Abstract
Mycoplasma leucyl-tRNA synthetases (LeuRSs) have been identified in which the connective polypeptide 1 (CP1) amino acid editing domain that clears mischarged tRNAs are missing (Mycoplasma mobile) or highly degenerate (Mycoplasma synoviae). Thus, these enzymes rely on a clearance pathway called pretransfer editing, which hydrolyzes misactivated aminoacyl-adenylate intermediate via a nebulous mechanism that has been controversial for decades. Even as the sole fidelity pathway for clearing amino acid selection errors in the pathogenic M. mobile, pretransfer editing is not robust enough to completely block mischarging of tRNALeu, resulting in codon ambiguity and statistical proteins. A high-resolution X-ray crystal structure shows that M. mobile LeuRS structurally overlaps with other LeuRS cores. However, when CP1 domains from different aminoacyl-tRNA synthetases and origins were fused to this common LeuRS core, surprisingly, pretransfer editing was enhanced. It is hypothesized that the CP1 domain evolved as a molecular rheostat to balance multiple functions. These include distal control of specificity and enzyme activity in the ancient canonical core, as well as providing a separate hydrolytic active site for clearing mischarged tRNA.
Keywords: protein evolution, quality control, statistical proteins, aminoacylation, translation
Aminoacyl-tRNA synthetases (aaRSs) catalyze the first step of protein synthesis. Each aaRS covalently attaches amino acid to its cognate tRNA to set the genetic code (1, 2). Faithful translation in modern cells imposes a great challenge for aaRSs as they select cognate substrate amino acids from a pool of structurally similar amino acids (1, 2). As such, about half of the 20 aaRSs have developed editing mechanisms to clear mischarged tRNA products before the noncognate amino acids can be incorporated into newly synthesized proteins at the ribosome.
Robust editing to achieve proteome fidelity has been commonly thought to be essential to the cell (1, 2). Indeed, fidelity can be critical for microbe and mammalian cell viability (3–5), as well as the prevention of neurological disease (5). However, in host-dependent pathogens, we and others have identified aaRS-dependent mechanisms that foster translational infidelity resulting in statistical proteins (6, 7). We hypothesize that some organisms have adapted to promote or tolerate threshold levels of statistical mistranslations. In these cases, multiple aaRS mechanisms that influence fidelity of the aminoacylation reaction can be affected through the evolution process.
All aaRS aminoacylation reactions (Eqs. 1 and 2) proceed in two steps with aminoacyl-adenylate as the intermediate:
Editing reactions have evolved to clear mistakes at both steps of the aaRS catalyzed reaction (8). In the posttransfer editing pathway (Eq. 3), mischarged tRNA is hydrolyzed in an aaRS domain that is distinct from the synthetic aminoacylation domain (9–16). In addition, editing of mischarged tRNA can occur in trans by independent proteins that function as tRNA-specific deacylases (17, 18):
The origin of pretransfer editing (Eq. 4) that hydrolyzes the adenylate intermediate has been much more controversial:
The editing domain has been proposed to be critical to pretransfer editing (19), in addition to its contribution to the posttransfer editing pathway. Both tRNA-dependent (20–23) and tRNA-independent (22, 24) pretransfer editing activities have been reported. Also, clearance of the adenylate by selective ejection from the enzyme’s active site into the aqueous environment has been suggested (24). It is possible that these diverse fidelity mechanisms could be used selectively or combinatorially by different aaRSs to clear noncognate products (8, 25).
In leucyl- (LeuRS), isoleucyl- (IleRS), and valyl-tRNA synthetases (ValRS), the hydrolytic active sites of posttransfer editing have been mapped to their homologous connective polypeptide 1 (CP1) domains (9, 26). The CP1 domain is inserted into the Rossmann fold that comprises the synthetic aminoacylation site via two β-strands (27). In the tRNA-bound editing complex, the LeuRS CP1 domain is extended away from the canonical aminoacylation core (28). However, in the aminoacylation complex that was recently solved for Escherichia coli LeuRS, the CP1 descends upon the aminoacylation active site to interact directly and indirectly with the 3′ end of the tRNA (29).
In LeuRS from the Mycoplasma mobile pathogen, this otherwise universally conserved CP1 domain is completely missing (6). Hence, M. mobile LeuRS produces tRNA mischarged with noncognate structurally related aliphatic amino acids, resulting in statistical proteins (6). Related LeuRSs from Mycoplasma synoviae and Mycoplasma agalactiae, which are poultry and sheep pathogens, respectively, have CP1 domains that are highly degenerate and lack any signature sequences that comprise the conventional hydrolytic active site (6) (Fig. S1).
Herein, we present a high-resolution X-ray crystal structure demonstrating that M. mobile LeuRS overlaps with the canonical structures of other LeuRSs. Using this M. mobile LeuRS core as a scaffold, we created a series of LeuRS hybrids with CP1 domains from different origins as well as from IleRS and ValRS (30). Remarkably, we determined that the addition of the CP1 domain enhances pretransfer editing activity and fidelity for each of these diverse hybrid models. This contrasts with deletion of CP1 editing domains from E. coli and Saccharomyces cerevisiae mitochondria LeuRSs, which activated a latent tRNA-dependent pretransfer editing activity that suppresses tRNA mischarging (21). We hypothesize, then, that the CP1 domain has evolved to function as a molecular rheostat to enhance fidelity by idiosyncratically potentiating substrate specificity (30) while balancing hydrolytic mechanisms that are associated with either the aminoacylation core or the separate editing domain.
