioning as a protein phosphatase-1 (PPP1)-adaptor protein. PPP1R9B-mediated PPP1 targeting of DCX induces dephosphorylation of the residue Ser297. PPP1R9B and DCX colocalize at the growth cones where MT and actin dynamically interact. Dephosphorylation of Ser297 of DCX by PPP1R9B/PPP1 is important for DCX distribution at neurite tips during neuronal migration.65
Tubulin – TUBA1A, TUBB2B, TUBB3
Given that PAFAH1B1, DCX and YWHAE are all MT-associated proteins, it is not a surprising finding that tubulin mutations themselves, including TUBA1A, TUBB2B and TUBB3 can cause neuronal migration disorders and cortical malformations.
TUBA1A
TUBA1A (tubulin alpha-subunit 1A isoform) was first reported to have key roles in cortical development in both mouse and human.66 The mouse model, first described in an ENU mutagenesis study, displayed hippocampal disorganization and behavioral deficits with no overt cortical phenotype, while in human TUBA1A mutation-harboring patients, cortical laminar organization was disrupted and hypoplastic cerebellum and brainstem regions were also observed. Mutations found in the mouse and humans were located either within GTP binding pocket or structurally close to the motif participating in beta-tubulin subunit interaction, suggesting that GTP-dependent incorporation of alpha- and beta-tubulin in heterodimer was impaired and resulted in defective MT polymerization.66 A subsequent study demonstrated that tubulin-related cortical malformations have a very broad phenotypic spectrum in human patients.67
TUBB2B/TUBB3
Heterozygous mutation in human TUBB2B (tubulin beta-subunit 2B isoform) causes asymmetric polymicrogyria (PMG), characterized by multiple small gyri separated by shallow sulci. Halting of neuronal migration within IZ in Tubb2b KD rat embryo brains suggests that TUBB2B contributes to neuronal migration during development.68 More recently, other beta-tubulin TUBB3 (tubulin beta-subunit 3 isoform) human heterozygous mutations were identified that lead to cortical malformation and perturbation of axon guidance by altering MT dynamics.69
RELN (Reelin)
Human and mouse genetics
RELN (Reelin), a large extracellular matrix glycoprotein (~400kDa), is expressed and secreted by Cajal-Retzius cells at the MZ layer of the developing brain. Mutation of the human RELN gene located on chromosome 7q22 causes an autosomal recessive form of lissencephaly syndrome with cerebellar hypoplasia as well as malformations in the hippocampus and brainstem.70
The mouse Reeler mutant (RELN mutation) was the first mouse mutant described with neuronal migration defects and widely studied. Homozygous Reeler mice display ataxia, cerebellar hypoplasia and inverted ‘outside-in’ cortical layering, composed of superficial early-born neurons and deep positioning later-born neurons (opposite to the normal CP layering).71,72 The absence of a well-defined MZ layer is another characteristic of Reeler mouse cortex due to over-migration of neurons. This observation suggests that RELN functions as a ‘stop’ signal of migrating neurons.73 The finding that RELN induces the detachment of migrating neuron from radial glial fibers74 is consistent with the idea that RELN acts as a ‘stop and detach’ signal during radial migration.75
MT regulation
RELN binds to the transmembrane receptors VLDLR (very low density lipoprotein receptor) and LRP8 (low density lipoprotein-related receptor 8, formerly known as APOER2). (see VLDLR/LRP8(APOER2) section). RELN enhances the interaction between RELN receptors and DAB1, an intracellular adaptor protein and further induces the phosphorylation of DAB1 by SRC family kinases (SRC, FYN) activation. Upon RELN binding, tyrosine phosphorylation of DAB1 activates the downstream PI3K pathway that mediates PI3K-AKT-GSK3beta signaling and consequently the CDK5-dependent phosphorylation of its substrates, MAPT (microtubule-associated protein tau) and MTAP1B is increased.76,77 Through this phosphorylation cascade, RELN-DAB1 regulates MT stability and dynamics in migrating neurons. Consistent with this, Dab1 KO mice display a Reeler-like phenotype with neuronal migration defects.78 Intriguingly, P-DAB1 also binds to PAFAH1B1, a key regulator of MT/dynein in phosphorylation-dependent manner. Compound heterozygous of Dab1/Pafah1b1 mouse mutant (Dab1+/−; Pafah1b1ko/+) display more severe cortical migration defects than single heterozygous mutants, suggesting important crosstalk between RELN and PAFAH1B1 signaling pathways in vivo and in vitro35 (Figure 5).
