Abstract
Sialic-acid-binding immunoglobulin-like lectin (Siglec5) is a carbohydrate-binding surface receptor expressed on neutrophils, monocytes and B cells in human lymphoid and myeloid cell lineages. Existing structural and functional data fail to define the clear ligand specificity of Siglec5, though like other Siglec family members, it binds a variety of complex carbohydrates containing a sialic acid at the non-reducing terminus. Prokaryotic expression of this protein has proven challenging due to disulfide bonds and Asn-linked glycosylation. We developed an expression and purification protocol that uses an on-column strategy to refold E. coli expressed protein that produced a high yield (2 mg / L) of the single N-terminal Siglec5 carbohydrate recognition domain (CRD). A 2D heteronuclear single-quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectrum showed this material was folded, and a secondary structure prediction based on the assigned chemical shifts of backbone atoms was consistent with a previously determined x-ray model. NMR chemical shift mapping of Siglec5 binding to three carbohydrate ligands revealed similarities in binding interfaces and affinities. In addition, the role of alternate protein conformations identified by NMR in ligand binding is discussed. These studies demonstrate the Siglec5 CRD alone is sufficient for binding sialylated carbohydrates and provide a foundation for further investigation of Siglec5 structure and function.
Keywords: on-column refolding, sialoside binding, glycoprotein, carbohydrate recognition, NMR chemical shift perturbation
Introduction
Sialic-acid-binding immunoglobulin-like lectins (Siglecs) are a group of cell surface receptors and signaling molecules [1]. These proteins are the targets of therapeutics designed to treat leukemia and autoimmune disorders owing to the restricted tissue expression profiles of many siglecs on specific myeloid and lymphoid cells [2]. Structural information on the nature of protein-carbohydrate interaction is potentially useful in the design of these therapeutics. However, such information is scarce, partly because of the difficulty in preparing suitable protein samples, and partly because of the diversity of potential ligands that need to be investigated. There are a few crystal structures of siglecs with carbohydrates in the binding site, but not all ligands are compatible with crystallization, and NMR offers an alternative source of structural information that is particularly useful when ligands are diverse and a large set of ligands needs to be investigated. Here we present a protein expression and refolding protocol suitable for the preparation of siglec NMR samples, along with preliminary NMR data on ligand interactions for the carbohydrate binding domain of one siglec, siglec-5.
Siglecs are type I membrane proteins with an N-terminal carbohydrate recognition domain (CRD), a single transmembrane helix, and a small cytosolic extension (Fig. 1). The CRD binds sialic acids, a group of nine carbon acidic sugars found at the termini of many cell-surface carbohydrates (glycans) in mammals [3, 4]. The CRD, in addition to other extracellular domains, is normally glycosylated with glycans terminated in sialic acid, something that is important for interactions of pairs of siglecs within the same membrane (cis interactions) as well as interactions between siglecs and between siglecs and other glycosylated proteins on proximate membranes (trans interactions). The cytoplasmic extension carries immunoreceptor tyrosine-based inhibitory motifs (ITIMs). Clustering of siglecs in cis interactions (Fig. 1B) is suggested to dampen intracellular immune signaling through these motifs. Some siglecs also recognize multiple microbial pathogens, including bacteria and viruses, and are believed to function as endocytic receptors as a part of an innate immune response [5, 6].
Figure 1.
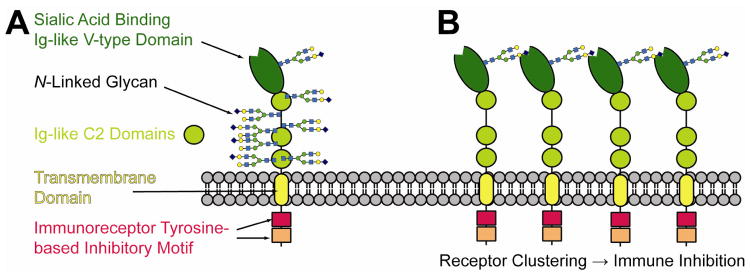
Human Siglec5 is comprised of four extracellular domains, a transmembrane region and two intracellular inhibitory domains (A). The extracellular portion of the molecule is heavily glycosylated. (B) Intramolecular interactions are thought to be an important component of the immunomodulatory properties of siglec family proteins.
