SUMMARY
The most common type of carbohydrate-transport system in Streptococcus mutans is the phosphoenolpyruvate (PEP)-sugar phosphotransferase system (PTS). We previously showed that fourteen PTSs exist in S. mutans UA159 (Ajdic et al., 2002). Several studies have shown that microorganisms growing in biofilms express different genes as compared to their planktonic counterparts. In this study, we showed that one PTS of S. mutans was expressed in sucrose-grown biofilms. Furthermore, the same PTS was also responsible for the transport and metabolism of disaccharide nigerose (3-O-α-D-glucopyranosyl-D-glucose). Additionally, the results indicate that the studied PTS might be involved in the transport and metabolism of carbohydrates synthesized by glucosyltransferase B (GtfB) and glucosyltransferase C (GtfC) of S. mutans. To our knowledge, this is the first report that shows PTS transport of a disaccharide (and possibly extracellular oligosaccharides) with α-1,3 linkage.
Keywords: S. mutans, Biofilm, Transporter, Carbohydrate, Microarray
INTRODUCTION
Streptococcus mutans is a major causative agent of human dental caries. The host’s diet is very important for the cariogenesis of S. mutans, as carbohydrate metabolism by this bacterium plays a key role in the formation of caries. S. mutans is able to metabolize a wide range of carbohydrates that may originate in the diet. If the diet is rich in sugars, especially sucrose, the end product of the sugar metabolism is mostly lactic acid that can lead to demineralization of tooth enamel. Therefore, sugar transport and metabolism by this bacterium are directly related to the onset and development of dental caries.
The most common type of carbohydrate-transport system in bacteria is the phosphoenolpyruvate (PEP)-sugar phosphotransferase system (PTS). Each PTS consists of two nonspecific energy-coupling components, enzyme I (EI), a heat-stable protein (HPr), and a sugar-specific multiprotein or multidomain permease known as enzyme II (EII). In most cases, subunits IIA and IIB are located in the cytoplasm, while subunits IIC and IID (when present) act as a membrane channel (Postma et al., 1993, Vadeboncoeur and Pelletier, 1997, Deutscher et al., 2006). The source of energy for carbohydrate transport is provided by phosphoenolpyruvate (PEP). During the process of carbohydrate PTS transport, the phosphoryl group on PEP is transferred to the imported sugar via several PTS proteins. First, phosphoryl group is transferred to EI. EI then transfers the phosphoryl group to a histidine residue on HPr. From HPr the phosphoryl is transferred to EIIA, and then to EIIB. Finally, EIIB phosphorylates carbohydrate as it crosses the plasma membrane through the transmembrane EIIC, forming sugar-phosphate. PTSs are responsible for binding, transmembrane transport, and phosphorylation of numerous sugar substrates. This holds true for S. mutans as well (Zeng and Burne, 2010, Zeng and Burne, 2009, Abranches et al., 2006, Abranches et al., 2003). We previously showed that fourteen PTSs exist in S. mutans UA159 (Ajdic et al., 2002, Ajdic and Pham, 2007).
Following initial colonization of the hard surfaces in the oral cavity, many bacteria including S. mutans, synthesize exopolysaccharides (EPS) and exooligosaccrarides (EOS) that result in the formation of a stable biofilm. EPS and EOS are produced by the action of extracellular glucansucrases (EC 2.4.1.5), also called glucosyltransferases (GTFs) and fructosyltransferase Ftf (EC 2.4.1.10). GTFs are enzymes that catalyze the transfer of glucosyl units formed following the cleavage of sucrose to a growing α-glucose chain, (Henrissat, 1998). The fructose moiety of sucrose is liberated during this process. Similarly, Ftf utilizes the fructose moiety of sucrose to synthesize EPS called fructan (Gibbons and Houte, 1975, Hamada and Slade, 1980). S. mutans expresses three GTFs: GtfB, GtfC and GtfD (formerly GtfI, GtfSI and GtfS) that produce different sizes and structures of EPS, called glucan. The nature of the linkages between glucosyl units determines its water solubility (Aoki et al., 1986, Hanada and Kuramitsu, 1988, Hanada and Kuramitsu, 1989). A low content of α-1,3 linkages and high content of α-1,6 linkages is associated with greater solubility of EPS, which is designated as dextran (Kuramitsu, 1975, Walker, 1978, Monchois et al., 1999). Reversed proportion of α-1,3 and α-1,6 linkages results in formation of water-insoluble glucose polymers called mutan. In addition to glucan production, GTFs can transfer glucose to fructose, in which case sucrose isomers are formed (Monchois et al., 2000). EPS produced by extracellular sucrose-utilizing enzymes of S. mutans contribute to the pathogenicity (Gibbons and Houte, 1975, Hamada and Slade, 1980, Loesche, 1986, Munro et al., 1991, Schroeder et al., 1989) of this organism in multiple ways including bacterial adherence (Schilling and Bowen, 1992, Hamada et al., 1984) and accumulation on the tooth surface (Freedman and Tanzer, 1982, Larrimore et al., 1983), and extracellular storage of carbohydrates that can be utilized during periods of nutrient deprivation (Bowen and Koo, 2011) (Gibbons, 1968b, Manly and Richardson, 1968, Wood, 1967).
Dental plaque, the biofilm that develops on the tooth surface, is a complex community of microorganisms. When the diet of the human host is rich in carbohydrates, the most prevalent organisms present are acidogenic and aciduric bacteria such as S. mutans (Bowden et al., 1979, Bowden, 1999, Bradshaw et al., 1989, Burne, 1998). Several studies have shown that microorganisms growing in biofilms express different genes as compared to their planktonic counterparts (Burne et al., 1997, Costerton et al., 1987, Costerton et al., 1995). In this study, we showed that previously uncharacterized PTS (locus SMU.100-105) of S. mutans was expressed in sucrose-grown biofilm and in planktonic cultures grown in disaccharide nigerose (3-O-α-D-glucopyranosyl-D-glucose). Additionally, the results indicate that this PTS might be also involved in the transport and metabolism of carbohydrates synthesized by glucosyltransferase B (GtfB) and glucosyltransferase C (GtfC) of S. mutans. To our knowledge, this is the first report that shows PTS transport of a disaccharide (and possibly EOS) with α-1,3 linkage.
METHODS
Bacterial strains and culture conditions
S. mutans UA159 was routinely grown in Todd-Hewitt broth with 0.3% Yeast Extract (THY) supplemented, when needed, with antibiotics at the following concentrations: kanamycin (500 μg per ml), erythromycin (5 μg per ml). For microarrays and qRT-PCR, we used “chemically defined” medium (CDM) that has been successfully implemented in our previously published studies (Ajdic and Pham, 2007). For RNA isolation, cells were grown in CDM supplemented with ultrapure 0.5% carbohydrate. List of the carbohydrates used in this study is presented in Table 2. The environmental conditions used for these experiments were 5% CO2, 37°C, pH 7.0. Due to cell aggregation that occurs in the sucrose-growth medium, the optical density of cultures grown in sucrose was estimated by growing a duplicate culture in CDM supplemented with glucose. All cultures used for qRT-PCR or microarray analyses were grown to a mid-exponential growth phase (OD600~0.65), except when stated otherwise.
Table 2.