Figure 5.
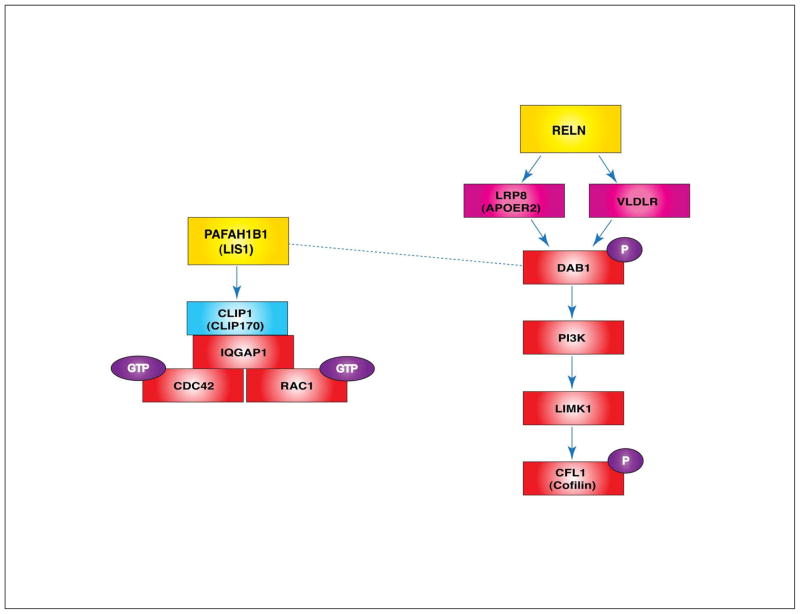
PAFAH1B1 and RELN-mediated actin regulation. Formation of the PAFAH1B1-CLIP1-IQGAP1-CDC42/RAC1 complex stabilizes and sustains the GTPase activities of CDC42 and RAC1. RELN, a large glycoprotein secreted from the MZ area in the cortex, directly binds to lipoprotein receptors such as VLDLR and LRP8 (APOER2). This binding recruits the DAB1 adaptor protein to the membrane where DAB1 is phosphorylated. P-DAB1 activates PI3K-LIMK1 signaling and LIMK1 phoshorylates CFL1 (cofilin), an actin-severing protein, which keeps CFL1 in an inactive state. This further stabilizes F-actin stress fibers in migrating neurons. Since the dynamic regulation of the actin cytoskeleton is required for neuronal motility, a balance between actin polymerization and depolymerization is essential for neuronal migration processes. (Dashed line: direct interaction between PAFAH1B1 and P-DAB1, Blue protein: MT regulator, red proteins: actin regulators)
Actin regulation
RELN-induced activation of the DAB1-PI3K pathway elevates LIMK1 (LIM-domain containing protein kinase 1) kinase activity, which phosphorylates CFL1 (formerly known as cofilin), an F-actin-severing protein. Since P-CFL1 (phosphorylated cofilin) is an inactive form, the RELN-DAB1-PI3K-LIMK1-P-CFL1 signaling cascade increases the stability of F-actin stress fibers and consequently reduces actin cytoskeleton turnover. Therefore, Reeler mutants fail to transmit a DAB1-PI3K-LIMK1 signal and display reduced P-CFL1 levels in migrating neurons, resulting in misregulation of the actin cytoskeleton75 (Figure 5). P-CFL1 may be enriched in the leading processes when migrating neurons reach the MZ layer. By elevating P-CFL1, RELN stabilizes the actin cytoskeleton and helps anchor the leading processes to the basal lamina of developing cortex. Therefore, reduced P-CFL1 in the absence of RELN results in destabilization of actin dynamics in the leading processes and leads to somal translocation failure. The Reeler mutant mouse cortex display defects in somal translocation at the final stage of migration near the MZ, suggesting that the RELN pathway mediates a ‘detach and go (somal translocation)’ signal in this stage of radial migration.75
ARX
ARX (aristaless-related homeobox) is an X-linked gene that encodes a transcription factor required for interneuron function and neuronal migration.87 Missense and truncation mutations of ARX lead to X-linked lissencephaly with abnormal genitalia (XLAG), and patients display epilepsy and severe mental retardation. Mouse Arx KO mutants similarly display neuropathological phenotypes such as GABAergic interneuron dysfunction due to tangential migration defects.88 Arx KO mice display disorganized pyramidal neuronal layering, suggesting ARX may play important role not only in tangential migration but radial migration as well.87,88 Since ARX can act as a