Human Siglec5 (CD170) is expressed in neutrophils, monocytes and at a low level in B cells [1]. Siglec5, like other siglecs, is thought to self-associate in a cis interaction [7] forming an immunosuppressive barrier that must be disrupted to elicit a cellular immune response. Siglec5 has been proposed to bind the acute-phase protein, α1-acid glycoprotein, as a native trans ligand [8], but also was shown to bind bacterial pathogenesis factors, including group B streptococcal cell-wall-anchored β protein [9, 10] and the sialylated lipopolysaccharide of Neisseria meningitidis [5]. Despite these reports, a clear definition of the molecular details and binding signatures of carbohydrate ligands are lacking. Unlike some siglecs with well-defined recognition motifs, Siglec5 appears to associate weakly with a broad range of small sialylated carbohydrates showing no clear preference [3]. To achieve a high degree of specificity, Siglec5 substrate recognition may invoke distal glycan contact or contact with the underlying polypeptide. This greatly expands the range of ligands to be investigated, and to date, few of these investigations have been carried out.
A structural model of the two N-terminal Siglec5 domains was previously solved by x-ray crystallography [11]. Both domains are Ig-like in structure with a V-type CRD followed by a C2 domain; they show a unique interdomain disulfide bond. Remarkably, the complexes of Siglec5 bound to α2–3 or α2–6 sialoglycosides showed similar dissociation constants and binding poses, despite the differences in linkage to the underlying galactose residue. This again suggests that if specificity is achieved, contacts beyond the three carbohydrate residue tested must be utilized. Even a very low affinity secondary site combined with the moderate affinity of the primary sialic acid binding site would combine at a theoretical maximum of the product of the two dissociation constants and dramatically enhance ligand affinity.
To provide a platform for probing Siglec5 ligand binding, we developed, and present here, a new E. coli-based expression, purification and refolding system suitable for use with nuclear magnetic resonance (NMR) applications. Siglec5 obtained by this method was characterized structurally by assigning the backbone resonances and establishing consistency with secondary structure elements seen in the crystal structure. Perturbation of the chemical shifts of select resonances was then used to functionally characterize the protein and determine the location and affinity of multiple sialic acid ligands. The results of these experiments are also presented.
Materials and Methods
All materials, unless otherwise noted, were purchased from Sigma-Aldrich (St. Louis, MO). Stable isotope-enriched compounds were purchased from Cambridge Isotopes (Cambridge, MA).
DNA construct for Siglec5
DNA encoding the two N-terminal domains (residues P19 to S233; becoming construct numbers P21 to S235) of human Siglec5 (UniProt DB: O15389) was codon optimized (GenScript, Piscataway, NJ) for E. coli expression and cloned into a pET28b (EMD Millipore, Billerica, MA) expression vector using the NdeI and XhoI restriction endonucleases (New England Biolabs, Ipswich, MA) following the 6xHis tag and thrombin protease site. The single CRD expression construct (human Siglec5 residues P19 to A141; construct residues P21-A143) was generated starting with this two domain construct by introducing a stop codon following A141 and changing C36 to A (A143 and C38A in the construct) using the QuikChange® (Agilent, Santa Clara, CA) system. Vector sequences were verified by DNA sequencing.
Expressing human Siglec5 in E. coli
Vectors were transformed into an E. coli expression strain (BL21*) which was then grown to OD600 ~ 0.4 at 37 °C in 1 L M9 minimal medium [12] supplemented with 15N ammonium chloride and / or 13CU glucose. At this point isopropyl β-D-1-thiogalactopyranoside was added to 0.5 mM and the culture was grown for 16–18 h at 18°C. Cells with a final OD600 ~ 3.5 were harvested by centrifugation.
