Abstract
Purpose
To identify a prognostic gene signature for HPV-negative OSCC patients.
Experimental Design
Two gene expression datasets were used; a training dataset from the Fred Hutchinson Cancer Research Center (FHCRC) (n=97), and a validation dataset from the MD Anderson Cancer Center (MDACC) (n=71). We applied L1/L2-penalized Cox regression models to the FHCRC data on the 131–gene signature previously identified to be prognostic in OSCC patients to identify a prognostic model specific for high-risk HPV-negative OSCC patients. The models were tested with the MDACC dataset using a receiver operating characteristic analysis.
Results
A 13-gene model was identified as the best predictor of HPV-negative OSCC-specific survival in the training dataset. The risk score for each patient in the validation dataset was calculated from this model and dichotomized at the median. The estimated 2-year mortality (± SE) of patients with high risk scores was 47.1 (±9.24)% compared with 6.35 (± 4.42)% for patients with low risk scores. ROC analyses showed that the areas under the curve for the age, gender, and treatment modality-adjusted models with risk score (0.78, 95%CI: 0.74-0.86) and risk score plus tumor stage (0.79, 95%CI: 0.75-0.87) were substantially higher than for the model with tumor stage (0.54, 95%CI: 0.48-0.62).
Conclusions
We identified and validated a 13-gene signature that is considerably better than tumor stage in predicting survival of HPV-negative OSCC patients. Further evaluation of this gene signature as a prognostic marker in other populations of patients with HPV-negative OSCC is warranted.
Keywords: gene signature, prognosis, HPV-negative, OSCC
Introduction
Recent studies are now starting to reveal the molecular sub-classifications of head and neck squamous cell carcinomas (HNSCC) which may help inform treatment options and prognosis. For example, HPV-driven oropharyngeal tumors are associated with better treatment response and prognosis compared to HPV-negative oropharyngeal tumors [1]. On the other hand, HPV-negative tumors, which typically arise in older patients with a history of extensive tobacco use, are more likely to carry TP53 mutations [2]. Immunohistochemical staining for p16, a surrogate marker of Human Papillomavirus (HPV) type 16 status [3, 4], with or without HPV testing, is now being applied clinically to oropharyngeal tumors to aid in assessing treatment responsiveness. In contrast, while HPV presence in the oropharynx portends better survival, we need better biomarkers to help prognosticate HPV-negative oral cavity and oropharyngeal tumors. Over the past decade, many studies have documented the use of gene expression signatures based on transcriptomic profiling to improve prognostication in many cancers. Specifically, gene expression tests, such as MammaPrint [5] and Oncotype DX [6, 7], are used clinically to guide treatment decisions for breast and colon cancer patients, respectively. We previously identified and validated a 131-gene signature (representing 108 unique known genes) that can distinguish clinically normal oral epithelium, oral dysplasia, and oral squamous cell carcinoma (OSCC) that were consisted of 70% oral cavity cancer and 30% oropharyngeal cancer [8]. Furthermore, we found that this signature was associated with OSCC-specific survival [9]. In the current study, we sought to validate the association of the 131-gene signature with survival in an independent population, and to extend our findings to determine whether a subset of this signature could aid in predicting the survival of HPV-negative OSCC patients.
Materials and Methods
Datasets
Expression profile datasets from the Fred Hutchinson Cancer Research Center (FHCRC) and from the University of Texas M.D. Anderson Cancer Center (MDACC) were used. The FHCRC dataset, used for discovery, comprised the gene expression profiles of 167 tumor samples from OSCC patients and 45 normal oral mucosa samples from patients without oral cancer, all treated at the University of Washington Medical Center, the Harborview Medical Center, or the Veterans Affairs Puget Sound Health Care System during 2003 to 2007. The FHCRC dataset also contained information on patient demographics, medical and lifestyle history, vital status, cause of death, and tumor characteristics, including tumor site, stage, and HPV status. The specimen collection, storage protocols and assays for specimen processing, and generation and normalization of raw expression data were performed as previously described [8]. The MDACC dataset, used for validation, contained the gene expression profiles of 103 samples generated from residual tumor tissue, taken from the MDACC Head and Neck Tumor Bank. Demographic characteristics, tumor site, stage, vital status, and cause of death were abstracted from the medical record and the MDACC Tumor Registry. The gene expression and other data from these tissue samples were analyzed under protocol DR09-0664 and de-identified data were shared with the FHCRC under MT2010-6749. Among 103 samples from the MDACC dataset, 74 were cancer samples from OSCC patients, 24 were matched normal oral samples from OSCC patients, and 5 were normal oral samples from other cancer patients treated in the MDACC Head and Neck Center. The gene expression data obtained from the MDACC were extracted and normalized, also using the gcRMA algorithm but as implemented in Partek® Genomics Suite™ software. The microarray data can be found in the Gene Expression Omnibus database under the accession number GSE41613 and GSE42743
Statistical Methods
Validation of the 131-gene signature association with survival
We have previously reported that the Principal Component (PC) scores calculated from the 131 genes were associated with OSCC-specific and overall survival [9]. To validate these results in the MDACC dataset, we performed a Principal Component Analysis (PCA) of the 131 genes for all 103 samples using R software version 2.8.1, function prcomp. We dichotomized the first PC score at the median, and compared the high and low PC score groups using Kaplan Meier curves for overall survival and cumulative incidence curves for OSCC-specific mortality using R packages survival and cmprsk. We then performed a Cox proportional hazard regression analysis comparing the group with high and low PC score.