transcriptional repressor and activator depending on promoter specificity, Arx mutation is accompanied by downregulation or upregulation of downstream target genes such as Lmo1, Ebf3, and Shox2 to influence the regionalization process of the developing brain.89,90 Genome-wide Chip-ChIP (chromatin immunoprecipition) promoter analysis from Arx mutant cells revealed that ARX binds to the promoters of genes in regulatory pathways important for axonal guidance and neurite extension91 suggesting that ARX is involved in cytoskeletal control of migration of post-mitotic neurons by transcriptional regulation.
Conclusion
Cytoskeletal remodeling of actin/MTs and the dynamic regulation of transport on these cellular structures are critical processes to induce neuronal polarization from the leading process to the trailing process and N-C coupling that drives nuclear and neuronal migration during CNS development. Several tubulin isoforms (TUBA1A, TUBB2B, TUBB3) are key components of neuronal MT arrays and DCX is tightly associated with MTs in migrating neurons. Loss of these proteins produces imbalances in MT dynamics, which causes lissencephaly or other cortical brain malformation in human. PAFAH1B1 (LIS1) is a key regulator of MT stability and dynamics by modulating cytoplasmic dynein localization and motor function, while YWHAE binds to P-NDEL1 to promote its function in dynein localization. Concomitant deletion of PAFAH1B1 and YWHAE genes in chromosome 17p13.3 is the cause of MDS in human, suggesting that misregulation of MT stability and dynein dysfunction are the causes of MDS in human. Since deletion of CRK, an actin-regulatory protein on chromosome 17p, also occurs in MDS, actin dynamics may also play a role in this severe phenotype. Interestingly, PAFAH1B1, DCX, and RELN have central functions in the interplay between actin and MT cytoskeletons by affecting different downstream cellular pathways. PAFAH1B1 specifically activates RAC1/CDC42 GTPase activity to promote F-actin polymerization. The binding of RELN to its receptors, VLDLR/LRP8 (APOER2) recruits DAB1 adaptor protein to stabilize F-actin. Since P-DAB1 also interacts with PAFAH1B1, while DCX interacts with LRP8 through binding to MAPK8IP1 (JIP-1), the effects of PAFAH1B1/DCX on MT dynamics converge to RELN-VLDLR/LRP8-DAB1-actin regulatory pathway, which allows crosstalk between the actin and MT cytoskeletons. PAFAH1B1, DCX, and RELN are important organizational nodes to coordinate neuronal migration. Lastly, ARX mutation in human results in interneuron dysfunction and this gene is implicated in tangential migration of GABAergic neurons in the developing brain. The finding that ARX binds to the promoters of genes in regulatory pathways important for axonal guidance and neurite extension91, suggests that ARX is involved in cytoskeletal control of migration of post-mitotic neurons by transcriptional regulation.