Purification and Refolding
Attempts to refold a high yield of the two domain construct using a published protocol [11] were not successful. A refolding protocol was therefore adapted from one used by Volkman and coworkers for chemokine refolding [13] and used with both the one and two domain constructs as described in this report. In our specific procedure all steps were conducted either on ice or in a 4 °C cold room with pre-chilled buffers. Cells were resuspended in 50 mM tris (hydroxymethyl) aminomethane, 100 mM sodium chloride, pH 8.0 and lysed with three passages through a French pressure cell at 18,000 psi. Inclusion bodies were separated from the cell supernatant by centrifugation (45 min at 40,000 x g). The resulting pellet was washed twice by dispersing pelleted inclusion bodies with a Dounce-type hand homogenizer in 50 mM tris, 10 mM ethylenediaminetetraacetic acid (EDTA), 2% triton X100, 500 mM sodium chloride, 2M urea, pH 8.0 and once with 50 mM tris, 100 mM sodium chloride, pH 8.0 with recovery by centrifugation after each wash step. Washed inclusion bodies were resuspended in 50 mM tris, 10 mM imidazole, 300 mM sodium chloride, 6M guanidine hydrochloride, 10 mM beta-mercaptoethanol, pH 8.0. Insoluble material was removed by centrifuging for 30 min at 40,000 x g. The supernatant was applied dropwise to 75mL of superflow Ni++ NTA resin (Qiagen, Valencia, CA) suspended in 75 mL of the previous buffer under constant mechanical stirring. 75 mL of resin is a relatively large volume, but lesser amounts reduced yield; it could be that protein dispersal within a large matrix volume reduced the potential for aggregation of intermediate folding states. This mixture was then gently poured into an empty column and the liquid fraction allowed to flow through by gravity. 200 mL of Wash 1 (20 mM tris, 100 mM sodium chloride, 1% triton X100, 10 mM beta-mercaptoethanol, pH 8.0) was passed over the column at a low flow rate (~1 drop s−1). When complete, 200 mL of wash 2 (20 mM tris, 100 mM sodium chloride, 5 mM beta-cyclodextran, 1 mM reduced glutathione, 0.5 mM oxidized glutathione, pH 8.0) was passed in the same way. These column washes together took between three and four hours to complete. Wash 3 (100 mL of 20 mM tris, 500 mM sodium chloride, pH 8.0) was then passed over the column in the same manner. The column was eluted with 1.5x total column volume of 25 mM tris, 300 mM sodium chloride, 300 mM imidazole, pH 8.0 in 10 mL fractions. Fractions judged to contain Siglec5 by gel electrophoresis were dialyzed overnight against 2 L of 50 mM glutamate, 50 mM arginine, 150 mM sodium chloride and 10 mM imidazole, pH 6.5 [14]. Protein was concentrated with 3kDa-cutoff centrifugal spin concentrator units (EMD Millipore). Protein recoveries were quantified using A280 (ε=31400 M−1 cm−1) on a Nanodrop® spectrophotometer (Thermo Scienitific, Waltham, MA).
NMR Spectroscopy
The backbone assignment experiments were collected on Varian 900 (VNMRS console), 800 and 600 (Inova consoles) MHz spectrometers equipped with cryogenically cooled, triple resonance probes. The backbone assignment pulse sequences were supplied by as part of the BioPack distribution (Agilent). NMR spectra were collected at 15 °C using protein concentrations of 0.2–0.4 mM in a buffer containing 50 mM glutamate, 50 mM arginine, 150 mM sodium chloride, 10 mM imidazole and 10% D2O, pH 6.5. Sequence-specific backbone resonance assignments were determined using HNCO, HNCA, HN(CO)CA, HN(CA)CB, and HN(COCA)CB experiments. NMR data were processed using NMRPipe [15] and analyzed using NMRViewJ [16]. Chemical shifts have been deposited in the BioMagResBank (18704). To assess the success of refolding observed chemical shifts were compared to chemical shifts predicted using the SHIFTX2 program [17] and the X-ray structure 2zg1 [11]. The secondary structure prediction was obtained from TALOS+ [18] using the chemical shift information.
Carbohydrate binding measurements
Titrations of different carbohydrates were performed by resuspending the analyte into the NMR buffer (listed above) at high concentration and diluting into the protein solution. Chemical shift perturbations of Siglec5 CRD (~100 μM) amide resonances observed during a titration with carbohydrate ligands were assessed using a series of 2D 15N-heteronuclear single quantum coherence spectra. Proton and nitrogen chemical shifts of amides were summed with scaling as suggested by Farmer et al. [19] and plotted against ligand concentration. Assuming the binding is weak and all points collected have a small fraction of ligand bound, an equation describing a Langmuir binding isotherm can be used to extract dissociation constants (fractional shift= 1/(1+(KD /(ligand concentration). Structural figures were prepared using PyMol (Schrodinger Scientific) based upon published structures (pdb – 2zg1, 2zg2, 2zg3) [11].