Development of a 13-gene signature prediction model for survival in HPV-negative OSCC patients
To build a prediction model for OSCC-specific survival unique to the HPV-negative OSCC patients, we performed regularized survival analysis using the data from 97 high-risk HPV-negative OSCC patients in the FHCRC dataset. High-risk HPV that we tested included HPV type 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 as previously described [10]. To select the most useful predictors among the 131 genes, we employed the L1-penalized (Lasso) Cox proportional hazard regression model (R package glmnet) [11]. In this model, the L1 penalty parameter controls the sparsity (number of non-zero parameters, or number of selected predictors) of the final estimated model. The larger this parameter is, the fewer the predictors will be selected. We utilized 10-fold cross validation partial likelihood scores to choose the optimal L1 penalty parameter. Specifically, we first randomly split the data into ten groups of roughly equal size. Then, for a given candidate L1 penalty parameter, we used data from nine out of the 10 groups to fit the model and calculated the corresponding partial likelihood score of the fitted model on the remaining group, and we repeated this for every possible collection of nine groups The final performance score of each candidate penalty parameter was the summation of the log partial likelihood scores from all 10 cross-validate iterations. We examined a sequence of candidate L1 penalty parameters and selected the one returning the best cross-validation performance score. We then refitted the model utilizing all the data with the optimal L1 penalty parameter. The resulting model included 13 genes. While L1-penalized regressions are helpful for variable selection, the coefficient estimation from such models tends to bias towards zero when a large degree of regularization (large value of L1 penalty parameter) is employed to identify the best model. Therefore, in order to obtain good coefficient estimation for the 13 selected transcripts in the Cox regression model, we further fit a L2-penalized Cox regression model [12]. Compared to the ordinary Cox regression model, the L2-penalized version provides good tradeoff between biases and variances and usually improves the prediction error. Again, the amount of L2-penalty applied was determined through a 10-fold cross validation. With the estimated coefficients from the selected L2-regresssion model, a continuous risk score was generated for each subject based on the expression of the 13 genes. We then fit three Cox regression models, each with the following independent variables: (I) age, gender, treatment, 13-gene risk score; (2) age, gender, treatment, stage; and (3) age, gender, treatment, stage, 13-gene risk score. Tumor stage was dichotomized as AJCC stage I/ II and III/ IV, treatment was dichotomized as unimodality and multimodality, and age was categorized as <50, 50-59, 60-69, and ≥ 70 years.
Validation of the 13-gene risk score-based prediction models for oral cavity cancer-specific survival in the MDACC dataset
HPV status was not available in the MDACC dataset. We therefore included only oral cavity cancer patients (n=71) to determine the 13 genes’ performance in predicting oral cavity cancer-specific survival. First, for each patient with oral cavity cancer in MDACC data set, we calculated a risk score based on his/her tumor expression levels of the 13 genes. We then applied the Cox regression models developed earlier to predict oral cavity cancer-specific survival for the MDACC patients. A two-year oral cavity cancer-specific survival receiver operating characteristic (ROC) curve was generated for each model using R package CompRisksROC, which accounts for competing risks [13], and the area under the curve (AUC) was calculated to quantify the predictive ability of each model. Leave-10-out jackknife was employed to calculate 95% confidence interval (CI) for the AUC estimates. In addition, we divided the patients into two groups using the risk score median as the cutoff. The cumulative incidence curve for oral cavity cancer-specific mortality was plotted for each group, and the equality of the cumulative mortality between the risk groups was tested using R package cmprsk, which implements the K-sample test described by Gray [14].