Although these previous studies have unraveled some key functions of several genes during neuronal migration in human and mouse, there are still many questions to be answered. It is likely that there is coordinate regulation between neuronal migration and neurogenesis/neuronal differentiation processes. Since many neuronal migration genes also encode centrosome-associated proteins (NDE1, NDEL1 and PAFAH1B1), cell cycle-dependent proliferation of NPs and the differentiation of daughter neurons and/or cell fates may be controlled by these centrosomal proteins. Recent studies of the human WDR62 gene demonstrated that deficiency of this centrosome-associate protein also causes micro-lissencephaly.92,93,94 Emerging evidence strongly supports the notion that subcellular components other than centrosome (critical for N-C coupling) participate in neuronal migration processes during cortical development. These candidates include gap junction proteins, adhesion molecules and proteins involved in vesicle trafficking or recycling pathways. However, it is not clear whether these proteins contribute to neuronal migration through actin/MT cytoskeletal regulation or via distinct processes. To further understand the exact molecular and cellular mechanisms underlying neuronal migration during mammalian cerebral cortex development, additional in vivo and in vitro studies are needed for the accurate mapping of genetic and physical interactions in protein-protein networks and the proper positioning of individual protein components within detailed signaling pathways. Future studies of neuronal migration will likely explore many aspects of cytoskeletal regulation in migratory processes to further understand neuropathological pathways responsible for neurodevelopmental brain malformation disorders caused by neuronal migration defects in human. Delineation of these pathways will aid in the identification of potential new therapeutic targets to ultimately cure these devastating diseases.
Figure 4.
NDE1 and NDEL1, key proteins integrating several signals in neuronal migration. NDE1 and NDEL1 are two mammalian homologues of Aspergillus nidulans nudE. Upon CDK5 (cyclin-dependent kinase 5)-mediated phosphorylation of NDEL1, P-NDEL1 binds to YWHAE (formerly known as 14-3-3 epsilon). Isolated lissencephaly sequence (ILS) is caused by the haploinsufficiency of human PAFAH1B1 (LIS1) gene. Simultaneous chromosomal deletion of the regions including YWHAE and PAFAH1B1 in human causes Miller Dieker syndrome (MDS), a severe case of lissencephaly with craniofacial malformation. Human NDE1 heterozygous mutations result in micro-lissencephaly.
Figure 6.
CDK5 phosphorylation substrates during neuronal migration. When CDK5 is activated by binding of CDK5R1 (p35)/CDK5R2 (p39), this kinase phosphorylates multiple substrates in migrating neurons. Among those substrates, DCX, NDEL1, MTAP1B and PTK2 (FAK) are MT-regulating proteins. A very interesting phosphorylation substrate of CDK5 is CDKN1B (p27kip1). P-CDKN1B suppresses RHOA GTPase activity. RHOA-ROCK-LIMK1-CFL1 and RHOA-ROCK-MLC signaling pathway have been implicated in actin cytoskeletal remodeling during neuronal migration. CDK5 indirectly affects these actin-regulating signaling pathways by modulating CDK5N1B function. (Blue proteins: MT regulators, red proteins: actin regulators)
Table 1.
| Gene | Chromosomal Locus in Human | Human Clinical Presentation | Migration Defects in Mouse | Regulation Mechanisms |
|---|---|---|---|---|
| PAFAH1B1 | 17p13.3 | ILS | Abnormal lamination of cortex and hippocampus | MT/Dynein Actin |
| YWHAE | 17p13.3 | MDS (with LIS1 deletion) | Abnormal lamination of cortex and hippocampus | MT |
| NDE1 | 16p13.11 | Micro-lissencephaly | Microcephaly Moderate lamination defects in cortex |
MT |
| DCX | Xq22.3-q23 | ILS (male) SBH (female) |
Mild disorganization of hippocampus, Modest cortical lamination defects | MT Actin |
| Tubulin (TUBA1A, TUBB2B, TUBB3) | TUBA1A, 12q13.12 TUBB2B, 6p25.2 TUBB3, 16q24.3 |
ILS with cerebellar hypoplasia TUBB2B – Asymmetric PMG |
Abnormal lamination of cortex | MT |
| RELN (Reelin) | 7q22 | ILS with cerebellar hypoplasia | Inverted layering of CP in cortex | MT Actin |
| VLDLR [LRP8 (APOER2), Mutations not found in human] | VLDLR, 9p24 | ILS with cerebellar dysfunction | Inverted layering of CP in cortex | MT Actin |
| ARX | Xp21.3 | XLAG | Absence of cortical interneurons and lamination defects | Transcriptional regulation |
The genes implicated in neuronal migration. (ILS: isolated lissencephaly sequence, MDS: Miller-Dieker syndrome, SBH: subcortical band heterotopia, PMG: polymicrogyria, XLAG: X-linked lissencephaly with abnormal genitalia. CP: cortical plate, MT: microtubule)
Contributor Information
Hyang Mi Moon, Biomedical Sciences Graduate Program, Department of Pediatrics, Institute for Human Genetics, University of California, San Francisco, San Francisco, CA, USA.