Disaccharides used in this study
| Disaccharide | Units | Bond | Growth | PTSBio Transcription |
Reference |
|---|---|---|---|---|---|
| Kojibiose | two glucose monomers | α(1-2) | no | N/A | this study |
| Nigerose | two glucose monomers | α(1-3) | yes | yes | this study |
| Maltose | two glucose | monomers α(1-4) | yes | no | (Ajdic &Pham, 2007) |
| Isomaltose | two glucose monomers | α(1-6) | no | N/A | this study |
| Sucrose | glucose + fructose monomers |
α(1-2) | yes | yes | this study |
| Turanose | glucose + fructose monomers |
α(1-3) | no | N/A | this study |
| Maltulose | glucose + fructose monomers |
α(1-4) | no | N/A | this study |
| Leucrose | glucose + fructose monomers |
α(1-5) | no | N/A | this study |
| Isomaltulose | glucose + fructose monomers |
α(1-6) | no | N/A | this study |
Growth curves
Growth curves of S. mutans UA159 were generated using a Bioscreen C Analyzer, version 2.4 (Oy Growth Curves AB Ltd., Finland). The cultures were incubated at 37°C for 24-48 h in CDM supplemented with the appropriate carbohydrate. The wideband 420-580 nm filter that is less sensitive to color changes of the media was used to detect the optical density of the cultures.
Biofilm batch cultures
Biofilm batch cultures were grown in polystyrene plates for 48 h in diluted CDM (0.5 X CDM) supplemented with 10 mM sucrose. The biofilms were seeded with 1:100 dilution of the 16 h planktonic bacterial culture grown in THY and washed twice with PBS pH 7.4. The environmental conditions used for these experiments were 5% CO2, 37°C, pH 7.0. The growth media was replaced with a fresh one after 24 h and 3 h before cell collection to assure expression of carbohydrate transporters (the 3 h incubation was determined empirically). The cells were then collected in TriReagent (Invitrogen) for RNA isolation.
Construction of the mutants
The mutants ftf−, gtfD−, gtfBC−, gtfB−, gtfC−, gtfB−Cc and nigB were constructed using the Overlap extension PCR method (Chalker et al., 2001). To construct each mutant, approximately 1000 bp upstream and downstream regions (including ~100 bp of the 5′-end and 3′-end of each ORF, respectively) was PCR-amplified. List of the primers used in this study is presented in Table 3. A fragment containing an antibiotic gene (erythromycin - erm or kanamycin - kan cassette) was amplified with specific primers (Table 3). The three fragments were mixed and amplified with corresponding primers (Table 3). The resulting PCR product was used for transformation of S. mutans UA159. Proper construction of the mutants was verified by PCR and sequencing. Non-polar cassettes were used for construction of the mutants (Zeng and Burne, 2008, Zeng and Burne, 2009), and the transcription of the downstream genes was checked by qRT-PCR.
Table 3.
Primers used in this study
| gtfB-P1 | TAC ATT GAC TCC CCT AAT CTT CTT G |
| gtfB-P2_erm | TTT TTG TTC ATG TAA TCA CTC CTT CCG CAG TTT ATA ACG CAC TTT CTT GTC |
| gtfB-P3_erm | ATA ATT CTA TGA GTC GCT GCC GAC TTC AGT TAT TAC GAT GCT AAC TCT GGA |
| gtfB-P4 | GAC CTG ATT ATA TTG AGC AAA GCT G |
| gtfC-P1 | CGT CAT GTT TGA AGG TTT CTC TAA T |
| gtfC-P2_kan | CGG TAT AAT CTT ACC TAT CAC CTT AGA CTT CCT GAA AGA GAG GTC AAA |
| gtfC-P3_kan | TCT AAA AGT TCG CTA GAT AGG GTT GGA TTC TTT GAC AAT TTC TTT AGA TT |
| gtfC-P4 | AAA TTC TTG ATT GCT TTT ATT TCC TC |
| gtfBC-P1 | TTT GTG GGA TAG TTT TGT TTT TAT CA |
| gtfBC- P2_kan |
CGG TAT AAT CTT ACC TAT CAC CTT CAC CCA TCT TTT CTT TAC TTT ACG |
| gtfBC- P3_kan |
TCT AAA AGT TCG CTA GAT AGG GTT GGA TTC TTT GAC AAT TTC TTT AGA TT |
| gtfBC-P4 | AAA TTC TTG ATT GCT TTT ATT TCC TC |
| gtfD-P1 | AAA TAT GCT GTT CTT TTT GCT AAC G |
| gtfD-P2_kan | CGG TAT AAT CTT ACC TAT CAC CTC CAG TGC TTT TTA ACC TTG TAC ATT |
| gtfD-P3_kan | TCT AAA AGT TCG CTA GAT AGG GTA ACA TGG TTT ACA ACA AAG TCG TC |
| gtfD-P4 | TCC ACT GAA TAA TTT CAC CTA CCT C |
| ftf-P1 | GAA AAA GCT CAT CCA GAT ATT TTC A |
| ftf-P2_kan | CGG TAT AAT CTT ACC TAT CAC CTC CCA AAA TTT CCC TTT CTT ATA CAT |
| ftf-P3_kan | TCT AAA AGT TCG CTA GAT AGG GAT TAG CTC TTT TCA GTG CTT TCT GT |
| ftf-P4 | TAG GAA TTA GCC GAC CTT CTT ATT T |
| bioB-P1 | TTA TGA AGA TGC TCT TGA ATG TAT CG |
| bioB-P2_erm | CAA ATC AAA CAA ATT TTG GGC CCG GTC TTT GAT CTA CTC TAG CCT CAA CAA |
| bioB-P3_erm | ATA ATT CTA TGA GTC GCT GCC GAC TTC AAT ATA TTC CGG ATG ATT CTG TAA |
| bioB-P4 | TAT ATA ATG CTG AAC CCA TGA ACA TC |
| erm-Fwd-P | CCG GGC CCA AAA TTT GTT TGA TTT |
| erm-Fwd | GAA GGA GTG ATT ACA TGA ACA AAA A |
| erm-Rev | AGTCGGCAGCGACTCATAGAAT |
| kan-Fwd | AGG TGA TAG GTA AGA TTA TAC CG |
| kan-Rev | CCC TAT CTA GCG AAC TTT TAG A |
erm-Fwd-P, PCR amplification with this primer included erm promoter
erm-Fwd (no P), PCR amplification with this primer did not included erm promoter
The gtfB-Cc mutant was constructed using the same primers utilized for gtfB− mutant (gtfB-P1, P2_erm, P3_erm, P4), except the erm-Fwd-P was used for amplification of the erm cassette resulting in the deletion of the gtfB and constitutive transcription of the gtfC gene. To verify that the gtfB-Cc mutant strain was properly constructed, transcription of the gtfB and gtfC genes was analyzed by qRT-PCR and compared to those of the WT and gtfBC− mutants. In the gtfB-Cc, the gtfB showed no transcription whereas gtfC was fully transcribed in aggregation growth mode at OD600=0.65.
RNA extraction and cDNA synthesis for microarray analysis
RNA extraction and cDNA synthesis were performed as previously published by our group (Ajdic and Pham, 2007). Briefly, the cultures for RNA isolation were grown at 37°C to an OD600 of approximately 0.65 in CDM containing 0.5% of the desired sugar. The cells were disrupted with a bead beater and the RNA was purified using the Ambion RiboPureTM -Bacteria Kit. Isolated RNA was treated with DNAse I, and the absence of DNA confirmed by PCR. cDNA was generated from 10 μg of RNA with SuperScript II reverse transcriptase (Invitrogen). DNA was fragmented with DNAse I using our previously published method (Ajdic and Pham, 2007). Quality and size (50-200 nucleotides) of the fragmented cDNA was evaluated using a Bioanalyzer 6000 (Agilent Technologies) followed by labeling with Biotin-ddUTP using the BioArray Terminal Labeling Kit.