Validation of 13 genes expression using Nanostring nCounter™ Gene Expression assay
The expressions of the 13 genes were further validated using a custom-designed NanoString nCounter™ Gene Expression Assay (Nanostring Technologies, Seattle USA). The assay is designed to provide a sensitive and reproducible method for detecting gene expression with the use of molecular barcodes called nCounter Reporter Probes in a multiplex format without reverse transcription or amplification [15-17]. A custom designed nCounter GEX codeset was developed for the 13 genes along with seven housekeeping genes (ALAS1, GAPDH, RPL27, RPS18, SDHA, TBP, and TUBA1B) previously demonstrated to be stably expressed in HNSCC [18]. When possible, the probes in the codeset were targeted near the sequence region of the Affymetrix probe target region. We used the Nanostring nCounter™ Gene Expression assay to test RNA samples from 36 fresh tumors preserved in RNAlater™ that were used in the generation of the signature and compared the NanoString results with the Affymetrix results. Approximately 100 ng of total RNA was processed on a nCounter™ Analysis System as per NanoString Gene Expression Assay standard protocol. To control for assay efficiency, six spike-in RNA controls were included in each sample ranging from 0.125-128 fM. All samples were processed at Nanostring Technologies in Seattle, Washington USA. To obtain the transcript counts, the raw data are first normalized using the geometric mean of the counts (POSA-F) from spiked-in synthetic RNA internal positive controls to account for systematic variation in reporter purification and imaging from lane-to-lane. These normalized counts are further corrected for mRNA input by using the reference gene counts in each assay. Our data showed that RPL27, RPS18, TBP, and ALAS1 had the smallest standard deviation and %CV across all samples. Thus we normalized the transcript counts against the average of these four housekeeping genes. We then used Pearson correlation to test the correlation between gene expression level from Affymetrix arrays and Nanostring nCounter™ Gene Expression assay.
Results
Patient characteristics
Selected characteristics of the 97 high-risk HPV-negative OSCC patients from the FHCRC and 71 oral cavity cancer patients from the MDACC studies are presented in Table 1. The majority of the patients from both institutions were older than 50 years old. The proportions of males and of current smokers were comparable between the institutions. There was a higher proportion of advanced stage cancers than early stage cancers and a higher proportion of patients treated with multimodality than unimodality in both institutions. The OSCC patients from the two institutions were followed for similar lengths of time: median of 26.3 months (range 0.2 to 64.9 months) at the FHCRC and median of 26.9 months (range 0.2 to 92.6 months) at the MDACC, among patients who were known to be alive at the end of the respective studies.
Table 1.
Selected characteristics of OSCC patients in the FHCRC and the MDACC dataset
| FHCRC (n=97) | MDACC (n=71) | |||
|---|---|---|---|---|
| n | % | n | % | |
| Age | ||||
| 19-49 | 22 | 22.7 | 20 | 28.2 |
| 50-59 | 28 | 28.9 | 15 | 21.1 |
| 60-69 | 27 | 27.8 | 19 | 26.8 |
| 70-88 | 20 | 20.6 | 17 | 23.9 |
| Gender | ||||
| Male | 66 | 68.0 | 55 | 77.5 |
| Female | 31 | 32.0 | 16 | 22.5 |
| Smoking status | ||||
| Never/former | 50 | 51.6 | 43 | 60.6 |
| Current | 47 | 48.5 | 28 | 39.4 |
| AJCC stage | ||||
| I | 30 | 30.9 | 3 | 4.2 |
| II | 11 | 11.3 | 16 | 22.5 |
| III | 15 | 15.5 | 15 | 21.1 |
| IV | 41 | 42.3 | 37 | 52.1 |
| Tumor site | ||||
| Oral cavity | 86 | 88.7 | 71 | 100.0 |
| Oropharynx | 11 | 11.3 | 0 | 0.0 |
| Extracapsular extension | ||||
| Yes | 18 | 18.6 | 25 | 35.2 |
| No | 79 | 81.4 | 46 | 64.8 |
| Vital status | ||||
| Alive | 46 | 47.4 | 31 | 43.7 |
| OSCC death | 30 | 30.9 | 22 | 31.0 |
| Death: other causes | 14 | 14.4 | 10 | 14.1 |
| Death: unknown cause | 7 | 7.2 | 8 | 11.3 |
| Treatment | ||||
| Unimodality | 43 | 44.3 | 24 | 33.8 |
| Multimodality | 53 | 54.6 | 46 | 64.8 |
| Unknown | 1 | 1 | ||
Survival analysis of the 131-gene signature using the MDACC dataset
Figure 1 shows Kaplan-Meier curves of overall survival and cumulative incidence curves for OSCC-specific mortality for the high and low first PC score groups. Patients with high PC scores had significantly poorer overall survival compared to the patients with low first PC scores (p = 0.0075). At three years, the estimated proportion dying (± SE) from all causes for patients with high PC scores was 57.6 (± 10.4)% compared with 20.3 (± 7.4)% for the patients with low PC scores. The cumulative OSCC-specific mortality was also significantly higher for the patients with high PC scores compared to the patients with low PC scores (p = 0.031). The estimated cumulative OSCC-specific mortality (± SE) at three years for patients with high PC scores was 44.6 (± 8.7)% compared with 24.1 (± 9.8)% for patients with high PC scores. Cox proportional hazard regression analyses showed that patients with high PC scores, compared with patients with low PC scores, had significantly higher overall and OSCC-specific mortality, with hazard ratios (HRs) of 2.98 (95% CI: 1.53-5.81) and 3.41 (95% CI: 1.38-8.40), respectively. The HRs remained significant after adjusting for age, gender, and tumor stage. The respective adjusted HRs were 2.64 (95% CI: 1.34-5.22) and 3.12 (95% CI: 1.24-7.87) for overall and OSCC-specific mortality.