Anthony Wynshaw-Boris, Email: WynshawborisT@peds.ucsf.edu, Department of Pediatrics, Institute for Human Genetics, Eli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research, University of California, San Francisco, San Francisco, CA, USA.
References
- 1.Dobyns WB, Reiner O, Carrozzo R, Ledbetter DH. Lissencephaly. A human brain malformation associated with deletion of the LIS1 gene located at chromosome 17p13. JAMA. 1993;270:2838–2842. doi: 10.1001/jama.270.23.2838. [DOI] [PubMed] [Google Scholar]
- 2.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 3.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 4.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 6.Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiner O, Sapoznik S, Sapir T. Lissencephaly 1 linking to multiple diseases: mental retardation, neurodegeneration, schizophrenia, male sterility, and more. Neuromolecular Med. 2006;8:547–565. doi: 10.1385/NMM:8:4:547. [DOI] [PubMed] [Google Scholar]
- 10.Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 11.Friocourt G, Marcorelles P, Saugier-Veber P, Quille ML, Marret S, Laquerriere A. Role of cytoskeletal abnormalities in the neuropathology and pathophysiology of type I lissencephaly. Acta Neuropathol. 2011;121:149–170. doi: 10.1007/s00401-010-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 13.Gambello MJ, Darling DL, Yingling J, Tanaka T, Gleeson JG, Wynshaw-Boris A. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paylor R, Hirotsune S, Gambello MJ, Yuva-Paylor L, Crawley JN, Wynshaw-Boris A. Impaired learning and motor behavior in heterozygous Pafah1b1 (Lis1) mutant mice. Learn Mem. 1999;6:521–537. doi: 10.1101/lm.6.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleck MW, Hirotsune S, Gambello MJ, Phillips-Tansey E, Suares G, Mervis RF, Wynshaw-Boris A, McBain CJ. Hippocampal abnormalities and enhanced excitability in a murine model of human lissencephaly. J Neurosci. 2000;20:2439–2450. doi: 10.1523/JNEUROSCI.20-07-02439.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris NR, Efimov VP, Xiang X. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998;8:467–470. doi: 10.1016/s0962-8924(98)01389-0. [DOI] [PubMed] [Google Scholar]
- 17.Aumais JP, Tunstead JR, McNeil RS, Schaar BT, McConnell SK, Lin SH, Clark GD, Yu-Lee LY. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci. 2001;21:RC187. doi: 10.1523/JNEUROSCI.21-24-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cappello S, Monzo P, Vallee RB. NudC is required for interkinetic nuclear migration and neuronal migration during neocortical development. Dev Biol. 2011;357:326–335. doi: 10.1016/j.ydbio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase [corrected] Nature. 1994;370:216–218. doi: 10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- 20.Tokuoka SM, Ishii S, Kawamura N, Satoh M, Shimada A, Sasaki S, Hirotsune S, Wynshaw-Boris A, Shimizu T. Involvement of platelet-activating factor and LIS1 in neuronal migration. Eur J Neurosci. 2003;18:563–570. doi: 10.1046/j.1460-9568.2003.02778.x. [DOI] [PubMed] [Google Scholar]
- 21.Sapir T, Elbaum M, Reiner O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 23.Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- 24.Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 25.Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- 26.Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspi M, Atlas R, Kantor A, Sapir T, Reiner O. Interaction between LIS1 and doublecortin, two lissencephaly gene products. Hum Mol Genet. 2000;9:2205–2213. doi: 10.1093/oxfordjournals.hmg.a018911. [DOI] [PubMed] [Google Scholar]