Results and Discussion
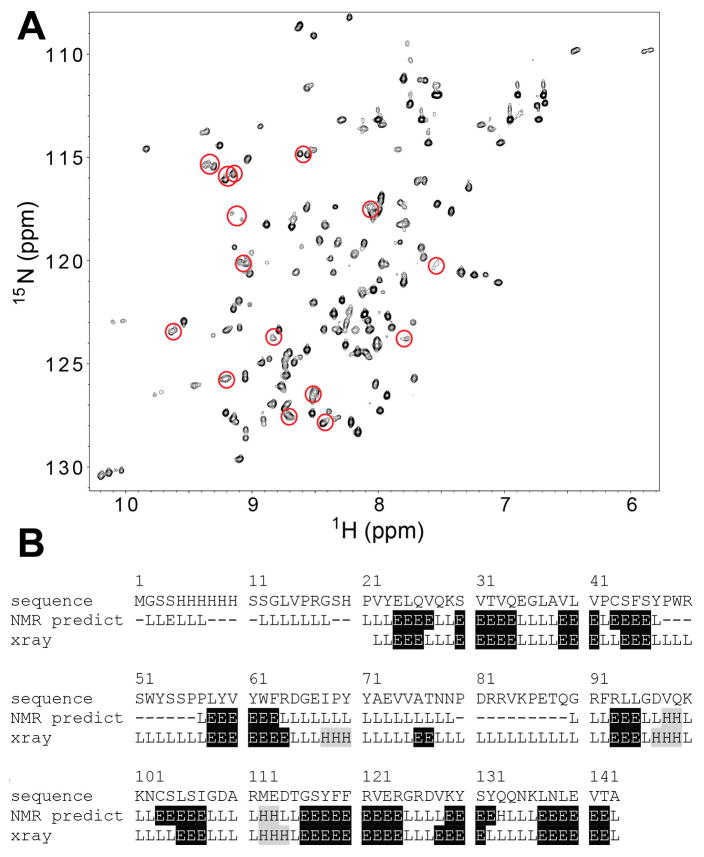
Refolding disulfide-stabilized and natively highly glycosylated human proteins following E. coli expression represents a fundamental challenge facing structural and functional studies. Exposed cysteines frequently oxidize to form crosslinked intermolecular aggregates rather than intramolecular disulfide bonds. The absence of glycosylation during E. coli expression also may contribute to improper folding geometries. To mitigate at least problems associated with disulfide bonds, an on-column refolding approach [13] was adopted, and substantial yields of high purity human Siglec5 constructs representing both the N-terminal Ig-like V-type CRD and the two N-terminal Ig-like V-type and C2 domains were obtained (yields ~2 mg / L). However, 2D NMR spectra from both constructs were poorly dispersed and the Siglec5 two domain construct precipitated slowly from solution (data not shown). Thrombin-catalyzed cleavage of the N-terminal tag improved the spectrum of the two domain construct to a small degree, but failed to extend the protein lifetime in solution (data not shown). The construct designed to express only the N-terminal CRD, with the interdomain disulfide bond disrupted by mutation (C38A), yielded a high level of expression in E. coli and after refolding a highly pure, soluble protein, was obtained as shown in Figure 2 (2mg / L expression and soluble to about 400μM). A 2D 15N heteronuclear single quantum coherence (HSQC) spectrum was well dispersed and showed approximately the number of peaks expected for the CRD (Fig. 3A).
Figure 2.
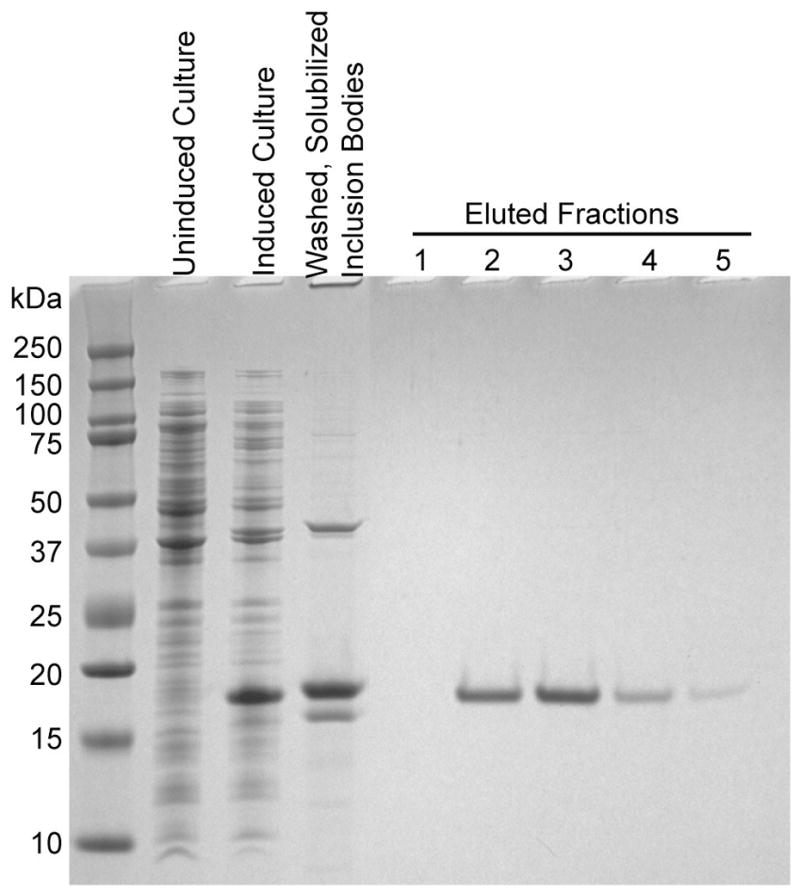
Siglec5 was refolded from E. coli inclusion bodies using an on-column refolding technique. The resulting protein was highly pure as seen in the eluted fractions.