Results
M. synoviae LeuRS Naturally Produces Mischarged tRNALeu.
Hydrolytic CP1 editing domains are ubiquitously conserved in LeuRS, IleRS, and ValRS across all three domains of life (27). Previously, we identified three unique cases of Mycoplasma LeuRSs that had altered or missing CP1 domains (6). The M. mobile CP1 domain is completely absent, resulting in statistical substitutions in the proteome. M. synoviae and M. agalactiae contained highly degenerated CP1 domains that we hypothesized also lacked posttransfer editing (Fig. S1).
To test the mischarging activity of M. synoviae LeuRS, the gene was synthesized using optimized codon use frequencies for expression in E. coli and also to convert triplet TGA stop codons (which are used to encode tryptophan in M. synoviae) to TGG. The monomeric LeuRS was purified by affinity chromatography via an N-terminal six-histidine tag. The enzyme robustly aminoacylated in vitro transcribed M. mobile tRNALeu (Fig. 1A). As we predicted, M. synoviae LeuRS displayed significant mischarging activity (Fig. 1B), similar to M. mobile LeuRS, which is missing its CP1 domain (6). Likewise, the degenerated CP1 domain failed to deacylate mischarged Ile-tRNALeu (Fig. 1C). We hypothesize that M. synoviae LeuRS represents an intermediate evolutionary step on a path to the elimination of the CP1 domain as found in M. mobile LeuRS.
Fig. 1.
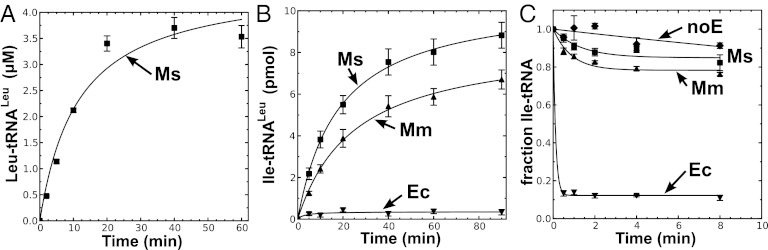
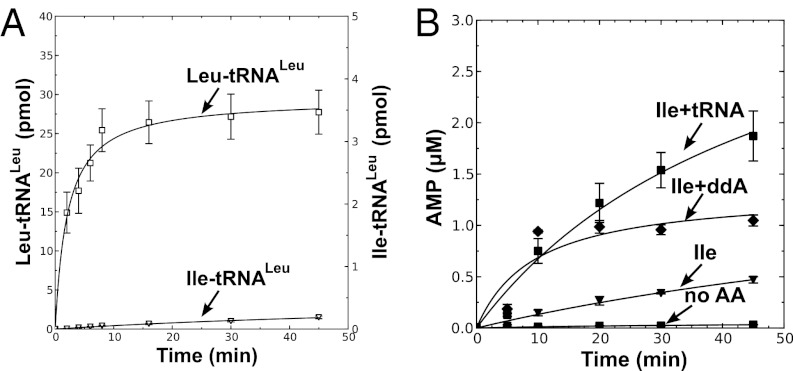
M. synoviae LeuRS mischarges tRNALeu. (A) Leucylation by M. synoviae LeuRS was carried out by using 21 µM [14C]leucine (50 μCi/mL), 4 µM tRNALeu, and 100 nM enzyme. (B) Isoleucine mischarging by M. synoviae LeuRS incorporated 40 µM [3H]isoleucine (166 μCi/mL), 4 µM tRNALeu, and 1 µM enzyme. M. mobile LeuRS was used as a positive control. (C) Deacylation reactions for M. synoviae LeuRS contained ∼6.5 µM [3H]Ile-tRNALeu and 100 nM enzyme. E. coli LeuRS was used as the positive control. Abbreviations are as follows: ■, M. synoviae LeuRS (Ms); ▲, M. mobile LeuRS (Mm); ▼, E. coli LeuRS (Ec); and ◆, no enzyme control (noE). Error bars represented SDs calculated from triplicated reactions.
Mycoplasma LeuRSs Maintain Weak Pretransfer Editing Activities.
E. coli LeuRS harbors a tRNA-dependent pretransfer editing activity that is unmasked when its CP1 domain is deleted (21). We also found that we could activate pretransfer editing in E. coli LeuRS by incorporating an unchargeable 2′-deoxyadenosine tRNALeu (2′dA-tRNA) (21). Thus, we hypothesized that the pathway to amino acid editing can shift between pre- and posttransfer editing mechanisms that are inherent to the enzyme (8, 25). Even though M. mobile has statistical substitutions at leucine codons, we sought to determine whether the pretransfer editing pathway was activated in this naturally occurring protein where the CP1 domain is eliminated. Likewise, we have also tested pretransfer editing in M. synoviae LeuRS where the CP1 domain was present but highly degenerate and yields mischarged tRNAs.
We incorporated unchargeable tRNA analogs that contained a 2′dA-tRNA or dideoxyadenosine-tRNA (ddA-tRNA) at the 3′ acceptor stem end into amino acid-dependent ATP hydrolysis assays. Because 2′dA-tRNA and ddA-tRNA are missing the functional 2′ hydroxyl group that is necessary for aminoacylation to occur, AMP formation in these reactions would reflect cycles of amino acid activation and hydrolysis that are representative of pretransfer editing (31, 32).