Figure 1.
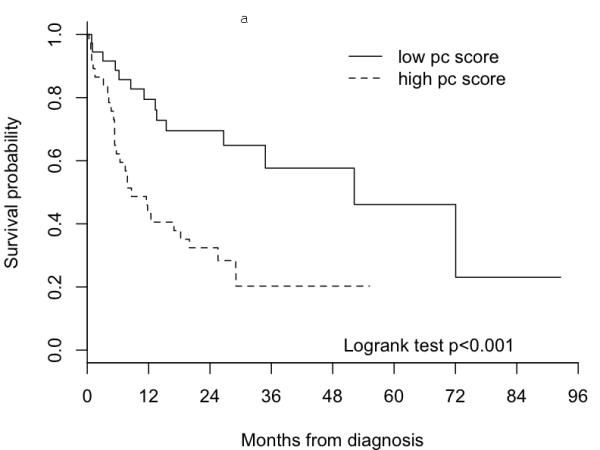
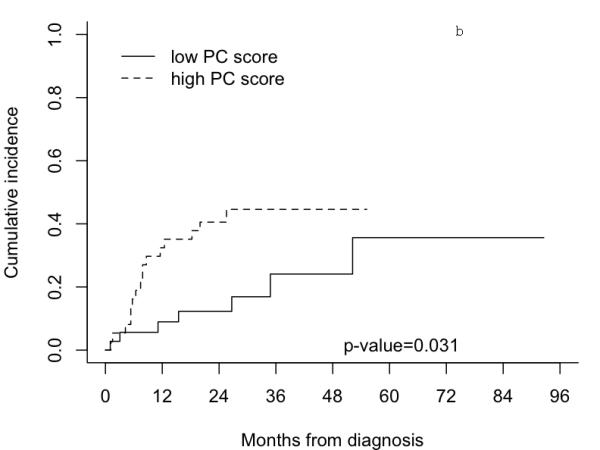
Survival curves of 74 OSCC patients in the MDACC dataset by group dichotomized by median of the first principal component derived from the 131-gene signature. Figure 1a) Kaplan-Meier survival curves for all causes mortality. Figure 1b) Cumulative incidences curves for OSCC-specific mortality.
Development and validation of a 13-gene prediction model for the survival of HPV-negative OSCC patients
Using a L1-penalized Cox proportional hazard regression method on the FHCRC dataset, we identified a prediction model with 13 genes. The gene symbols and coefficients from the L2-penalized Cox regression model of the 13 genes are presented in Table 2.
Table 2.
Thirteen genes used in the model for prediction of survival from oral cavity cancer
| Probe ID | Gene Symbol | Prediction model coefficient* |
Fold- Change** |
|---|---|---|---|
| 1552283_s_at | ZDHHC11 | 0.000443 | 0.30 |
| 1554008_at | OSMR | 0.231391 | 2.54 |
| 1568765_at | SERPINE1 | 0.383316 | 2.34 |
| 201108_s_at | THBS1 | 0.205039 | 2.35 |
| 201497_x_at | MYH11 | -0.206855 | 0.27 |
| 205200_at | CLEC3B (TNA) | -0.131867 | 0.37 |
| 207517_at | LAMC2 | -0.008777 | 2.80 |
| 209900_s_at | SLC16A1 | 0.171380 | 2.08 |
| 210797_s_at | OASL | 0.216877 | 3.76 |
| 222344_at | NREP | 0.199328 | 1.72 |
| 230104_s_at | TPPP | -0.124634 | 0.47 |
| 240000_at | LIPI | 0.864964 | 0.95 |
| 242417_at | LOC283278 | -0.438477 | 0.67 |
For the genes with positive coefficients, a higher expression level was associated with a higher risk of OSCC-specific mortality; and, for the genes with negative coefficients, a lower expression level was associated with a higher risk of OSCC-specific mortality.