Quantitative Real-time (qRT) PCR
Quantification of specific transcript was accomplished by the comparative CT method using the Bio-Rad MyiQ Real-time PCR Detection System. The primers amplified 100-110 bp specific fragments. Efficiency of amplification was confirmed by analyzing melting curves of each amplicon. All qRT-PCR reactions were performed using SYBR Green master mix (Bio-Rad) with specific primers and cDNA (that was prepared following the protocol for cDNA preparation for microarray except, in this case, DNAse I fragmentation and biotin labeling were omitted). Samples in which SuperScript II was omitted during cDNA synthesis were used as a negative control. The qRT-PCR amplification with primers to the gyrA was used for normalization. Non-template controls were included to confirm the absence of primer-dimer formation. All samples, including non-template controls, were performed in triplicate.
The comparison of gene expression in various conditions was accomplished by the comparison of CT values automatically generated by the MyiQ software. The relative comparison of expression of the gene of interest (goi) in two different conditions (goi1 and goi2) was presented as fold change (FC). All FC values of the genes of interest were normalized to gyrA since our previous data showed that it produced little variation of expression in mid-exponential growth phase (OD600~0.65) (Ajdic and Pham, 2007). Previously published mathematical models (Malke et al., 2006, Pfaffl, 2001) for relative quantifications of qRT-PCR data were performed.
Expression Microarray (EM)
A whole genome custom GeneChip Antisense Expression Microarray chip, designed in collaboration with Affymetrix (Santa Clara, CA), was used in this study. The technical details about the chip were previously published (Ajdic and Pham, 2007).
Hybridization, washing and scanning of the microarray chips were performed according to the procedures described by Affymetrix (www.affymetrix.com) and our previously published protocols (Ajdic and Pham, 2007). The GeneChip® Operating Software (GCOS) version 1.4 analysis program (Affymetrix, Santa Clara, CA) was used to analyze gene expression and expression clustering, respectively. The data were compared using GCOS batch analyses. Normalization of all probe sets was done by GCOS. The software computed a normalization value: Trimmed Mean Signalbaseline=(Normalization Value) X (Trimmed Mean Signalexperiment). The cut off score for the analysis was two-fold.
Relevant microarray data was confirmed by qRT-PCR.
Microarray accession numbers
Microarray data are available at the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under platform and series accession numbers GPL4769 and GSE35605, respectively.
RESULTS
Global transcriptional analysis of S. mutans sugar transporters in sucrose-grown biofilm
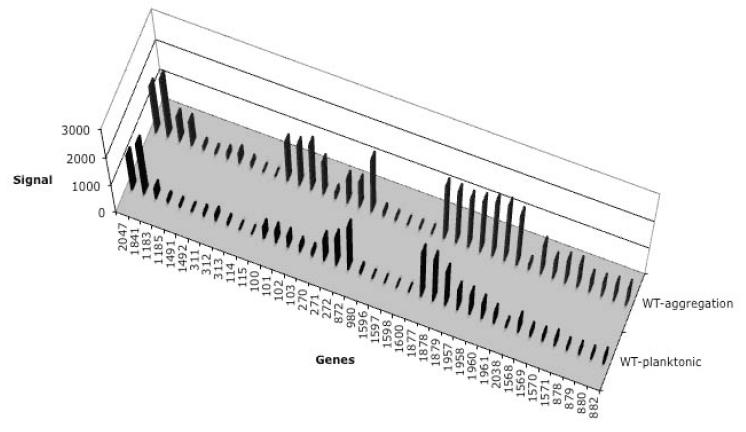
In a previous study, we determined global differential transcription profiles of S. mutans UA159 for 13 carbohydrates when in a planktonic mode of growth (Ajdic and Pham, 2007). In this study, differential expression patterns of carbohydrate transporters were analyzed in S. mutans biofilms grown in sucrose for 48 hours. Transcription profiles of the biofilm grown cells were generated by microarrays and compared to our previous microarray results for planktonic cells (Ajdic and Pham, 2007). The transcription patterns of two carbohydrate transporters were significantly different in the biofilm as compared to those in planktonic grown cultures. The genes (SMU.1957-1961) encoding PTSFru/Man specific for fructose and mannose, and a locus consisting of 6 genes (SMU.100-105), organized as a putative operon, were up-regulated in biofilms (Figure 1). Annotation data (Ajdic et al., 2002) suggested that this latter putative operon encoded enzymes for PTS transport and metabolism of an unknown substrate. Because the genes of this locus were transcribed in sucrose-grown biofilm, we named the putative operon OBio and the PTS it encodes PTSBio.
Fig. 1.
Differential transcription of the genes for PTS Enzyme II and ABC transporters following growth of S. mutans UA159 in biofilm. Microarray results are presented as vertical bars. The numerical ORF designation (as presented in NCBI genome database) for genes encoding sugar transporters is presented on the X-axis. Genes encoding PTSBio are numbered as 100-103. Complete ORF legend is presented in Supplemental material. Value of the normalized expression signal is presented on the Y-axis. Transcription of the gene gyrA (SMU.1114) served as an endogenous control because it did not show significant variation of transcription in the conditions compared. Two or more biological replicates were analyzed in each experiment for each condition, thus we present the average results. Conditions: WT-planktonic, cells grown in CDM culture supplemented with 28 mM glucose; WT-biofilm, cells grown in depleted CDM supplemented with 10 mM sucrose.
Transcription of the PTSBio was also studied in glucose or fructose-grown biofilms using qRT-PCR. S. mutans formed very fragile biofilm if grown in defined medium supplemented with sugars other than sucrose. Expression of the PTSBio was very low in glucose or fructose-grown biofilms (Supplemental Figure 1) as compared to the same in sucrose-grown biofilms. Specifically, its transcription was approximately 5-fold lower in glucose and 7-fold lower in fructose-grown biofilm. Therefore, the PTSBio may not play an important role in biofilm metabolism in the absence of sucrose.
Computational analysis of the OBio
The OBio locus was analyzed using two ORF-calling programs, Glimmer (Salzberg et al., 1998, Delcher et al., 1999) originally used for annotation of the S. mutans UA159 genome database and the National Center for Biotechnology Information ORF finder (www.ncbi.nml.nih.gov). Results revealed the presence of six ORFs organized in a putative operon. Domain and motif search for the predicted ORFs suggested the presence of a four-component transport system of the PTS family encoding EII subunits (EIIB, EIIC, EIID, and EIIA), an enzyme involved in carbohydrate metabolism, and a transcriptional regulator of the LacI repressor family. The putative operon structure is shown in Figure 2. The PTS subunits consisted of EIIB belonging to the PTS_IIB_mannose superfamily (cd00001; pfam03830), a cytoplasmic component of PTS necessary for the uptake of carbohydrate across the cytoplasmic membrane and its phosphorylation; EIIC of the EII_sorbose superfamily (pfam03609), a membrane subunit; EIID of the AGA superfamily (mannose/fructose/sorbose family) (pfam03613); and EIIA of the mannose superfamily (pfam03610), a cytoplasmic subunit that receives a phosphoryl group from HPr and transfers it to EIIB which phosphorylates the substrate. The putative enzyme showed high similarity to α-glucosidase / glycosyl hydrolases (cd06595; pfam02065) of the GH31 superfamily. Enzymes of this family cleave a terminal carbohydrate moiety from a substrate that varies in size, depending on the enzyme. The transcriptional regulator contained specific structural features that included a small DNA-binding domain with a helix-turn-helix (HTH) motif at the N-terminus (cd01392). Analysis of the C terminus revealed a regulatory ligand-binding domain for oligomerization and for effector binding, and an approximately 18-amino acid linker connecting these two functional domains. (cd06291). For LacI-like transcriptional regulators, the ligands are sugars.