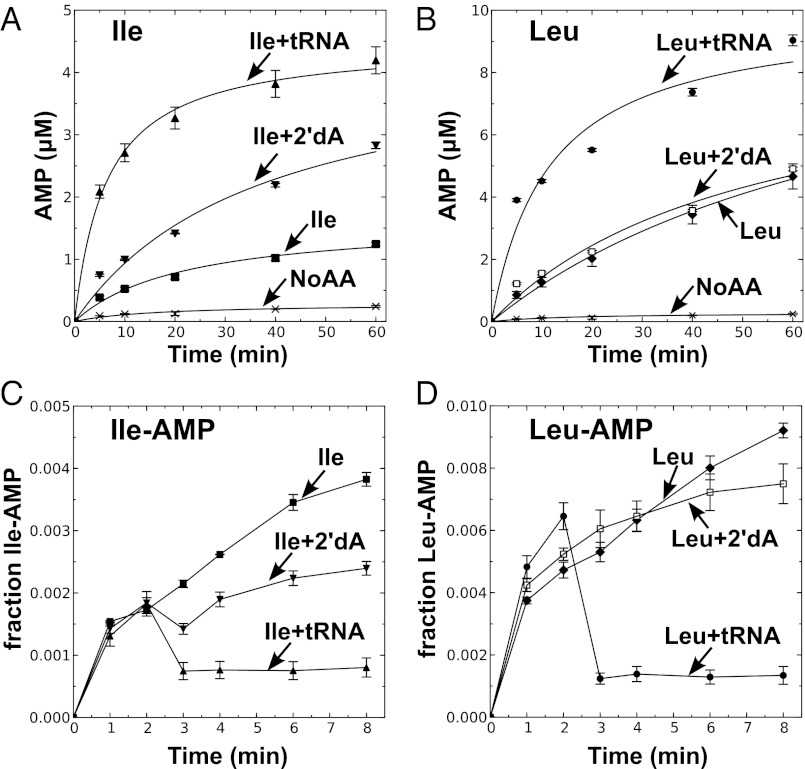
The 2′dA-tRNA clearly stimulated AMP production by M. mobile LeuRS (Fig. 2A) and M. synoviae LeuRS (Fig. S2A) in the presence of isoleucine, but not the cognate leucine substrate (Fig. 2B and Fig. S2B). We also determined that this pretransfer editing activity cleared isoleucyl-adenylate when 2′dA-tRNA or ddA-tRNA was added (Fig. 2C and Fig. S2C). In comparison, leucyl-adenylate was stable when Mycoplasma LeuRS and a modified tRNA was present (Fig. 2D and Fig. S2D). Thus, the synthetic core of Mycoplasma LeuRS harbors a bona fide tRNA-dependent pretransfer editing activity. We hypothesized that Mycoplasma relies on this pretransfer editing activity to maintain a threshold level of fidelity that is required by this pathogen, albeit it is not sufficient to fully suppress mischarging and protect the proteome from statistical substitutions.
Fig. 2.
M. mobile LeuRS exhibits pretransfer editing activity. Reaction mixtures contained 10 μM M. mobile tRNALeu or a tRNA analog with 2′-deoxyadenosine substituted for the A76 nucleotide (2′dA-tRNA) and 1 μM M. mobile LeuRS. Amino acid-dependent AMP formation is measured by TLC of reaction aliquots from ATPase reactions with 5 mM isoleucine (A) or 5 mM leucine (B). Aminoacyl-adenylate was measured with 5 mM isoleucine (C) or 5 mM leucine (D). Fractions indicated by the y axis represent the intensity of the spot representing [32P]adenylate divided by the total intensity of 32P in the lane. Abbreviations are as follows: ■, isoleucine only (Ile); ▲, isoleucine and tRNA (Ile+tRNA); ▼, isoleucine and 2′dA-tRNA (Ile+2'dA); ◆, leucine only (Leu); ●, leucine and tRNA (Leu+tRNA); □, leucine and 2′dA-tRNA (Leu+2'dA); and ×, no amino acid control (No AA). Error bars represent SDs derived from triplicated reactions.
X-Ray Crystal Structure of M. mobile LeuRS-LeuAMS Is Similar to Other LeuRSs.