Figure 3.
(A) A heteronuclear 1H-15N correlation experiment of the purified, unliganded Siglec5 CRD showed well-dispersed resonances. Peaks showing peak doubling in the triple-resonance experiments are circled. (B) A secondary structure prediction based on chemical shifts of the assigned backbone resonances was highly similar to the secondary structural elements observed in an x-ray crystallography model of Siglec5 (pdb 2zg1) [11]. The amino acid numbers in this figure and the text refer to the position within the construct, not the native human Siglec5 (UniProt DB: O15389). ‘E’ represents beta sheet elements, ‘H’ alpha helices and ‘L’ loops. Unassigned amino acids are denoted with dashes (−).
NMR chemical shifts, being sensitive to local torsion angles and the chemistry of the immediate environment (pH, hydrophobicity, charge, etc.) contain a wealth of information. To access this information crosspeaks observed in the HSQC spectrum were assigned by triple-resonance protein NMR techniques [20]. As a result, a majority of the HN (86%), N (81%), C′ (81%), Cα (83%), and Cβ (79%) resonances were assigned, not including the N-terminal Met, the His6 Tag and thrombin cleavage site residues. Cysteine Cα and Cβ chemical shifts were consistent with disulfide bond formation [21]. Residues in two regions of the protein sequence were not assignable due to the loss of resonance intensity (Fig 3B). These regions correspond to two extended loops in the models derived from x-ray diffraction data, and do not appear to have extensive secondary structure or contacts with the body of the protein. Loop motion on the μs-ms timescale could account for the loss of observable signals in this region [20].
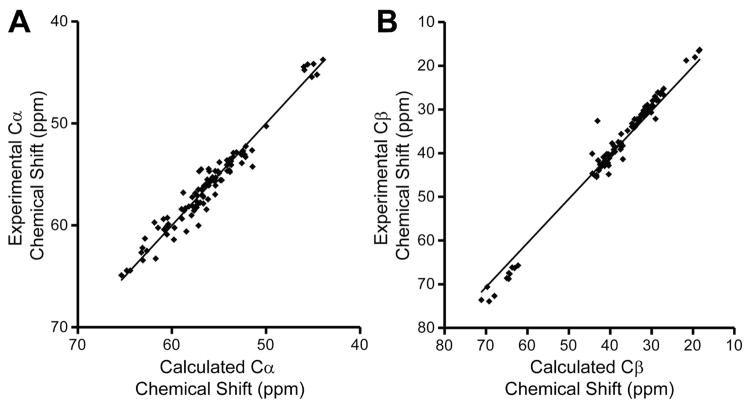
A comparison of backbone dihedral angles predicted from chemical shifts of the CRD-only construct to those of this domain in the published Siglec5 structure revealed a high level of similarity. Backbone chemical shifts are sensitive to phi and psi torsion angles, and can be indexed to provide dihedral angle estimates [18]. β-sheet, α-helices and loop regions predicted from the NMR data are remarkably similar to those observed in x-ray models [11] indicating a high degree of secondary structure similarity (Fig 3B). Furthermore, backbone chemical shifts predicted [17] from the x-ray crystallography structure (pdb 2zg1) [11] were highly correlated with experimental chemical shifts (R2 values of 0.95 (Cα), 0.97 (Cβ), 0.70 (C′), 0.64(N), and 0.40 (HN). These predictions take into account additional nonsequential effects on chemical shift. The predictions for Cα and Cβ shifts are more robust and the correlations for these are shown in Figure 4.