Fig. 2.
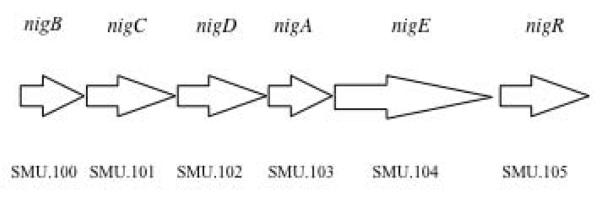
Schematic presentation of putative operon (OBio) encoding proteins for PTSBio and metabolic enzymes. Arrows represent the genes. Genes nigB, nigC, nigD and nigA encode putative EIIB, EIIC, EIID and EIIA components of the PTSBio, respectively. Gene nigE encoding an enzyme (annotated as putative α-glucosidase/glycosyl hydrolase) and a transcriptional regulator (nigR) are also part of this putative operon. The numerical ORF designation (as deposited to NCBI genome database) for genes encoding PTSBio is presented under the arrows.
Expression of the PTSBio is coordinated with EPS/EOS production
S. mutans cultures grown in defined medium supplemented with sucrose exhibit cell-to-cell aggregation. The aggregation is promoted by the extracellular EPS/EOS produced by S. mutans. We will refer to this mode of growth as ‘aggregation growth mode”. The transcription of the genes encoding the PTSs of UA159 was analyzed by microarrays following this growth condition (Figure 3). The genes encoding the PTSBio were transcribed in aggregation growth mode (Figure 3). However, when the cultures were grown in the defined medium supplemented with sucrose and dextranase, sucrose-induced cell aggregation was abolished. The genes encoding the PTSBio were not transcribed in the presence of dextranase (Figure 3). So it was evident that the genes encoding the PTSBio responded to the formation of EPS/EOS produced from sucrose. Hence, sucrose migh not be the primary substrate for the PTSBio.
Fig. 3.
Differential transcription of the genes for PTS Enzyme II and ABC transporters following growth of S. mutans UA159 in sucrose. Description of the microarray data presentation is explained in Fig. 1. Conditions: WT-planktonic, cells grown in CDM supplemented with 0.5% sucrose and 10 U/ml dextranase; WT-aggregation, cells grown in CDM supplemented with 0.5% sucrose.
GtfB and GtfC are also required for PTSBio expression
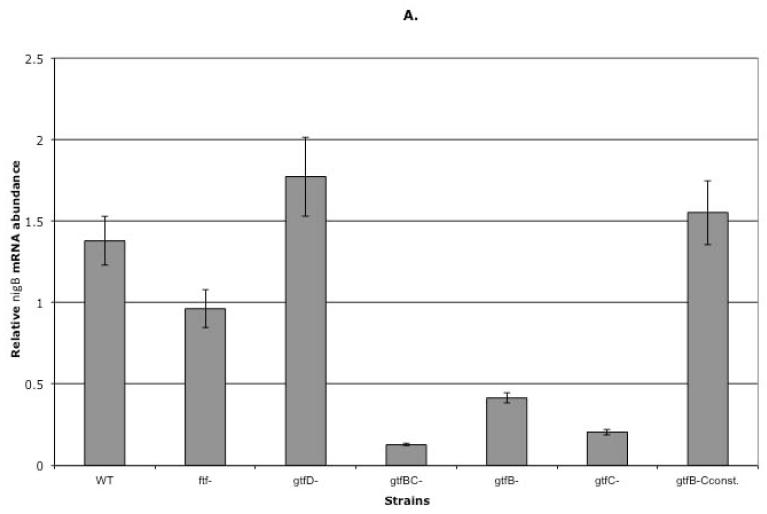
S. mutans produces three glucosyltransferases (GtfD, GtfB and GtfC encoded by gtfD, gtfB and gtfC, respectively) and one fructosyltransferase (Ftf encoded by ftf) that are responsible for synthesis of glucans and fructans, respectively. Glucans can be water-soluble or water-insoluble (Kuramitsu, 1975, Walker, 1978, Monchois et al., 1999). Water-soluble glucans and fructans are considered to be extra-cellular sugar storage (Gibbons, 1968b, Manly and Richardson, 1968, Wood, 1967). Because PTSBio was expressed in sucrose-dependent biofilm and sucrose-dependent aggregation growth mode we hypothesized that its expression was induced by oligosaccharides and/or polysaccharides synthesized from sucrose moieties (glucose and/or fructose). To test our hypothesis, genes encoding enzymes responsible for water-soluble glucan or fructan synthesis were deleted from the genome of UA159. Specifically, single gtfD− and ftf− mutants were constructed by replacing these genes with non-polar antibiotic cassettes (as stated in Methods). Transcription of the first gene (nigB) of the OBio was analyzed in the mutants using qRT-PCR and compared to that of the WT, following an aggregation mode of growth. The results revealed minor change of the nigB transcription in either mutant (1.4 and 1.3 fold difference in ftf− and gtfD− single mutants, respectively) as compared to the WT (Figure 4A). Data suggested that the activity of GtfD and Ftf was not required for the PTSBio transcription.
Fig. 4.
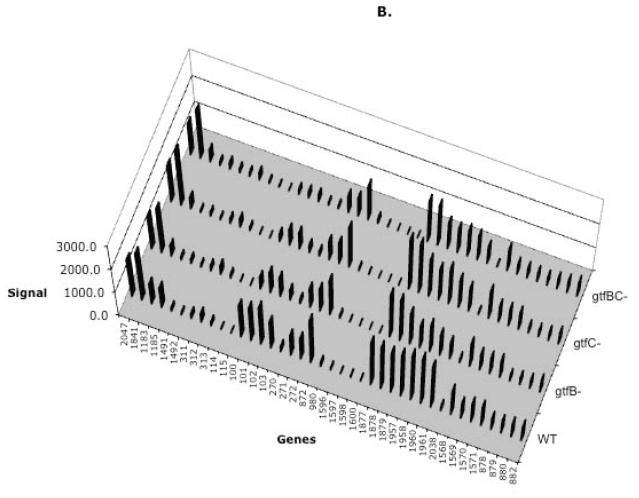
A) Transcription of the nigB in the WT, ftf and gtf mutants. Gene transcription was analyzed by qRT-PCR in the WT and the mutant strains following an aggregation mode of growth. mRNA was isolated from the tested strains during a mid-exponential phase of growth (OD600=0.65). The Real-time PCR results were presented as relative mRNA abundance (vertical columns). Standard deviation is presented for each result. B) Differential transcription of the genes for PTS Enzyme II and ABC transporters following growth of gtfB−, gtfC− and gtfBC− mutants. Description of the microarray data presentation is explained in Fig. 1.
Our next question was whether expression of Gtfs responsible for production of water-insoluble glucans was necessary for transcription of OBio. To answer this question, a gtfBC− double mutant was constructed using antibiotic replacement strategy. Again, transcription of the nigB was analyzed in this mutant using qRT-PCR and compared to that of the WT following an aggregation mode of growth. Transcription of nigB was 10.8 fold lower in the gtfBC− mutant (Figure 4A) indicating that either GtfB or GtfC or both were required for expression of the PTSBio. Finally, to find out if one or both of these enzymes contribute to the expression of the studied transporter, their genes were individually replaced with antibiotic cassettes resulting in construction of the gtfB− and the gtfC− mutants. Transcription of the nigB was analyzed in both mutants using qRT-PCR. As shown in Figure 4A, transcription of the nigB was 3.3 fold lower in gtfB−and 6.7 fold lower in gtfC− as compared to that of the WT indicating that the products of these enzymes contribute to the differential regulation of the nigB.