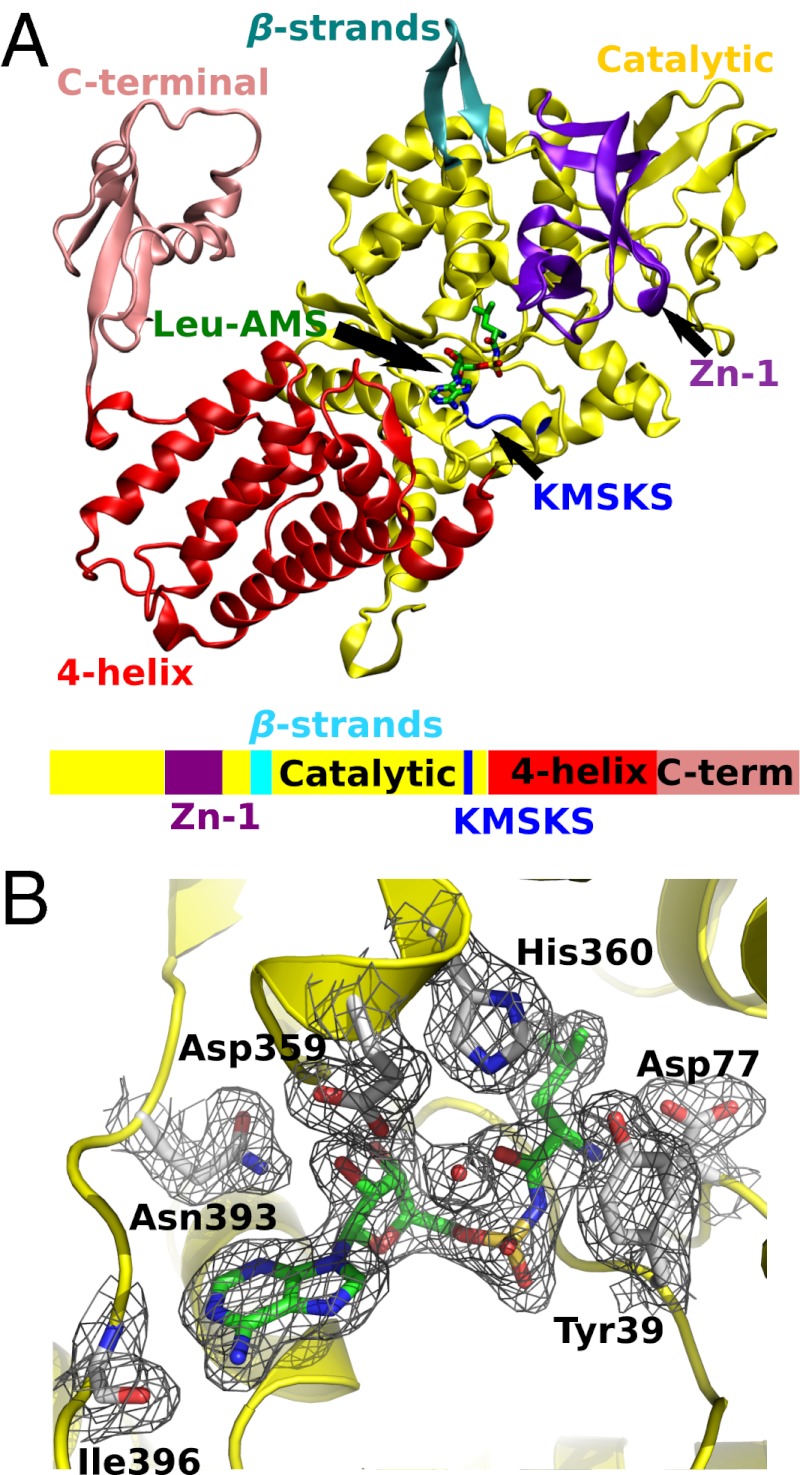
M. mobile LeuRS was cocrystallized as a dimer in two conformations with a leucyl-sulfamoyl-adenylate analog (Leu-AMS) of leucyl adenylate in the space group R32 and diffracted to 2.07 Å. One of the conformers (Fig. 3A) had weak density representing the mobile C-terminal domain, which was absent in the other conformer. Although tRNA was present in the crystallization medium, neither of the LeuRS conformers contained bound tRNA.
Fig. 3.
X-ray crystal structure of M. mobile LeuRS Leu-AMS complex. (A) A ribbon diagram of the protomer with electron density of C-terminal domain is colored as follows: catalytic domain, yellow; four-helix bundle domain, red; C-terminal domain, pink; Zn binding domain, purple; KMSKS loop, blue; two linking β-strands, cyan. The Leu-AMP analog, Leu-AMS, is shown in stick model. A color-coordinated cartoon of the primary sequence is shown below the structure. (B) The 2Fo-Fc electron density map of the aminoacylation active site is contoured at 1.0 σ (black mesh). The Leu-AMS is highlighted in green, and interacting amino acid residues are labeled and shown in gray.
The overall structure of the M. mobile LeuRS enzyme is similar to structures determined for Thermus thermophilus LeuRS (27) (TtLeuRS, PDB ID code 1H3N) and E. coli LeuRS (29) (EcLeuRS, PDB ID code 4AQ7), despite that the native M. mobile LeuRS is missing CP1 and leucine-specific domains (6) (Fig. S3). This supports the modular nature of aaRS expansion and provides a glimpse of the aminoacylation core in its ancient state. In T. thermophilus and E. coli LeuRSs, two β-strands link the core of the enzyme to the CP1 domain that is responsible for posttransfer editing. Significantly, although M. mobile LeuRS is missing its CP1 domain, the two β-strands (residue Gly226 to Lys237), now connected by a dipeptide (Asp-Gly), are retained and structurally conserved. These overlap with the N-terminal β-strand Arg226 to Gly229 and C-terminal β-strand Lys412 to Tyr415 from E. coli LeuRS. The structure model for the C-terminal domain is based on weak, but unambiguous, electron density present in difference Fourier maps. The C-terminal domain is linked to the core of the enzyme by a flexible tether (11) and interacts with the corner of the L-shaped tRNA. Thus, stabilization of the position of this mobile domain in either the editing or aminoacylation complex requires tRNA binding (29).
The aminoacylation core of the protein is intact, and the adenylate analog Leu-AMS is bound to the enzyme active site (Fig. 3B). M. mobile LeuRS and T. thermophilus LeuRS (27) rely upon similar sets of amino acid residues to bind Leu-AMS in their active sites (Fig. S3A), suggesting that the adenylate is stabilized by conserved interactions within the canonical LeuRS core. The flexible KMSKS loop responsible for ATP binding partially covers the active site entrance in a manner similar to the semiopen conformation of this signature sequence in E. coli and T. thermophilus LeuRSs (27, 29). Compared with the E. coli LeuRS aminoacylation complex (Fig. S3C), a conserved tyrosine residue undergoes significant conformational change upon tRNA binding. The phenyl ring of Tyr39 in M. mobile LeuRS adopts a conformation similar to that of Tyr43 in T. thermophilus LeuRS (27) (Fig. S3B) to protect the adenylate from nonproductive hydrolysis. In contrast, in the E. coli LeuRS aminoacylation complex, the side chain of the homologous tyrosine is flipped outward by about 120° to accommodate binding of the terminal tRNA A76 (29) (Fig. S3D).