Fold-changes comparing patients with high risk score to patients with low risk score, based on the original expression scale
Figure 2 shows cumulative incidence curves for oral cavity cancer-specific mortality by high and low risk scores in the MDACC dataset. Patients with high risk scores had significantly higher oral cavity cancer-specific mortality than patients with low risk scores (p = 0.0253). The estimated proportions of patients dying from oral cavity cancer (± SE) at two years for high and low risk scores were 47.1 (± 9.24) % and 6.35 (± 4.42)%, respectively. The unadjusted HR from a Cox proportional hazard regression analysis comparing patients with high risk scores and low risk scores was 3.61 (95% CI: 1.45 – 9.01, p = 0.0059). The HR adjusting for age, gender, tumor stage, and treatment modality was also significant (HR 3.72, 95% CI: 1.37 – 10.10, p = 0.0099). The ROC curves for survival from oral cavity cancer patients at two years for three prediction models (stage, stage plus risk score, and risk score) adjusting for age, gender, and treatment modality are shown in Figure 3. The AUC for the prediction model with tumor stage was 0.54 (95% CI: 0.48-0.62). Incorporating a term for the 13-gene risk score into the model with tumor stage substantially and significantly improved the ability to predict survival from oral cavity cancer at two years from 0.54 to 0.79 (95% CI: 0.75-0.87, p<0.001). The risk score itself, without tumor stage, also had a significantly higher predictability of survival in oral cavity cancer patients than the tumor stage; the AUC for the model with risk score was 0.78 (95% CI: 0.74-0.86) compared with 0.54 for the model with tumor stage (p<0.001).
Figure 2.
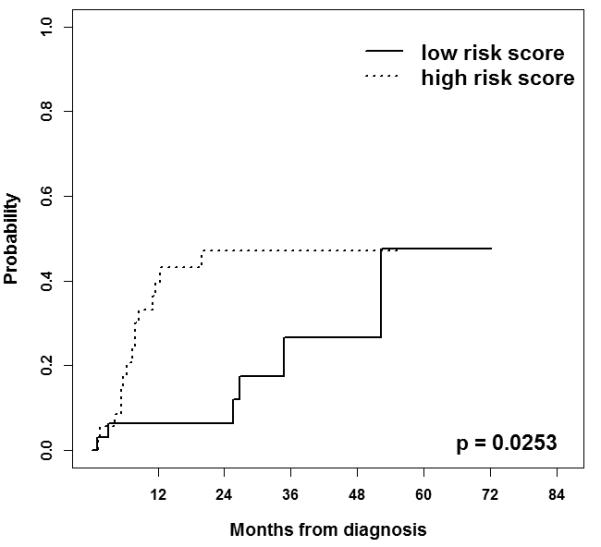
Cumulative incidence curves for oral cavity cancer-specific mortality of the 71 patients with oral cavity cancer in the MDACC dataset by group dichotomized by median of the risk score.
Figure 3.
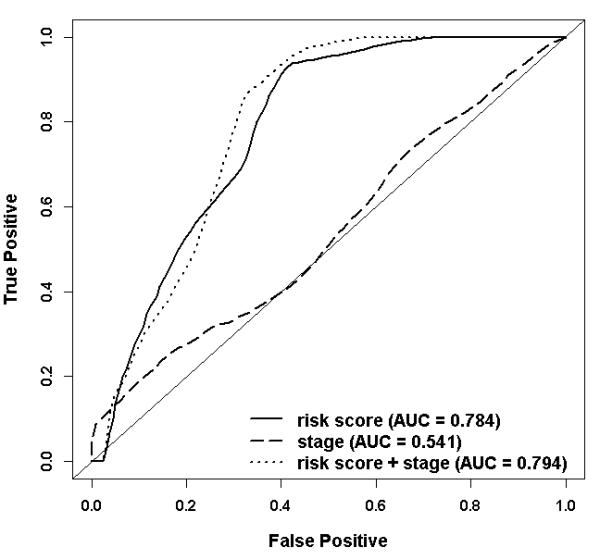
ROC curves comparing the ability to predict two years oral cavity cancer-specific survival of the three prediction models adjusted for age, gender, and treatment in the 71 oral cavity cancer patients from the MDACC dataset. The jackknife p-value testing the AUCs between the model containing term for “risk score” and the model containing term for “stage” is <0.001; the p-value comparing the models containing terms for both “risk score and stage” and the model containing the term “stage” is <0.001.