To analyze if additional carbohydrate transporters were differentially transcribed in the gtfB−, gtfC− and gtfBC− mutants, microarray analysis was performed. As shown in Figure 4B, transcription patterns for the genes encoding carbohydrate transporters were comparable in the mutants as compared to that of the WT, except for the genes encoding PTSBio which were highly transcribed only in the WT.
Constitutive transcription of the gtfC triggers transcription of OBio
Several attempts to clone gtfB and gtfC genes on a plasmid for complementation of the corresponding deleted genes failed. Plasmid constructs exhibited deletions upon transformation, which resulted in considerably lower or complete lack of gene expression. To circumvent this problem, a genomic transcriptional fusion of the gtfC was constructed. In this construct, the gtfB gene was deleted from the genome and the gtfC gene was expressed from the constitutive erm promoter (Claverys et al., 1995). This mutant was named gtfB-CC. To verify proper transcription of the gtfB and gtfC genes in the constructed strain, qRT-PCR was performed as explained in the Methods. Transcription of the nigB was analyzed in the gtfB-CC mutant by qRT-PCR following aggregation growth mode (OD600=0.65). The results showed that nigB transcription was similar to that of the WT (Figure 4A) suggesting that the constitutive transcription of the gtfC in the absence of gtfB was sufficient to induce full transcription of the OBio in the mid-exponential phase of growth.
Substrate(s) for the PTSBio accumulates in aggregation mode of growth and in biofilm
Cell aggregates form quickly at the OD600=0.25-0.3 during the cultivation of the WT in CDM supplemented with sucrose. However, aggregation of the gtfB− and gtfC− mutants was delayed as compared to that of the WT. The gtfC− mutant formed visible aggregates at OD600=0.35-0.4, whereas aggregation of the gtfB− mutant cells was apparent around OD600=0.4-0.45. No aggregation was noticed in the gtfBC− mutant cells throughout growth. Because of the variation in timing of the glucan production in the mutants and the WT, we have also analyzed transcription of the nigB in the late stage (OD600=0.9) of the aggregation-growth mode by qRT-PCR. As presented in Figure 5, the nigB transcription was similar in the gtfB− (1.2 fold difference) and slightly lower in the gtfC− mutant (1.57 fold difference) as compared to that of the WT following this growth condition. However, transcription of the nigB was more than 4 fold lower in the gtfBC− double mutant.
Fig. 5.
Transcription of the nigB in the gtfB− and gtfC− mutants during late exponential phase of aggregation mode of growth. All samples were collected at an OD600~0.9. Gene transcription was analyzed by Real-time PCR. Transcription of the nigB in the WT and the gtfBC mutant was used as the positive and negative controls, respectively. The Real-time PCR results were presented as relative mRNA abundance (vertical columns). Standard deviation is presented for each result.
Additionally, transcription of all UA159 carbohydrate transporters (including the PTSBio) was analyzed in the gtfB− and gtfC− mutants grown in biofilms. This analysis was performed by microarrays and the results for selected genes (SMU.100-103) were confirmed by qRT-PCR (data not shown). Results showed that the transcriptional profiles of the studied genes in the mutants were similar to that of the WT following biofilm growth (Supplemental Figure 2). Furthermore, expression of the PTSBio was also analyzed by microarrays in sucrose-grown biofilms of the WT in which the biofilms were incubated in fresh medium supplemented with glucose during the last three hours of biofilm growth. Transcription of the PTSBio in glucose-incubated biofilms was similar to that of the sucrose-incubated biofilms (Supplemental Figure 2).
Collectively, these data suggest that the substrate(s) for the PTSBio accumulate in the presence of sucrose, GtfB and GtfC. We can also conclude that the substrate(s) accumulation in the presence of sucrose and in the absence of one glucosyltransferase (GtfB or especialy GtfC) is delayed.
What is the substrate for PTSBio?
GTFs of different microorganisms can produce sucrose isomers such as leucrose, turanose, maltulose, and isomaltulose (Table 3) (Monchois et al., 2000). We hypothesized that GtfB and GtfC of UA159 might be able to produce sucrose isomers that could be taken-up by the PTSBio. Therefore, UA159 was grown in leucrose, turanose, maltulose, or isomaltulose as a sole carbohydrate source. The results revealed that UA159 did not grow in CDM supplemented with any of these sucrose isomers, suggesting that they were neither substrate for the PTSBio, nor for any other carbohydrate transporter of this strain.
As mentioned previously, GTFs of S. mutans produce α-linked glucose EOS and EPS from sucrose. The majority of the linkages found in the EPS and EOS are α(1-6) and α(1-3) although others, such as α(1-4) and α(1-2) can also be present (Monsan et al., 1995, Khalikova et al., 2005). Because our data showed that activity of GtfB and GtfC is required for expression of the PTSBio, we hypothesized that this transporter could be responsible for uptake of disaccharides and/or oligosaccharides containing glucose molecules linked by the above mentioned bonds. To check this, growth of UA159 and transcription of the PTSBio were analyzed in CDM supplemented with 0.5% disaccharides consisting of two glucose molecules linked by the bonds found in EOS produced by S. mutans. Disaccharides chosen for this analysis were: kojibiose α(1-2), nigerose α(1-3), and isomaltose α(1-6) (Table 3). Maltose, a disaccharide in which glucose molecules are linked by an α(1-4) bond, was excluded from the analysis because our previous data showed that maltose supported growth of UA159, but the PTSBio was not expressed in the presence of maltose (Ajdic and Pham, 2007). The strain UA159 did not grow in kojibiose and isomaltose, but it did grow in nigerose. Furthermore, nigB, the first gene of the putative operon encoding the PTSBio was transcribed following growth in nigerose, as shown by qRT-PCR (Figure 6A). To find out if additional carbohydrate transporters of UA159 were differentially transcribed following growth in nigerose, microarray analysis was performed. As shown in Figure 6B, transcription patterns for the genes encoding carbohydrate transporters were similar following growth in glucose or nigerose, except for the genes encoding PTSBio, which were highly transcribed in nigerose, only. Additionaly, maltodextrin and multiple sugar (MSM) ABC transport systems were also slightly upregulated, which may indicate that they were also involved in nigerose uptake. Considering that both ABC transporters are primarely responsible for oligosacharide transport, alternative explanation might be that nigerose serves as a primer for oligosaccharide(s) synthesis which are then transported via these two ABC systems.
Fig. 6.
A) Transcription of the nigB following growth in nigerose. Cells were grown in planktonic culture to mid-exponential phase (OD600=0.65) in CDM supplemented with nigerose or glucose. Gene transcription was analyzed by qRT-PCR. The qRT-PCR results were presented as relative mRNA abundance (vertical columns). Standard deviation is presented for each result. B) Differential transcription of the genes for PTS Enzyme II and ABC transporters following growth of UA159 in nigerose or glucose. The same cDNA samples analyzed by qRT-PCR were used for microarrays. Description of the microarray data presentation is explained in Fig. 1.