- 30.Jimenez-Mateos EM, Wandosell F, Reiner O, Avila J, Gonzalez-Billault C. Binding of microtubule-associated protein 1B to LIS1 affects the interaction between dynein and LIS1. Biochem J. 2005;389:333–341. doi: 10.1042/BJ20050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, et al. LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol Cell Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kholmanskikh SS, Koeller HB, Wynshaw-Boris A, Gomez T, Letourneau PC, Ross ME. Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat Neurosci. 2006;9:50–57. doi: 10.1038/nn1619. [DOI] [PubMed] [Google Scholar]
- 34.Kholmanskikh SS, Dobrin JS, Wynshaw-Boris A, Letourneau PC, Ross ME. Disregulated RhoGTPases and actin cytoskeleton contribute to the migration defect in Lis1-deficient neurons. J Neurosci. 2003;23:8673–8681. doi: 10.1523/JNEUROSCI.23-25-08673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assadi AH, Zhang G, Beffert U, McNeil RS, Renfro AL, Niu S, Quattrocchi CC, Antalffy BA, Sheldon M, Armstrong DD, et al. Interaction of reelin signaling and Lis1 in brain development. Nat Genet. 2003;35:270–276. doi: 10.1038/ng1257. [DOI] [PubMed] [Google Scholar]
- 36.Lo Nigro C, Chong CS, Smith AC, Dobyns WB, Carrozzo R, Ledbetter DH. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum Mol Genet. 1997;6:157–164. doi: 10.1093/hmg/6.2.157. [DOI] [PubMed] [Google Scholar]
- 37.Chong SS, Pack SD, Roschke AV, Tanigami A, Carrozzo R, Smith AC, Dobyns WB, Ledbetter DH. A revision of the lissencephaly and Miller-Dieker syndrome critical regions in chromosome 17p13. 3. Hum Mol Genet. 1997;6:147–155. doi: 10.1093/hmg/6.2.147. [DOI] [PubMed] [Google Scholar]
- 38.Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, et al. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 39.Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, Gross A, Martin CL, Allanson J, Pilz DT, Olney AH, et al. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet. 2003;72:918–930. doi: 10.1086/374320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 41.Park TJ, Curran T. Crk and Crk-like play essential overlapping roles downstream of disabled-1 in the Reelin pathway. J Neurosci. 2008;28:13551–13562. doi: 10.1523/JNEUROSCI.4323-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park TJ, Boyd K, Curran T. Cardiovascular and craniofacial defects in Crk-null mice. Mol Cell Biol. 2006;26:6272–6282. doi: 10.1128/MCB.00472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuki T, Pramatarova A, Howell BW. Reduction of Crk and CrkL expression blocks reelin-induced dendritogenesis. J Cell Sci. 2008;121:1869–1875. doi: 10.1242/jcs.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 45.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, et al. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am J Hum Genet. 2011;88:536–547. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakircioglu M, Carvalho OP, Khurshid M, Cox JJ, Tuysuz B, Barak T, Yilmaz S, Caglayan O, Dincer A, Nicholas AK, et al. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am J Hum Genet. 2011;88:523–535. doi: 10.1016/j.ajhg.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, Miyagawa S, Hiraiwa N, Yoshiki A, Wynshaw-Boris A, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pawlisz AS, Mutch C, Wynshaw-Boris A, Chenn A, Walsh CA, Feng Y. Lis1-Nde1-dependent neuronal fate control determines cerebral cortical size and lamination. Hum Mol Genet. 2008;17:2441–2455. doi: 10.1093/hmg/ddn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Youn YH, Pramparo T, Hirotsune S, Wynshaw-Boris A. Distinct dose-dependent cortical neuronal migration and neurite extension defects in Lis1 and Ndel1 mutant mice. J Neurosci. 2009;29:15520–15530. doi: 10.1523/JNEUROSCI.4630-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hippenmeyer S, Youn YH, Moon HM, Miyamichi K, Zong H, Wynshaw-Boris A, Luo L. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68:695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 54.des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 55.Corbo JC, Deuel TA, Long JM, LaPorte P, Tsai E, Wynshaw-Boris A, Walsh CA. Doublecortin is required in mice for lamination of the hippocampus but not the neocortex. J Neurosci. 2002;22:7548–7557. doi: 10.1523/JNEUROSCI.22-17-07548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deuel TA, Liu JS, Corbo JC, Yoo SY, Rorke-Adams LB, Walsh CA. Genetic interactions between doublecortin and doublecortin-like kinase in neuronal migration and axon outgrowth. Neuron. 2006;49:41–53. doi: 10.1016/j.neuron.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka T, Koizumi H, Gleeson JG. The doublecortin and doublecortin-like kinase 1 genes cooperate in murine hippocampal development. Cereb Cortex. 2006;16 (Suppl 1):i69–73. doi: 10.1093/cercor/bhk005. [DOI] [PubMed] [Google Scholar]
- 58.Pramparo T, Libiger O, Jain S, Li H, Youn YH, Hirotsune S, Schork NJ, Wynshaw-Boris A. Global developmental gene expression and pathway analysis of normal brain development and mouse models of human neuronal migration defects. PLoS Genet. 2011;7:e1001331. doi: 10.1371/journal.pgen.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci. 2003;6:1277–1283. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- 60.Sapir T, Shmueli A, Levy T, Timm T, Elbaum M, Mandelkow EM, Reiner O. Antagonistic effects of doublecortin and MARK2/Par-1 in the developing cerebral cortex. J Neurosci. 2008;28:13008–13013. doi: 10.1523/JNEUROSCI.2363-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaar BT, Kinoshita K, McConnell SK. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 2004;41:203–213. doi: 10.1016/s0896-6273(03)00843-2. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka T, Serneo FF, Tseng HC, Kulkarni AB, Tsai LH, Gleeson JG. Cdk5 phosphorylation of doublecortin ser297 regulates its effect on neuronal migration. Neuron. 2004;41:215–227. doi: 10.1016/s0896-6273(03)00852-3. [DOI] [PubMed] [Google Scholar]
- 63.Gdalyahu A, Ghosh I, Levy T, Sapir T, Sapoznik S, Fishler Y, Azoulai D, Reiner O. DCX, a new mediator of the JNK pathway. EMBO J. 2004;23:823–832. doi: 10.1038/sj.emboj.7600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsukada M, Prokscha A, Ungewickell E, Eichele G. Doublecortin association with actin filaments is regulated by neurabin II. J Biol Chem. 2005;280:11361–11368. doi: 10.1074/jbc.M405525200. [DOI] [PubMed] [Google Scholar]
- 65.Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, Allen PB, Ellisman MH, Gleeson JG. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell. 2007;129:579–591. doi: 10.1016/j.cell.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, et al. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poirier K, Keays DA, Francis F, Saillour Y, Bahi N, Manouvrier S, Fallet-Bianco C, Pasquier L, Toutain A, Tuy FP, et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A) Hum Mutat. 2007;28:1055–1064. doi: 10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- 68.Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong SE, Shugart YY, Huang DT, Shahwan SA, Grant PE, Hourihane JO, Martin ND, Walsh CA. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 71.Caviness VS., Jr Neocortical histogenesis in normal and reeler mice: a developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 72.D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 73.Frotscher M. Cajal-Retzius cells, Reelin, and the formation of layers. Curr Opin Neurobiol. 1998;8:570–575. doi: 10.1016/s0959-4388(98)80082-2. [DOI] [PubMed] [Google Scholar]