Validation with Nanostring nCounter™ Gene Expression assay
We found good to excellent correlation between gene expression level from Affymetrix array and from Nanostring nCounter™ assay across samples for all probes except for LIPI. The correlation coefficient for each gene was as follow: 0.96 (OASL), 0.94 (SERPINE1), 0.92 (MYH11), 0.89 (THBS1), 0.86 (SLC16A1), 0.86 (LAMC2), 0.85 (OSMR), 0.80 (TPPP), 0.75 (ZDHHC11), 0.71 (NREP), 0.67 (LOC283278), 0.66 (CLEC3B), and 0.06 (LIPI).
Discussion
We previously reported on a 131-gene signature which identified OSCC patients with poor survival and that, when used in combination with AJCC stage, predicted survival better than AJCC stage alone [9]. In this study, we showed that this 131-gene signature is also predictive of OSCC survival in an independent dataset from the MDACC. Moreover, in keeping with the fact that HPV is already a useful biomarker for oropharyngeal carcinomas [19], and the that a survival gene signature would be most relevant for HPV-negative OSCC, we used the 131 genes to identify a subset of genes, which included 13 genes, that was most predictive of OSCC-specific survival in HPV-negative OSCC patients. We showed that this 13-gene signature is strongly associated with OSCC-specific survival in the MDACC dataset, and that OSCC-specific survival estimates using this signature were substantially better than those using AJCC stage alone. These findings represent the first development and validation of a survival gene signature for HPV-negative OSCC using independent datasets from two institutions and document the generalizability of our previous 131-survival gene signature for OSCC patients. If further validated in multicenter studies, the 13-gene signature may help physicians and patients make better personalized treatment choices. For example, patients with stage I/II disease that test in the high-risk category could be considered for treatment intensification with multimodality therapy, and vice versa for those with stage III/IV disease. In our previous study, we had used step-wise regression to identify the following subset of genes from the 131-gene signature that were most strongly associated with OSCC-specific survival while adjusting for age, sex, stage, HPV status, treatment intensity and comorbidity [9]: LAMC2, OSMR, SERPINE1, OASL, SLC16A1, KLF7, THBS1, HOMER3, GRP68, PDPN, AKRD35, CDH3, EPS8L1. In this study, we also sought to narrow our list of genes, but with two main analytical differences: 1) we restricted the cohort to HPV-negative OSCC patients in the training set, and 2) the methodology to identify the subset of 13 genes was substantially different. Still, six genes overlapped between these two analyses (LAMC2, OSMR, SERPINE1, THBS1, SLC16A1, and OASL) and all but one of these were among the genes most strongly associated with survival in our previous report. These genes all play a role in cell invasion and motility, cell-to-cell signaling, signal transduction, and proliferation, processes essential to metastasis and cancer progression [20-28]. Many of the other seven genes identified in this study have also been implicated in processes relevant to cancer. For example, a recent study identified tetranectin (CLEC3B) to be downregulated in the serum and saliva of metastatic versus primary OSCC [29]. NREP (P311) has been shown to be over-expressed in invasive glioma cells, localizes in focal adhesions, and is constitutively serine-phosphorylated; decreased phosphorylation has been associated with migration-activated glioma cells [30]. MYH11 is a member of the myosin family and we have previously identified myosin 5A as a potential biomarker of occult metastasis in OSCC [31]. Mutations in this gene have been found to be associated with intestinal, breast and prostate cancer [32, 33]. TPPP is thought to play a role in microtubule assembly and stabilization [34], and a gain in its chromosomal location 5p15.33 has been associated with high-grade bladder cancer [35]. Interestingly, ZDHHC11, in our 13 gene signature, shares this location with TPPP.
A limitation of our study is the lack of information on HPV status in the MDACC dataset that we used for validation. Since oral cavity tumors are much less likely to be associated with HPV infection [36, 37], we limit the validation set to oral cavity tumors only.
In conclusion, we have identified and validated a 13-gene expression signature that is strongly predictive of survival in HPV-negative OSCC patients. Importantly, 1) the signature adds substantial predictive information beyond stage, and 2) the small number of genes in the signature should make it more amenable to develop a cost-effective clinical test. Together, these data provide strong justification to proceed with multi-institutional trials to determine whether this signature is effective as a prognostic marker.
Statement of Translational Relevance.