Deletion of nigB
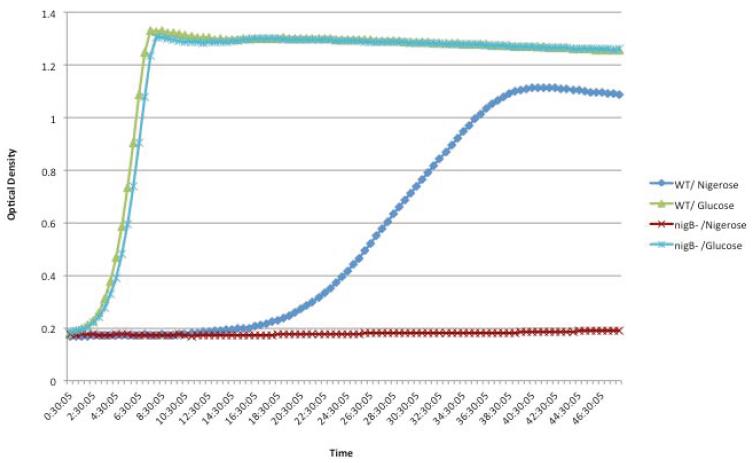
Deletion of any gene encoding an EII component of PTS results in an inactive transport system. Therefore, to verify the phenotype of the PTSBio we deleted the gene nigB encoding the EIIB component of the PTSBio. The mutant nigB− was constructed using overlap extension PCR method (as explained in Methods). The nigB− retained capacity to grow in glucose, but lost capability to grow in nigerose (Figure 7). These data are clearly showing that nigB gene is required for cell-growth in the presence of nigerose.
Fig. 7.
Growth of the WT and the nigB− mutant in nigerose or glucose. Samples were grown at 37°C for 48 hours in CDM supplemented with 0.5 % sugar.
DISCUSSION
In a previous study, we determined global differential transcription profiles of S. mutans UA159 for several monosaccharides, disaccharides, a β-glucoside, oligosaccharides and a sugar-alcohol (Ajdic and Pham, 2007). Our results revealed that PTSs were responsible for transport of most carbohydrates tested. In fact, all monosaccharides, disaccharides, β-glucoside, and sugar-alcohols we tested were transported by PTSs. In contrast, two ABC transporters were specific for oligosaccharides. All these experiments were done in planktonic cultures. In this study, differential expression patterns of carbohydrate transporters were analyzed in S. mutans biofilms and compared to those of planktonic cultures. Two carbohydrate transporters, PTSFru/Man and PTSBio (and accompanying enzymes) were differentially expressed in sucrose-grown biofilms as compared to those in sucrose- or glucose-grown planktonic cultures. Our previous studies showed that the PTSFru/Man (SMU.1957-1961) was fully induced in planktonic cultures grown in fructose or mannose (Ajdic and Pham, 2007). The same PTS was partially induced in planktonic cultures grown with sucrose in the presence of exogenously added dextranase (Ajdic & Pham, 2007). In this study, we showed that the PTSFru/Man was upregulated in biofilms and aggregation growth mode. The PTSFru/Man transports fructose and mannose (Ajdic and Pham, 2007, Zeng and Burne, 2010). Consequently, its induction was expected following growth in sucrose because sucrose serves as a substrate for glucosyltransferases (Hamada and Slade, 1980), fructosyltransferase (Hamada and Slade, 1980) and fructanase (DaCosta and Gibbons, 1968, Walker et al., 1983). These extracellular enzymes of S. mutans release the fructose moiety from sucrose, which is then taken up by fructose PTSs (PTSFru and PTSFru/Man). Interestingly, expression of the PTSFru/Man was much higher in biofilm and aggregation growth mode as compared to that in planktonic cultures grown in sucrose with dextranase, suggesting a lower concentration of fructose in the media during planktonic growth of S. mutans, perhaps due to a lower rate of fructose moiety liberation from sucrose in the presence of a high concentration of dextranase. Varieties of microorganisms synthesize extracellular dextranases and S. mutans is one of them (Guggenheim and Burckhardt, 1974). Dextranases catalyze the hydrolysis at the α-1,6-glycosidic bond of dextran and release of glucose or short oligosaccharides consisting of several glucose molecules (Khalikova et al., 2005, Walker, 1972). The activity of the S. mutans dextranase is apparently well synchronized with the production of other enzymes involved in extracellular sugar metabolism resulting in the accumulation of dextrans if sucrose is available. However, in the presence of a high concentration of dextranase added to the media at the beginning of the culture growth, newly synthesized dextran is rapidly cleaved (Walker, 1972, Hamada et al., 1975) which may result in disbalanced production of glucosyltransferase, fructosyltransferase and fructanase and consequently lower liberation of fructose from sucrose and lower expression of inducible PTSFru/Man.
The second transporter that exhibited full expression in biofilm and aggregation growth mode was PTSBio. Computational analysis of the OBio (SMU.100-105) suggested that this putative operon encoded proteins for PTS transport, metabolism and regulation of carbohydrate utilization. Our previous studies showed that the PTSBio was not expressed in planktonic cultures grown in 13 different sugars (Ajdic and Pham, 2007). Furthermore, PTSBio was not expressed in sucrose-grown planktonic cultures in the presence of exogenously added dextranase when this enzyme was added at the initial stage of growth. Oral glucans contain a large proportion of α-1,3-glycosidic bonds that are resistant to the action of dextranase. However, if dextranase is added at the initial stage of the glucan synthesis, this enzyme is able to suppress EPS production because initially the oligosaccharide chains are elongated only by the attachment of glucosyl residues with α-1,6-glycosidic linkages (Walker, 1972). Out results showed that if EPS production was suppressed by dextranase (despite of the presence of sucrose), PTSBio was not expressed. The result led us to conclude that PTSBio was not responsible for sucrose uptake in this experimental condition. Furthermore, we hypothesized that S. mutans utilized sucrose to synthesize an appropriate substrate that is then taken-up by PTSBio. S. mutans expresses four extracellular enzymes responsible for EPS synthesis: three glucosyltransferases (GtfD, GtfB and GtfC enzymes; EC 2.4.1.5) and fructosyltransferase (Ftf enzyme; EC 2.4.1.10). The GtfD synthesizes EPS in which the prevalent bonds are α-1,6-glycosyl linkages (glucans) (Hanada and Kuramitsu, 1989), whereas the GtfB and GtfC produce EPS rich in α-1,3-glycosyl linkages (mutans) (Aoki et al., 1986, Hanada and Kuramitsu, 1988). The Ftf is responsible for production of EPS (fructans) composed predominantly of α-2,1-linked fructosyl units (Birkhed et al., 1979, Ebisu et al., 1975). Because α-1,6-glycosyl linkages of glucan and α-2,1-fructosyl linkages of fructan can be cleaved to oligosaccharides by dextranase and fructanase respectively, these two types of EPS are considered extracellular carbohydrate storage (Gibbons, 1968b, Gibbons, 1968a, Manly and Richardson, 1968, Wood, 1967) and as such possible substrate for PTSBio. However, neither deletion of the ftf gene nor deletion of the gtfD gene showed significant effect on expression of PTSBio. Surprisingly, simultaneous deletion of the gtfB and gtfC genes completely abolished PTSBio transcription in the presence of sucrose during mid- and late-exponential phases of aggregation growth mode. Unfortunately, we could not analyze expression of the PTSBio in the double gtfBC− mutant in biofilm because this mutant does not form biofilm. Independent deletion of either gtfB or gtfC dramatically decreased transcription of this PTS in the exponential phase of culture growth, suggesting that the GtfB and GtfC utilize sucrose to synthesize the substrate that is then taken-up by the PTSBio. Furthermore, our results showed that if either of these enzymes was expressed, full transcription of the PTSBio was detected in the later stages of aggregation and biofilm growth, suggesting that either enzyme was capable of synthesizing the substrate, albeit with different timing. It is also possible that the modification of the substrate of one enzyme by the other produces the optimal carbohydrate molecule for which the PTSBio exhibits the highest affinity. Another interesting observation was expression of the PTSBio in the 48 h biofilm even if sucrose was replaced with glucose during the last 3 h of biofilm growth, suggesting that the substrate for this transporter accumulated in biofilm. Furthermore, transcription of OBio was not inhibited by glucose under studied experimental conditions. Therefore, most likely, OBio is not catabolically repressed by glucose. This is further supported by the absence of the CRE consensus sequence in the promoter region of the OBio.