CP1 Insertions Stimulate Pretransfer Editing in M. mobile LeuRS.
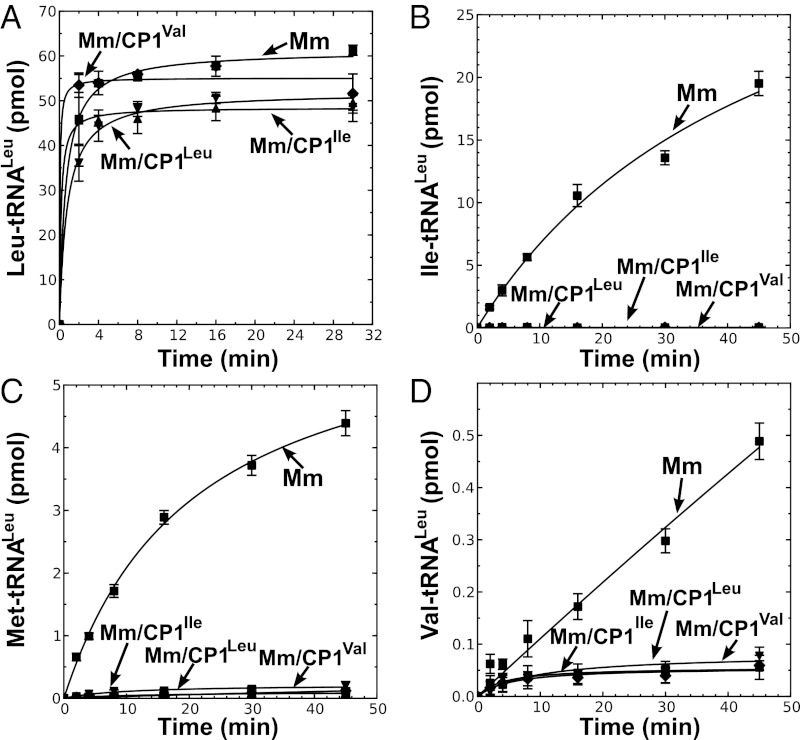
It has been proposed that a primitive translation machinery lacked modern specificity and fidelity mechanisms, resulting in statistical protein synthesis (33). As fidelity requirements increased during evolution, the canonical aminoacylation core was fused to a CP1 domain to accommodate more stringent amino acid specificity requirements. Despite its unusual lack of a CP1 domain, the solved crystal structure of M. mobile LeuRS supports that it has retained a canonical aminoacylation core as well as faithful interactions with the adenylate intermediate that mimics a typical LeuRS. Thus, we hypothesized that we can use M. mobile LeuRS as a scaffold to reconstruct evolutionary events that adapted the aaRS for increased accuracy. As such, the CP1 domain from E. coli LeuRS was fused to M. mobile LeuRS (MmLeuRS/CP1Leu) (30). We also constructed hybrids using the CP1 domains from IleRS (MmLeuRS/CP1Ile) and ValRS (MmLeuRS/CP1Val) (30). Each of the hybrid LeuRSs displayed leucylation activities comparable to the wild-type M. mobile enzyme (30) (Fig. 4A), supporting that the LeuRS core domain in these hybrid proteins retained its structural integrity. Significantly and regardless of their specificities, these inserted CP1 domains suppressed LeuRS mischarging of isoleucine (Fig. 4B), methionine (Fig. 4C), and valine (Fig. 4D).
Fig. 4.
Aminoacylation and misaminoacylation activities of M. mobile LeuRSs that contain hybrid CP1 domains. Aminoacylation of 4 μM tRNALeu in presence of (A) 21 μM [3H]leucine (318 μCi/mL), (B) 21 μM [3H]isoleucine (166 μCi/mL), (C) 20.2 μM [35S]methionine (115 μCi/mL), and (D) 20 μM [14C]valine (259 μCi/mL). Symbols used are as follows: ■, MmLeuRS (Mm); ▲, MmLeuRS/CP1Leu (Mm/CP1Leu); ▼, MmLeuRS/CP1Ile (Mm/CP1Ile); and ◆, MmLeuRS/CP1Val (Mm/CP1Val). Error bars represent SDs from triplicated reactions.