- 74.Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, Kreidberg JA, Anton ES. Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 75.Cooper JA. A mechanism for inside-out lamination in the neocortex. Trends Neurosci. 2008;31:113–119. doi: 10.1016/j.tins.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 76.Beffert U, Morfini G, Bock HH, Reyna H, Brady ST, Herz J. Reelin-mediated signaling locally regulates protein kinase B/Akt and glycogen synthase kinase 3beta. J Biol Chem. 2002;277:49958–49964. doi: 10.1074/jbc.M209205200. [DOI] [PubMed] [Google Scholar]
- 77.Gonzalez-Billault C, Del Rio JA, Urena JM, Jimenez-Mateos EM, Barallobre MJ, Pascual M, Pujadas L, Simo S, Torre AL, Gavin R, et al. A role of MAP1B in Reelin-dependent neuronal migration. Cereb Cortex. 2005;15:1134–1145. doi: 10.1093/cercor/bhh213. [DOI] [PubMed] [Google Scholar]
- 78.Howell BW, Gertler FB, Cooper JA. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 1997;16:121–132. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su SC, Tsai LH. Cyclin-Dependent Kinases in Brain Development and Disease. Annu Rev Cell Dev Biol. 2011;27:465–491. doi: 10.1146/annurev-cellbio-092910-154023. [DOI] [PubMed] [Google Scholar]
- 80.Pigino G, Paglini G, Ulloa L, Avila J, Caceres A. Analysis of the expression, distribution and function of cyclin dependent kinase 5 (cdk5) in developing cerebellar macroneurons. J Cell Sci. 1997;110 (Pt 2):257–270. doi: 10.1242/jcs.110.2.257. [DOI] [PubMed] [Google Scholar]
- 81.Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- 82.Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat Cell Biol. 2006;8:17–26. doi: 10.1038/ncb1338. [DOI] [PubMed] [Google Scholar]
- 83.Ozcelik T, Akarsu N, Uz E, Caglayan S, Gulsuner S, Onat OE, Tan M, Tan U. Mutations in the very low-density lipoprotein receptor VLDLR cause cerebellar hypoplasia and quadrupedal locomotion in humans. Proc Natl Acad Sci U S A. 2008;105:4232–4236. doi: 10.1073/pnas.0710010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D’Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T. Reelin is a ligand for lipoprotein receptors. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 85.Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 86.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 87.Bonneau D, Toutain A, Laquerriere A, Marret S, Saugier-Veber P, Barthez MA, Radi S, Biran-Mucignat V, Rodriguez D, Gelot A. X-linked lissencephaly with absent corpus callosum and ambiguous genitalia (XLAG): clinical, magnetic resonance imaging, and neuropathological findings. Ann Neurol. 2002;51:340–349. doi: 10.1002/ana.10119. [DOI] [PubMed] [Google Scholar]
- 88.Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 89.Fulp CT, Cho G, Marsh ED, Nasrallah IM, Labosky PA, Golden JA. Identification of Arx transcriptional targets in the developing basal forebrain. Hum Mol Genet. 2008;17:3740–3760. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Colasante G, Sessa A, Crispi S, Calogero R, Mansouri A, Collombat P, Broccoli V. Arx acts as a regional key selector gene in the ventral telencephalon mainly through its transcriptional repression activity. Dev Biol. 2009;334:59–71. doi: 10.1016/j.ydbio.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 91.Quille ML, Carat S, Quemener-Redon S, Hirchaud E, Baron D, Benech C, Guihot J, Placet M, Mignen O, Ferec C, et al. High-throughput analysis of promoter occupancy reveals new targets for Arx, a gene mutated in mental retardation and interneuronopathies. PLoS One. 2011;6:e25181. doi: 10.1371/journal.pone.0025181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nicholas AK, Khurshid M, Desir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topcu M, McDonald MT, Barry BJ, Felie JM, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]