What is the substrate for the PTSBio? We tested a series of disaccharides to address this question. Tested disaccharides (maltose, kojibiose, nigerose, and isomaltose) consisted of glucose moieties linked by α-glycosyl bonds found in S. mutans EPS. In addition, we also tested several sucrose isomers (leucrose, turanose, maltulose, and isomaltulose) known to be produced by Gtfs of different organisms. UA159 did not grow in disaccharides with α-1,6 (isomaltose) and α-1,2 (kojibiose) glycosyl linkages, nor did it grow in any of the sucrose isomers tested. In contrast, this strain grew in maltose (α-1,4 linkage) and maltotriose (Ajdic and Pham, 2007, Webb et al., 2008), but PTSBio was not expressed in the presence of these sugars (Ajdic and Pham, 2007). Among all tested sugars, nigerose (α-1,3 linkage) was the only disaccharide capable of inducing OBio. Furthermore, deletion of the nigB, the first gene of OBio, rendered the mutant incapable of growth in nigerose confirming that this sugar is a substrate for the PTSBio. To our knowledge, this is a first report showing that a disaccharide in which two glucose units linked by α-1,3 bond is utilized for growth by S. mutans. Our results suggest that disaccharide (and possibly oligosaccharides) consisting of α-1,3-linked glucose molecules synthesized by GtfB and GtfC from sucrose are substrates for the PTSBio, and as such serve as an extracellular carbohydrate storage in S. mutans biofilm.
Supplementary Material
Supplemental Fig. 1 Transcription of the nigB following biofilm growth in glucose, fructose or sucrose. Gene transcription was analyzed by qRT-PCR. The qRT-PCR results were presented as relative mRNA abundance (vertical columns).
Supplemental Fig. 2 Differential transcription of the genes for PTS Enzyme II and ABC transporters following growth of gtfB− and gtfC− mutants in biofilm. Biofilms were grown in diluted CDM supplemented with sucrose for 48 h or for 45 h plus 3 h in glucose (WT-glu). Description of the microarray data presentation is explained in Fig. 1.
Table 1.
Bacterial strains used in this study and their derivation.
| S. mutans Strains | Description | Reference |
|---|---|---|
| WT | UA159, serotype c strain, wild-type | P. Caufield |
| gtfB − | Δ gtfB; ErmR | This work |
| gtfC − | Δ gtfC; KanR | This work |
| gtfBC − | ΔgtfBC; KanR | This work |
| gtfD − | Δ gtfD; KanR | This work |
| ftf − | Δftf; KanR | This work |
| nigB − | ΔnigB; ErmR | This work |
ErmR, erythromycin resistance; KanR, kanamycin resistance.
ACKNOWLEDGMENTS
We thank Mpala Pilula and Gorana Savic for technical support and Dr. J. Banas and Dr. M. Sengupta for the critical reading of the manuscript. This work was supported by grant DE021424 (to D.A.) from the National Institute of Dental and Craniofacial Research.
References
- ABRANCHES J, CANDELLA MM, WEN ZT, BAKER HV, BURNE RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol. 2006;188:3748–56. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ABRANCHES J, CHEN YY, BURNE RA. Characterization of Streptococcus mutans strains deficient in EIIAB Man of the sugar phosphotransferase system. Appl Environ Microbiol. 2003;69:4760–9. doi: 10.1128/AEM.69.8.4760-4769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AJDIC D, MCSHAN WM, MCLAUGHLIN RE, SAVIC G, CHANG J, CARSON MB, PRIMEAUX C, TIAN R, KENTON S, JIA H, LIN S, QIAN Y, LI S, ZHU H, NAJAR F, LAI H, WHITE J, ROE BA, FERRETTI JJ. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–9. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AJDIC D, PHAM VT. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol. 2007;189:5049–59. doi: 10.1128/JB.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AOKI H, SHIROZA T, HAYAKAWA M, SATO S, KURAMITSU HK. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect Immun. 1986;53:587–94. doi: 10.1128/iai.53.3.587-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIRKHED D, ROSELL KG, GRANATH K. Structure of extracellular water-soluble polysaccharides synthesized from sucrose by oral strains of Streptococcus mutans, Streptococcus salivarius, Streptococcus sanguis and Actinomyces viscosus. Arch Oral Biol. 1979;24:53–61. doi: 10.1016/0003-9969(79)90175-4. [DOI] [PubMed] [Google Scholar]
- BOWDEN GH. Controlled environment model for accumulation of biofilms of oral bacteria. In: DOYLE RJ, editor. Biofilms. Academic Press; 1999. [DOI] [PubMed] [Google Scholar]
- BOWDEN GH, ELLWOOD DC, HAMILTON IR. Microbial ecology of the oral cavity. In: ALEXANDER M, editor. Advances in microbial ecology. Plenum Press; New York: 1979. [Google Scholar]
- BOWEN WH, KOO H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRADSHAW DJ, MCKEE AS, MARSH PD. Effects of carbohydrate pulses and pH on population shifts within oral microbial communities in vitro. J Dent Res. 1989;68:1298–302. doi: 10.1177/00220345890680090101. [DOI] [PubMed] [Google Scholar]
- BURNE RA. Oral streptococci... products of their environment. J Dent Res. 1998;77:445–52. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- BURNE RA, CHEN YY, PENDERS JE. Analysis of gene expression in Streptococcus mutans in biofilms in vitro. Adv Dent Res. 1997;11:100–9. doi: 10.1177/08959374970110010101. [DOI] [PubMed] [Google Scholar]
- CHALKER AF, MINEHART HW, HUGHES NJ, KORETKE KK, LONETTO MA, BRINKMAN KK, WARREN PV, LUPAS A, STANHOPE MJ, BROWN JR, HOFFMAN PS. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J Bacteriol. 2001;183:1259–68. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLAVERYS JP, DINTILHAC A, PESTOVA EV, MARTIN B, MORRISON DA. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–8. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- COSTERTON JW, CHENG KJ, GEESEY GG, LADD TI, NICKEL JC, DASGUPTA M, MARRIE TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–64. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- COSTERTON JW, LEWANDOWSKI Z, CALDWELL DE, KORBER DR, LAPPIN-SCOTT HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–45. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- DACOSTA T, GIBBONS RJ. Hydrolysis of levan by human plaque streptococci. Arch Oral Biol. 1968;13:609–17. doi: 10.1016/0003-9969(68)90139-8. [DOI] [PubMed] [Google Scholar]
- DELCHER AL, HARMON D, KASIF S, WHITE O, SALZBERG SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–41. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEUTSCHER J, FRANCKE C, POSTMA PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EBISU S, KATO K, KOTANI S, MISAKI A. Structural differences in fructans elaborated by streptococcus mutans and Strep. salivarius. J Biochem. 1975;78:879–87. doi: 10.1093/oxfordjournals.jbchem.a130993. [DOI] [PubMed] [Google Scholar]
- FREEDMAN ML, TANZER JM. Use of mutants to study the glucan-associated pathophysiology of Streptococcus mutans. In: SCHLESSINGER D, editor. Microbiology. American Society of Microbiology; Washington, D. C.: 1982. [Google Scholar]
- GIBBONS RJ. Formation and significance of bacterial polysaccharides in caries etiology. Caries Res. 1968a;2:164–71. doi: 10.1159/000259554. [DOI] [PubMed] [Google Scholar]