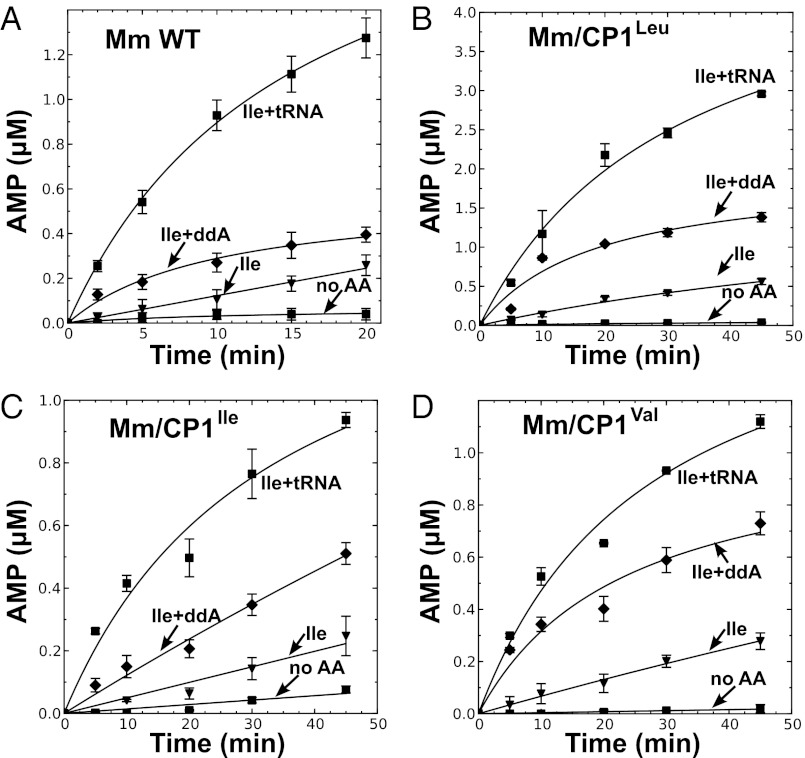
We asked whether the addition of these CP1 domains stimulated tRNA-dependent pretransfer editing of the M. mobile LeuRS active site. We performed amino acid-dependent ATP hydrolysis assays with M. mobile ddA-tRNALeu. Compared with wild-type M. mobile LeuRS (Fig. 5A), all three CP1 fusion enzymes displayed increased tRNA-dependent AMP production in the presence of isoleucine (Fig. 5 B–D). The cognate leucine substrate failed to increase tRNA-dependent AMP formation activity (Fig. S4 A–D). Significantly, although the amplitude of the stimulation of pretransfer editing varied depending on the hybrid, it occurred regardless of the origin and specificities of the fused CP1 domains.
Fig. 5.
CP1 addition to hybrid M. mobile LeuRS enhances pretransfer editing. Reaction mixture contained 1 μM enzyme, 10 μM tRNALeu, or tRNALeu with A76 replaced by dideoxyadenosine (ddA), 18.1 μM [α-32P]ATP (40 μCi/mL), and 2.5 mM isoleucine. The AMP formation activities are measured for Mm LeuRS (A), MmLeuRS/CP1Leu (Mm/ CP1Leu) (B), MmLeuRS/CP1Ile (Mm/CP1Ile) (C), and MmLeuRS/CP1Val (Mm/CP1Val) (D). Symbols used are as follows: ◆, reaction without amino acid present (no AA); ▲, isoleucine (Ile); ▼, isoleucine with tRNA (Ile+tRNA); and ■, isoleucine with ddtRNA (Ile+ddA). Error bars represent the SD values based on three separate experiments.
Degenerate M. synoviae CP1 Domain Confers Fidelity to M. mobile LeuRS via Enhanced Pretransfer Editing Activity.
Because M. synoviae LeuRS produces mischarged tRNAs (Fig. 1B) similar to M. mobile LeuRS, we asked whether insertion of an editing-defective degenerate CP1 domain would also influence pretransfer editing activity of M. mobile LeuRS. We generated a hybrid M. mobile LeuRS that contained the degenerated CP1 domain from M. synoviae LeuRS (MmLeuRS/MsLeu). This fusion enzyme robustly aminoacylated in vitro transcribed M. mobile tRNALeu (Fig. 6A). Surprisingly, the editing-defective M. synoviae CP1 domain also suppressed mischarging (Fig. 6A).
Fig. 6.
M. synoviae LeuRS CP1 addition to M. mobile LeuRS suppresses mischarging via enhanced pretransfer editing activity. (A) Aminoacylation and misaminoacylation activity of Mm/Ms LeuRS was carried out with 4 μM tRNALeu in the presence of 21 μM [3H]leucine (318 μCi/mL) with 1 μM enzyme or 21 μM [3H]isoleucine (97 μCi/mL) with 1 μM enzyme. Symbols used are as follows: □, Leu-tRNALeu and △, Ile-tRNALeu. (B) The AMP formation activity was measured for Mm/Ms LeuRS using 1 μM enzyme, 10 μM tRNALeu, or tRNALeu with A76 replaced by dideoxyadenosine (ddA-tRNA), 18.1 μM [α-32P]ATP, and 2.5 mM isoleucine. Symbols used are as follows: ◆, reaction without amino acid present (no AA); ▲, isoleucine (Ile); ▼, isoleucine with tRNA (Ile+tRNA); and ■, isoleucine with ddA-tRNA (Ile+ddA). Error bars represent the SD values based on three separate experiments.
We asked whether this increased fidelity could be attributed to increases in amino acid discrimination in the aminoacylation site (30) or enhanced pretransfer editing activity, or a combination of both mechanisms. Whereas the kcat was similar for leucine (11 ± 2 s−1) and isoleucine (9 ± 2 s−1), PPi exchange assays showed that the Km was, respectively, 0.03 ± 0.01 mM and 2.7 ± 0.7 mM, resulting in a discrimination factor of just 1/111. Thus, unlike fusion of the E. coli LeuRS and IleRS CP1 domains that enhanced amino acid discrimination of the M. mobile LeuRS (30), the MmLeuRS/MsLeu mimics fusion of the ValRS CP1 domain. As such, the chimeric Mycoplasma LeuRS exhibited an enhanced pretransfer editing activity (Fig. 6B) that cleared misactivated isoleucine, but not cognate leucine (Fig. S4E). Because pretransfer editing has been associated with the canonical core of LeuRS (21), we hypothesize that the linkage or connectivity between the aminoacylation active site and CP1 domain is more important to adenylate clearance as opposed to the nature of the independent CP1 domain.