Mounting evidence suggests that head and neck squamous cell carcinoma (HNSCC) is not a single disease but rather can be further subclassified molecularly. Subsite-specific etiologic and prognostic markers, such as HPV, are emerging. Patients with HPV-positive oropharyngeal carcinomas are known to have better treatment response and survival. In contrast, there are no currently defined molecular markers to assess treatment responsiveness for HPV-negative oral squamous cell carcinomas (OSCC). Recent works including our own have shown that gene expression profiling can identify subgroups of patients with poor survival. Thus, gene expression signatures show promise as clinical tools to help guide treatment intensification. In this study, we built on our previous molecular work to subclassify OSCC and have identified and validated a prognostic 13-gene signature that showed a higher ability than tumor stage in predicting survival for patients with HPV-negative OSCC. This is the first demonstration of a gene set that provides prognostic information beyond AJCC stage for this subgroup of tumors.
Acknowledgements
We wish to express our deepest appreciation to the study participants and their families for their contribution to this study. The study is supported by resources and the use of facilities at the Fred Hutchinson Cancer Research Center, University of Washington Medical Center, Harborview Medical Center and the Veterans Affairs Puget Sound Health Care System and the MD Anderson Cancer Center. We also thank Kevin R. Coombes, PhD of the MD Anderson Cancer Center for his mentorship to FCH and related contributions to this work.
Grant Support The study at FHCRC was supported by grants from the National Institutes of Health, National Cancer Institute (NIH NCI R01CA095419), National Center for Research Resources grant 1KL2RR025015-01, Amos Medical Faculty Development Program Award from the Robert Wood Johnson Foundation, and by institutional funds from the Fred Hutchinson Cancer Research Center. The study at the MD Anderson Cancer Center was supported by Specialized Program of Research Excellence in Head and Neck Cancer Grant P50CA97007 from the National Cancer Institute, “Clinician Investigator Program in Translational Research” K12CA088084, Clinical Research Program 2 L30CA117652-02A1, and THANC Foundation Young Investigator Award in Head and Neck Cancer.
References
- [1].Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–9. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- [2].Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96:998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- [3].Wittekindt C, Gultekin E, Weissenborn SJ, Dienes HP, Pfister HJ, Klussmann JP. Expression of p16 protein is associated with human papillomavirus status in tonsillar carcinomas and has implications on survival. Adv Otorhinolaryngol. 2005;62:72–80. doi: 10.1159/000082474. [DOI] [PubMed] [Google Scholar]
- [4].Rotnaglova E, Tachezy R, Salakova M, Prochazka B, Kosl’abova E, Vesela E, et al. HPV involvement in tonsillar cancer: prognostic significance and clinically relevant markers. Int J Cancer. 2011;129:101–10. doi: 10.1002/ijc.25889. [DOI] [PubMed] [Google Scholar]
- [5].Cardoso F, van’t Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70-gene profile: the MINDACT trial. Journal of Clinical Oncology. 2008;26:729–35. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- [6].Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- [7].Gray RG, Quirke, Handley K, Lopatin M, Magill L, Baehner FL, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611–9. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- [8].Chen C, Mendez E, Houck J, Fan W, Lohavanichbutr P, Doody D, et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2152–62. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mendez E, Houck JR, Doody DR, Fan W, Lohavanichbutr P, Rue TC, et al. A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin Cancer Res. 2009;15:1353–61. doi: 10.1158/1078-0432.CCR-08-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lohavanichbutr P, Houck J, Fan W, Yueh B, Mendez E, Futran N, et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180–8. doi: 10.1001/archoto.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of statistical software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- [12].Benner A, Zucknick M, Hielscher T, Ittrich C, Mansmann U. High-dimensional Cox models: the choice of penalty as part of the model building process. Biometrical Journal. 2010;52:50–69. doi: 10.1002/bimj.200900064. [DOI] [PubMed] [Google Scholar]
- [13].Saha P, Heagerty PJ. Time-dependent predictive accuracy in the presence of competing risks. Biometrics. 2010;66:999–1011. doi: 10.1111/j.1541-0420.2009.01375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. The Annals of Statistics. 1988;16:1141–54. [Google Scholar]