Figure 4.
The comparison of chemical shifts calculated with a crystal structure and the ShiftX2 program [17] and experimentally determined values shows a high degree of similarity for (A) Cα and (B) Cβ atoms.
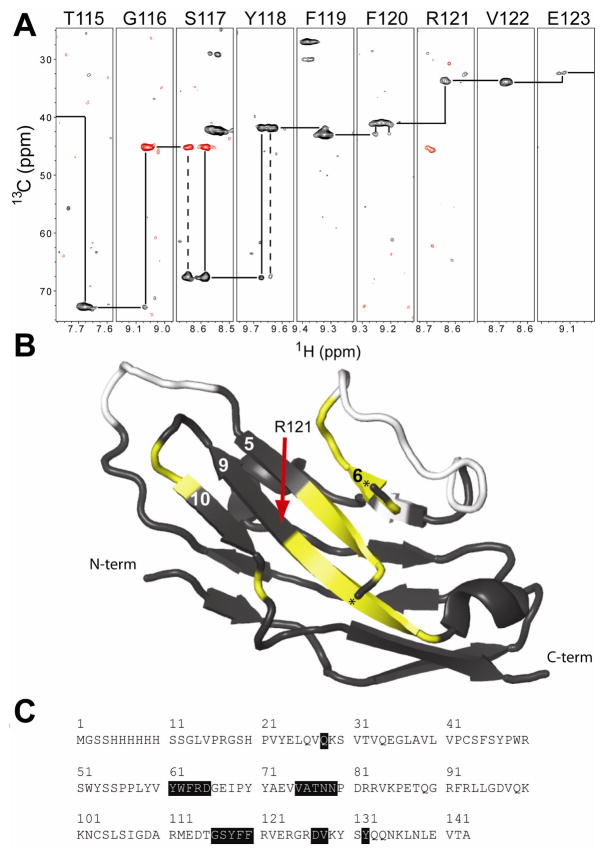
The NMR data did suggest the presence of one unusual structural feature for the CRD not reported in the x-ray crystallography models of the two domain constructs. A few amino acids displayed multiple, distinct, chemical shifts for the same atoms. Sequential backbone traces, as shown in Figure 5A, can be clearly seen to pass through regions with duplicate, discrete 1H, 15N, and to a lesser extent, 13C chemical shifts. It is not clear if the second domain stabilizes one conformation, or if crystal contacts excluded the crystallization of the alternate form that was not seen in x-ray crystallography models. Peak intensities in most cases appeared equal, suggesting equal populations (pairs circled in Figure 3). Furthermore, population distributions did not change during ligand titration. Five of these regions were identified, and once mapped onto a structural model co-localized to the central β-sheet adjacent to the sialic acid binding site (strands 5,6, 9&10 in Figure 5B). The remaining intradomain disulfide bridge is located on the other face of the molecule, thus it is unlikely these two Siglec5 forms are due to inconsistent disulfide formation. Alternatively, it is possible that the central β-strand (strand 9) can occupy two different conformations in the β-sheet, differing only in register. Interconversion of two forms differing in register would be expected to interconvert slowly (<1 s−1). It will be of value to determine the role of structural heterogeneity, both of this central β-sheet and the unassigned loop regions, in ligand binding and Siglec5 function.
Figure 5.
NMR experiments indicated alternate forms of Siglec5. (A) A sequence walk of an HN(CA)CB experiment showed alternative conformations and weak resonances on the beta strand formed by residues 115–123. Positive (negative) signals are shown with black (red) contours. (B) Such patterns were observed along many portions of the protein sequence. These are shown in yellow mapped on the black ribbon diagram of Siglec5 and in (C). Unassigned residues are shown in white. A small helix obscured the view of the central beta sheet and is not shown; the termini of this helix are marked with a “★.”