Discussion
Establishing and maintaining the genetic code requires faithful translation of the codon message to its cognate amino acid (34). In modern cells, this is facilitated by exquisite quality-control mechanisms that ensure error correction when necessary for misaminoacylation. Primitive translation machineries that coevolved in a chaotic environment with an emerging genetic code were likely error-prone and produced statistical proteins (33). Indeed, urzymes, a rationally designed minimal construct of a class I aaRS active site that retains catalytic activity, exhibit decreased specificity for cognate amino acids (35, 36). This supports that evolutionary addition of auxiliary domains enhanced substrate specificity.
Sequence- and structure-based phylogenetic analyses demonstrated that aaRSs originate from their canonical aminoacylation core (37, 38). Thus, these early translation mechanisms would have been dependent on aaRS progenitors composed simply of their core domains to aminoacylate tRNA. In the case of the class I aaRSs, the core is a Rossmann ATP binding fold that is distributed widely throughout many protein families. The class II aaRS core fold is much more rare (39–41). As the cell and its genetic code became more sophisticated, domains responsible for clearing errors were added in both aaRS classes to provide the necessary quality control for translation.
LeuRS, IleRS, and ValRS rely on a conserved CP1 tRNA deacylase domain to clear mischarged tRNA (9, 13) before they are released to elongation factor-Tu for delivery to the ribosome. Previously, we showed that we can remove the CP1 domain from yeast mitochondrial and E. coli LeuRS and maintain complete fidelity (21). In these two LeuRS cases, the CP1 domain acted as a rheostat to dampen pretransfer editing. Deletion of the CP1 domain unmasked a tRNA-dependent pretransfer editing mechanism for LeuRS that was associated with the canonical core of the enzyme. Pretransfer editing clears the misactivated adenylate before the noncognate amino acid can be transferred to the tRNA. Because of its association with the ancient aminoacylation core, we hypothesize, then, that pretransfer editing might have served as an ancient quality-control mechanism.
Herein, we demonstrated that two Mycoplasma LeuRSs also harbor pretransfer editing activity. In these two natural examples, however, the pretransfer editing activity is insufficient to protect mischarging and results in errors that are translated during protein synthesis. Remarkably, addition of the CP1 domain—regardless of its origin from a LeuRS, IleRS, or ValRS—again served as a molecular rheostat, but to increase pretransfer editing activity.
It is clear that Mycoplasma capitalized on the rheostat capabilities of the LeuRS CP1 domain to influence translational fidelity. It is possible that producing statistical proteins is strategically embraced by these Mycoplasma pathogens to evade host immune systems (6). This inherent pretransfer editing activity in the canonical core also supports that a primitive LeuRS (before CP1 domain insertion) likely contained this as a rudimentary fidelity mechanism that was sufficient to provide a threshold level of fidelity for an early cell as well as enough charged tRNALeu to allow an ancient organism to prosper.
The homologous CP1 domains have adapted for different specificities to clear noncognate amino acid, which idiosyncratically challenge the fidelity of LeuRS, IleRS, and ValRS. In LeuRS and ValRS, the CP1 domains have evolved to efficiently execute posttransfer editing. The balance of fidelity mechanisms that rely on post- or pretransfer editing can differ and shift (8, 25). A preponderance of posttransfer editing versus pretransfer editing can be dictated by a rapid aminoacyl-transfer step in the aminoacylation site (22, 23, 41). Likewise, the CP1 domain in IleRS relies upon pretransfer editing as its dominant editing pathway (42) because of a slow transfer rate (22).
Our recent work revealed that fusion of the CP1 domains to M. mobile LeuRS can improve amino acid selectivity for aminoacylation (30). Here, we showed that fusion of CP1 domains from different specificities and origins can stimulate pretransfer editing activity in M. mobile LeuRS, albeit at different levels. Regardless, this enhanced pretransfer editing suppresses mischarging independently (MmLeuRS/CP1Val and MmLeuRS/MsLeu) or synergistically with an enhancement of selectivity in the aminoacylation site (MmLeuRS/CP1Leu and MmLeuRS/CP1Ile) (30). It is possible that the CP1 insertion favored an active site conformation more suitable for pretransfer editing and amino acid selection. In many cases, this activity seems to be enhanced in the presence of tRNA (20, 21, 23). Thus, in the evolution of LeuRS, IleRS, and ValRS, the original insertion of the CP1 domain likely affected fidelity via a set of mechanisms. Indeed, we hypothesize that the CP1 domain addition used a rheostat-like function as a selective advantage to potentiate amino acid selection and pretransfer editing activity in the aminoacylation core, while introducing a posttransfer editing hydrolytic active site.
Materials and Methods
Crystallization and X-Ray Diffraction.
M. mobile LeuRS was crystallized in 0.1 M bis-Tris (pH 5.5), 0.6 M ammonium acetate, and 20% (vol/vol) PEG3350 at 4 °C, via hanging drop vapor diffusion. Drops combined 1 μL of mother liquor and 1 μL of protein solution at 10 mg/mL LeuRS mixed with 2 mM Leu-AMS and 40 μM M. mobile tRNALeu. Crystals diffracting to 2.07 Å were grown for 3 mo. The crystals were soaked for 10 s in mother liquor supplemented with 15% (vol/vol) glycerol, mounted on CrystalCap HT cryoloops (Hampton Research), and vitrified in liquid nitrogen.
Diffraction data were collected at an insertion device beam line (21-ID-F) using Mar 225 CCD detector (LS-CAT sector; Advanced Photon Source). Data were integrated and scaled using the HKL-2000 (43) package. Molecular replacement was carried out using E. coli LeuRS (PDB ID code 4AQ7) as a model with Phaser (44). Manual model rebuilding was carried out as described in SI Materials and Methods. Data collection and refinement statistics are provided in Table S1.