- [15].Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnology. 2011;11:46. doi: 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- [17].Northcott PA, Shih DJ, Remke M, Cho YJ, Kool M, Hawkins C, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123:615–26. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lallemant B, Evrard A, Combescure C, Chapuis H, Chambon G, Raynal C, et al. Reference gene selection for head and neck squamous cell carcinoma gene expression studies. BMC Molecular Biology. 2009;10:78. doi: 10.1186/1471-2199-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schenk S, Hintermann E, Bilban M, Koshikawa N, Hojilla C, Khokha R, et al. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMP-dependent mammary gland involution. J Cell Biol. 2003;161:197–209. doi: 10.1083/jcb.200208145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bajou K, Masson V, Gerard RD, Schmitt PM, Albert V, Praus M, et al. The plasminogen activator inhibitor PAI-1 controls in vivo tumor vascularization by interaction with proteases, not vitronectin. Implications for antiangiogenic strategies. J Cell Biol. 2001;152:777–84. doi: 10.1083/jcb.152.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pedersen H, Brunner N, Francis D, Osterlind K, Ronne E, Hansen HH, et al. Prognostic impact of urokinase, urokinase receptor, and type 1 plasminogen activator inhibitor in squamous and large cell lung cancer tissue. Cancer Res. 1994;54:4671–5. [PubMed] [Google Scholar]
- [23].Bornstein P. Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J Cell Biol. 1995;130:503–6. doi: 10.1083/jcb.130.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yee KO, Streit M, Hawighorst T, Detmar M, Lawler J. Expression of the type-1 repeats of thrombospondin-1 inhibits tumor growth through activation of transforming growth factor-beta. American Journal of Pathology. 2004;165:541–52. doi: 10.1016/s0002-9440(10)63319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–54. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- [26].Mosley B, De Imus C, Friend D, Boiani N, Thoma B, Park LS, et al. Dual oncostatin M (OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J Biol Chem. 1996;271:32635–43. doi: 10.1074/jbc.271.51.32635. [DOI] [PubMed] [Google Scholar]
- [27].Pinheiro C, Longatto-Filho A, Ferreira L, Pereira SM, Etlinger D, Moreira MA, et al. Increasing expression of monocarboxylate transporters 1 and 4 along progression to invasive cervical carcinoma. Int J Gynecol Pathol. 2008;27:568–74. doi: 10.1097/PGP.0b013e31817b5b40. [DOI] [PubMed] [Google Scholar]
- [28].Koukourakis MI, Giatromanolaki A, Bougioukas G, Sivridis E. Lung cancer: a comparative study of metabolism related protein expression in cancer cells and tumor associated stroma. Cancer Biology and Therapy. 2007;6:1476–9. doi: 10.4161/cbt.6.9.4635. [DOI] [PubMed] [Google Scholar]
- [29].Arellano-Garcia ME, Li R, Liu X, Xie Y, Yan X, Loo JA, et al. Identification of tetranectin as a potential biomarker for metastatic oral cancer. International Journal of Molecular Sciences. 2010;11:3106–21. doi: 10.3390/ijms11093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mariani L, McDonough WS, Hoelzinger DB, Beaudry C, Kaczmarek E, Coons SW, et al. Identification and validation of P311 as a glioblastoma invasion gene using laser capture microdissection. Cancer Res. 2001;61:4190–6. [PubMed] [Google Scholar]
- [31].Mendez E, Lohavanichbutr P, Fan W, Houck JR, Rue TC, Doody DR, et al. Can a metastatic gene expression profile outperform tumor size as a predictor of occult lymph node metastasis in oral cancer patients? Clinical Cancer Research. 2011;17:2466–73. doi: 10.1158/1078-0432.CCR-10-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Alhopuro P, Phichith D, Tuupanen S, et al. Unregulated smooth-muscle myosin in human intestinal neoplasia. Proc Natl Acad Sci U S A. 2008;105:5513–8. doi: 10.1073/pnas.0801213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Alhopuro P, Karhu A, Winqvist R, Waltering K, Visakorpi T, Aaltonen LA. Somatic mutation analysis of MYH11 in breast and prostate cancer. BMC Cancer. 2008;8:263. doi: 10.1186/1471-2407-8-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Acevedo K, Li R, Soo P, Suryadinata R, Sarcevic B, Valova VA, et al. The phosphorylation of p25/TPPP by LIM kinase 1 inhibits its ability to assemble microtubules. Exp Cell Res. 2007;313:4091–106. doi: 10.1016/j.yexcr.2007.08.012. [DOI] [PubMed] [Google Scholar]
- [35].Yamamoto Y, Chochi Y, Matsuyama H, Eguchi S, Kawauchi S, Furuya T, et al. Gain of 5p15.33 is associated with progression of bladder cancer. Oncology. 2007;72:132–8. doi: 10.1159/000111132. [DOI] [PubMed] [Google Scholar]
- [36].Isayeva T, Li Y, Maswahu D, Brandwein-Gensler M. Human papillomavirus in non-oropharyngeal head and neck cancers: a systematic literature review. Head and neck pathology. 2012;6(Suppl 1):104–20. doi: 10.1007/s12105-012-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lingen MW, Xiao W, Schmidt A, Jiang B, Pickard R, Kreinbrink P, et al. Low etiologic fraction for high-risk human papillomavirus in oral cavity squamous cell carcinomas. Oral Oncol. 2012 doi: 10.1016/j.oraloncology.2012.07.002. Epub ahead of print. [DOI] [PubMed] [Google Scholar]