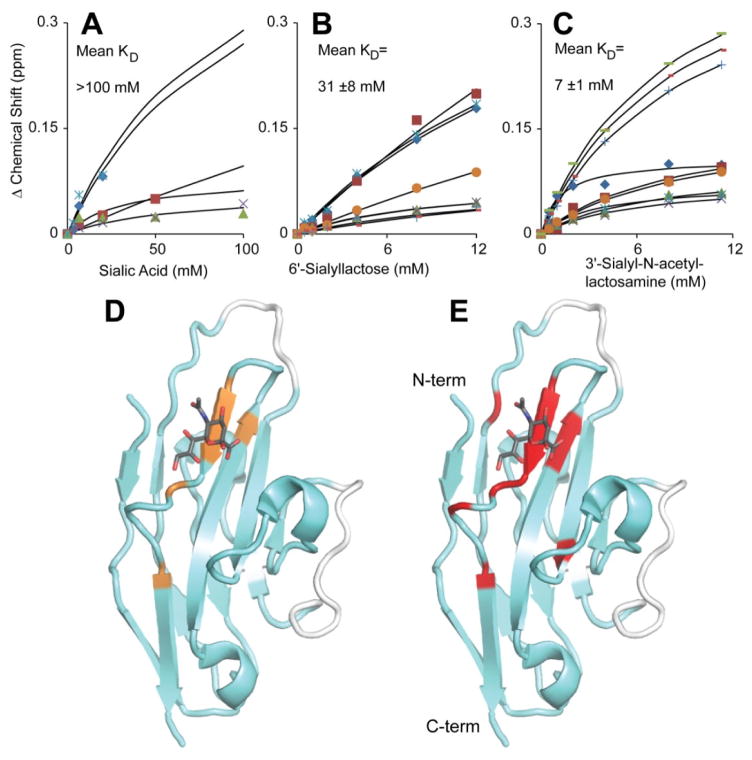
The affinity of the Siglec5 CRD for different carbohydrates was assessed by following the changes of crosspeak position in a 2D HSQC spectrum. The NMR cross peaks gradually changed position with increasing carbohydrate concentration, indicating these complexes are in fast exchange on the NMR timescale (data not shown). The CRD bound to 3′-sialyl-N-acetyllactosamine with an affinity similar to that previously reported for 3′-sialyllactose (7 ±3 mM vs 8.7 mM [11]; Figure 6). 6′-sialyllactose binding was significantly weaker, but in the same range reported earlier (31 ±8 mM vs 8.0 mM [11]). It is of interest to note that the previous values were determined with a two-domain Siglec5 version. The sialic acid terminating these trisaccharides, when used in isolation, bound more weakly with an estimated KD >100 mM, suggesting considerable binding energy is obtained from the presence of galactose and glucose (or N-acetylglucosamine) residues of the 3′- and 6′-sialosides. The anomeric configuration of sialic acid in solution is a mixture of α and β forms, unlike the 3′- and 6′-sialosides which are pure α, though it is unlikely this heterogeneity accounts for all of the difference in sialic acid binding vs sialoside binding. These binding data indicate the Siglec5 CRD alone is sufficient for sialoside binding. The role of the C2 domains is unclear, though these may extend the CRD away from abundant membrane-bound cell surface sialosides.
Figure 6.
NMR titrations showed the Siglec5 CRD binds free sialic acid (A), 6&-sialyllactose (B) and 3′-sialyl-N-acetyllactosamine (C). Binding curves were fit to each individual data series and then averaged to determine the mean KD ± standard deviation. The curves fit in (A) were generated using known end point values for the affected resonances based upon the fits in (B and C). (D) The sialic acid – Siglec5 interface shown in orange was determined by mapping the resonances shifted in (A) to the Siglec5 structure. The binding interfaces determined from (B) and (C) were indistinguishable and are shown as red residues plotted on the model in (E). A stick model of sialic acid is included (pdb 2zg1). Assigned resonances are shown with a cyan ribbon, unassigned with white.
The identities of crosspeaks shifted during the titrations were mapped onto a Siglec5 structural model to determine the binding interface (Fig. 6D and E). Although some residues may show an alteration in chemical shift due to an induced structural rearrangement resulting from binding, and not a direct contact with the ligand, the clustering of residues showing shifts adds confidence to the identification of a binding site. Mapping the 6′-sialyllactose and 3′-sialyl-N-acetyllactosamine interfaces revealed the same residues, which is surprising considering the different α2–6 and α2–3 sialic acid linkages, respectively. The sialic acid – Siglec5 interface shares many of these same residues. In addition to the absence of direct interactions with galactose and glucose residues, the less extensive region perturbed may be due to the relatively small crosspeak shifts observed during this titration (Fig. 6A and D). The location of these interfaces is consistent with the location of the binding site shown in a published report [11]. The position of the bound ligands shown in Fig. 6D and E are based on that report.