- GIBBONS RJ. Role of extracellular bacterial polysaccharides in the caries process. J Dent Res. 1968b;47:926–7. doi: 10.1177/00220345680470065601. [DOI] [PubMed] [Google Scholar]
- GIBBONS RJ, HOUTE JV. Bacterial adherence in oral microbial ecology. Annu Rev Microbiol. 1975;29:19–44. doi: 10.1146/annurev.mi.29.100175.000315. [DOI] [PubMed] [Google Scholar]
- GUGGENHEIM B, BURCKHARDT JJ. Isolation and properties of a dextranase from streptococcus mutans OMZ 176. Helv Odontol Acta. 1974;18:101–13. [PubMed] [Google Scholar]
- HAMADA S, KOGA T, OOSHIMA T. Virulence factors of Streptococcus mutans and dental caries prevention. J Dent Res. 1984;63:407–11. doi: 10.1177/00220345840630031001. [DOI] [PubMed] [Google Scholar]
- HAMADA S, MIZUNO J, MURAYAMA Y, OOSHIMA Y, MASUDA N. Effect of dextranase on the extracellular polysaccharide synthesis of Streptococcus mutans; chemical and scanning electron microscopy studies. Infect Immun. 1975;12:1415–25. doi: 10.1128/iai.12.6.1415-1425.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMADA S, SLADE HD. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–84. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANADA N, KURAMITSU HK. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect Immun. 1988;56:1999–2005. doi: 10.1128/iai.56.8.1999-2005.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANADA N, KURAMITSU HK. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect Immun. 1989;57:2079–85. doi: 10.1128/iai.57.7.2079-2085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENRISSAT B. Glycosidase families. Biochem Soc Trans. 1998;26:153–6. doi: 10.1042/bst0260153. [DOI] [PubMed] [Google Scholar]
- KHALIKOVA E, SUSI P, KORPELA T. Microbial dextran-hydrolyzing enzymes: fundamentals and applications. Microbiol Mol Biol Rev. 2005;69:306–25. doi: 10.1128/MMBR.69.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KURAMITSU HK. Characterization of extracellular glucosyltransferase activity of Steptococcus mutans. Infect Immun. 1975;12:738–49. doi: 10.1128/iai.12.4.738-749.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LARRIMORE S, MURCHISON H, SHIOTA T, MICHALEK SM, CURTISS R., 3RD In vitro and in vivo complementation of Streptococcus mutans mutants defective in adherence. Infect Immun. 1983;42:558–66. doi: 10.1128/iai.42.2.558-566.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOESCHE WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–80. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALKE H, STEINER K, MCSHAN WM, FERRETTI JJ. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int J Med Microbiol. 2006;296:259–75. doi: 10.1016/j.ijmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- MANLY RS, RICHARDSON DT. Metabolism of levan by oral samples. J Dent Res. 1968;47:1080–6. doi: 10.1177/00220345680470061301. [DOI] [PubMed] [Google Scholar]
- MONCHOIS V, VIGNON M, RUSSELL RR. Mutagenesis of asp-569 of glucosyltransferase I glucansucrase modulates glucan and oligosaccharide synthesis. Appl Environ Microbiol. 2000;66:1923–7. doi: 10.1128/aem.66.5.1923-1927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONCHOIS V, WILLEMOT RM, MONSAN P. Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol Rev. 1999;23:131–51. doi: 10.1111/j.1574-6976.1999.tb00394.x. [DOI] [PubMed] [Google Scholar]
- MONSAN P, PAUL F, AURIOL D. New developments in the application of enzymes to synthesis reactions. Peptides and oligosaccharides. Ann N Y Acad Sci. 1995;750:357–63. doi: 10.1111/j.1749-6632.1995.tb19980.x. [DOI] [PubMed] [Google Scholar]
- MUNRO C, MICHALEK SM, MACRINA FL. Cariogenicity of Streptococcus mutans V403 glucosyltransferase and fructosyltransferase mutants constructed by allelic exchange. Infect Immun. 1991;59:2316–23. doi: 10.1128/iai.59.7.2316-2323.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PFAFFL MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POSTMA PW, LENGELER JW, JACOBSON GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–94. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALZBERG S, DELCHER AL, FASMAN KH, HENDERSON J. A decision tree system for finding genes in DNA. J Comput Biol. 1998;5:667–80. doi: 10.1089/cmb.1998.5.667. [DOI] [PubMed] [Google Scholar]
- SCHILLING KM, BOWEN WH. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect Immun. 1992;60:284–95. doi: 10.1128/iai.60.1.284-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHROEDER VA, MICHALEK SM, MACRINA FL. Biochemical characterization and evaluation of virulence of a fructosyltransferase-deficient mutant of Streptococcus mutans V403. Infect Immun. 1989;57:3560–9. doi: 10.1128/iai.57.11.3560-3569.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VADEBONCOEUR C, PELLETIER M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev. 1997;19:187–207. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- WALKER GJ. Some properties of a dextranglucosidase isolated from oral streptococci and its use in studies on dextran synthesis. J Dent Res. 1972;51:409–14. doi: 10.1177/00220345720510022901. [DOI] [PubMed] [Google Scholar]
- WALKER GJ. Dextrans. Int. Rev. Biochem. 1978;16:75–126. [Google Scholar]
- WALKER GJ, HARE MD, MORREY-JONES JG. Activity of fructanase in batch cultures of oral streptococci. Carbohydr Res. 1983;113:101–12. doi: 10.1016/0008-6215(83)88222-6. [DOI] [PubMed] [Google Scholar]
- WEBB AJ, HOMER KA, HOSIE AH. Two closely related ABC transporters in Streptococcus mutans are involved in disaccharide and/or oligosaccharide uptake. J Bacteriol. 2008;190:168–78. doi: 10.1128/JB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOOD JM. The amount, distribution and metabolism of soluble polysaccharides in human dental plaque. Arch Oral Biol. 1967;12:849–58. doi: 10.1016/0003-9969(67)90107-0. [DOI] [PubMed] [Google Scholar]
- ZENG L, BURNE RA. Multiple sugar: phosphotransferase system permeases participate in catabolite modification of gene expression in Streptococcus mutans. Mol Microbiol. 2008;70:197–208. doi: 10.1111/j.1365-2958.2008.06403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENG L, BURNE RA. Transcriptional regulation of the cellobiose operon of Streptococcus mutans. J Bacteriol. 2009;191:2153–62. doi: 10.1128/JB.01641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENG L, BURNE RA. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol Microbiol. 2010;75:1145–58. doi: 10.1111/j.1365-2958.2009.07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1 Transcription of the nigB following biofilm growth in glucose, fructose or sucrose. Gene transcription was analyzed by qRT-PCR. The qRT-PCR results were presented as relative mRNA abundance (vertical columns).
Supplemental Fig. 2 Differential transcription of the genes for PTS Enzyme II and ABC transporters following growth of gtfB− and gtfC− mutants in biofilm. Biofilms were grown in diluted CDM supplemented with sucrose for 48 h or for 45 h plus 3 h in glucose (WT-glu). Description of the microarray data presentation is explained in Fig. 1.