Conclusions
This work has demonstrated a method to produce a single, non-glycosylated Siglec5 CRD with high yield and high purity that binds sialylated carbohydrates with affinities similar to a two domain construct. The conditions described here provide adequate protein stability for time-consuming NMR experiments and functional assays. The identified carbohydrate – Siglec5 interfaces were remarkably similar for different ligands, despite different linkages of the critical sialic acid to the underlying disaccharides. Furthermore, the backbone chemical shift assignment will serve to facilitate the future structural and functional investigations of Siglec5.
Highlights.
A fast method to recover properly folded bacterially-expressed Siglec5 in high yield (2mg/L) is reported.
The backbone NMR chemical shifts for the Siglec5 carbohydrate recognition domain are assigned.
Protein / carbohydrate binding interfaces are identified.
Acknowledgments
The authors thank Laura Morris for calculating Siglec5 chemical shifts based upon the x-ray crystallography models. This work was financially supported by the grants R01GM033225 and P41RR005351 from the National Institutes of Health. A.W.B. was supported by a NIH Kirschstein National Research Service Award (F32AR058084). The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- CRD
carbohydrate recognition domain
- HSQC
heteronuclear single-quantum coherence
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- NMR
nuclear magnetic resonance
- Siglec
sialic-acid-binding immunoglobulin-like lectin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nature Reviews Immunology. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 2.O’Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30:240–248. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMillan SJ, Crocker PR. CD33-related sialic-acid-binding immunoglobulin-like lectins in health and disease. Carbohydrate Research. 2008;343:2050–2056. doi: 10.1016/j.carres.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Varki A. Essentials of glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2009. [PubMed] [Google Scholar]
- 5.Jones C, Virji M, Crocker PR. Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol Microbiol. 2003;49:1213–1225. doi: 10.1046/j.1365-2958.2003.03634.x. [DOI] [PubMed] [Google Scholar]
- 6.Rempel H, Calosing C, Sun B, Pulliam L. Sialoadhesin expressed on IFN-induced monocytes binds HIV-1 and enhances infectivity. PLoS One. 2008;3:e1967. doi: 10.1371/journal.pone.0001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat Chem Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 8.Gunnarsson P, Levander L, Pahlsson P, Grenegard M. The acute-phase protein alpha 1-acid glycoprotein (AGP) induces rises in cytosolic Ca2+ in neutrophil granulocytes via sialic acid binding immunoglobulin-like lectins (siglecs) FASEB J. 2007;21:4059–4069. doi: 10.1096/fj.07-8534com. [DOI] [PubMed] [Google Scholar]
- 9.Carlin AF, Chang YC, Areschoug T, Lindahl G, Hurtado-Ziola N, King CC, Varki A, Nizet V. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 2009;206:1691–1699. doi: 10.1084/jem.20090691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordstrom T, Movert E, Olin AI, Ali SR, Nizet V, Varki A, Areschoug T. Human Siglec-5 inhibitory receptor and immunoglobulin A (IgA) have separate binding sites in streptococcal beta protein. J Biol Chem. 286:33981–33991. doi: 10.1074/jbc.M111.251728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuravleva MA, Trandem K, Sun PD. Structural implications of Siglec-5-mediated sialoglycan recognition. J Mol Biol. 2008;375:437–447. doi: 10.1016/j.jmb.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansson M, Li YC, Jendeberg L, Anderson S, Montelione BT, Nilsson B. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J Biomol NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 13.Veldkamp CT, Peterson FC, Hayes PL, Mattmiller JE, Haugner JC, 3rd, de la Cruz N, Volkman BF. On-column refolding of recombinant chemokines for NMR studies and biological assays. Protein Expr Purif. 2007;52:202–209. doi: 10.1016/j.pep.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golovanov AP, Hautbergue GM, Wilson SA, Lian LY. A simple method for improving protein solubility and long-term stability. J Am Chem Soc. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- 15.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 16.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 17.Han B, Liu YF, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. Journal of Biomolecular Nmr. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farmer BT, Constantine KL, Goldfarb V, Friedrichs MS, Yanchunas J, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nature Structural Biology. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 20.Cavanagh J. Protein NMR spectroscopy : principles and practice. Academic Press; Amsterdam ; Boston: 2007. [Google Scholar]
- 21.Sharma D, Rajarathnam K. C-13 NMR chemical shifts can predict disulfide bond formation. Journal of Biomolecular Nmr. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]