Enzyme Assays.
ATPase assays containing 50 mM Tris (pH 7.5), 10 mM MgCl2, 5 mM DTT, 10 μM M. mobile tRNALeu or unchargeable analog, 18 μM radiolabeled ATP, and 1 μM enzyme were initiated with 2.5 mM amino acid. Reaction aliquots (2 μL) were quenched by spotting on PEI-cellulose TLC plates. The plates were developed in 750 mM KH2PO4 (pH 3.5) and analyzed by phosphorimaging.
We adapted a method (45) to isolate adenylate. To accumulate adenylate, the ATPase assay reaction was allowed to react for 2 min, after which 10 μM of M. mobile tRNALeu or its unchargeable analog was added to the reaction buffer. Aliquots of 1.5 μL were quenched in 3 μL of 0.1% SDS and 50 mM sodium acetate (pH 5.0) and spotted on PEI-cellulose TLC plates, which were developed in 0.1 M ammonium acetate and 5% acetic acid at 25 °C and analyzed by phosphorimaging.
Supplementary Material
Acknowledgments
We thank Drs. Satish Nair and Raven Huang for providing advice on our X-ray crystallography experiments and Prof. Nair for useful comments on our manuscript. This work was supported by National Institutes of Health Grants GM063789 and P41RR05964 and National Science Foundation Grant MCB-0843611. The Advanced Photon Source is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Use of the Life Sciences Collaborative Access Team Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor, which supported this research program with Grant 085P1000817.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3ZIU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218374110/-/DCSupplemental.
References
- 1.Mascarenhas AP, Martinis SA, An S, Rosen AE, Musier-Forsyth K. 2008. Fidelity mechanisms of the aminoacyl-tRNA synthetases. Protein Engineering, eds RajBhandary UL, Koehrer C (Springer, Berlin), pp 153–200.
- 2.Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286(5446):1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 3.Karkhanis VA, Boniecki MT, Poruri K, Martinis SA. A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J Biol Chem. 2006;281(44):33217–33225. doi: 10.1074/jbc.M607406200. [DOI] [PubMed] [Google Scholar]
- 4.Karkhanis VA, Mascarenhas AP, Martinis SA. Amino acid toxicities of Escherichia coli that are prevented by leucyl-tRNA synthetase amino acid editing. J Bacteriol. 2007;189(23):8765–8768. doi: 10.1128/JB.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 6.Li L, et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA. 2011;108(23):9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocha R, Pereira PJB, Santos MAS, Macedo-Ribeiro S. Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen. Proc Natl Acad Sci USA. 2011;108(34):14091–14096. doi: 10.1073/pnas.1102835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinis SA, Boniecki MT. The balance between pre- and post-transfer editing in tRNA synthetases. FEBS Lett. 2010;584(2):455–459. doi: 10.1016/j.febslet.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin L, Hale SP, Schimmel P. Aminoacylation error correction. Nature. 1996;384(6604):33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 10.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23(23):4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu JL, Rho SB, Vannella KM, Martinis SA. Functional divergence of a unique C-terminal domain of leucyl-tRNA synthetase to accommodate its splicing and aminoacylation roles. J Biol Chem. 2006;281(32):23075–23082. doi: 10.1074/jbc.M601606200. [DOI] [PubMed] [Google Scholar]
- 12.Dock-Bregeon AC, et al. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol Cell. 2004;16(3):375–386. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Nureki O, et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280(5363):578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 14.Fukai S, et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103(5):793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 15.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol. 2005;12(10):915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 16.Mursinna RS, Martinis SA. Rational design to block amino acid editing of a tRNA synthetase. J Am Chem Soc. 2002;124(25):7286–7287. doi: 10.1021/ja025879s. [DOI] [PubMed] [Google Scholar]
- 17.An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279(41):42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 18.Ruan B, Söll D. The bacterial YbaK protein is a Cys-tRNAPro and Cys-tRNA Cys deacylase. J Biol Chem. 2005;280(27):25887–25891. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson TL, et al. Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol Cell. 2002;9(2):353–362. doi: 10.1016/s1097-2765(02)00449-5. [DOI] [PubMed] [Google Scholar]
- 20.Bishop AC, Beebe K, Schimmel PR. Interstice mutations that block site-to-site translocation of a misactivated amino acid bound to a class I tRNA synthetase. Proc Natl Acad Sci USA. 2003;100(2):490–494. doi: 10.1073/pnas.0237335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boniecki MT, Vu MT, Betha AK, Martinis SA. CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase. Proc Natl Acad Sci USA. 2008;105(49):19223–19228. doi: 10.1073/pnas.0809336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulic M, Cvetesic N, Perona JJ, Gruić-Sovulj I. Partitioning of tRNA-dependent editing between pre- and post-transfer pathways in class I aminoacyl-tRNA synthetases. J Biol Chem. 2010;285(31):23799–23809. doi: 10.1074/jbc.M110.133553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cvetesic N, Perona JJ, Gruic-Sovulj I. Kinetic partitioning between synthetic and editing pathways in class I aminoacyl-tRNA synthetases occurs at both pre-transfer and post-transfer hydrolytic steps. J Biol Chem. 2012;287(30):25381–25394. doi: 10.1074/jbc.M112.372151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hati S, et al. Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in “selective release” of noncognate amino acids. J Biol Chem. 2006;281(38):27862–27872. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar J, Martinis SA. Amino-acid-dependent shift in tRNA synthetase editing mechanisms. J Am Chem Soc. 2011;133(46):18510–18513. doi: 10.1021/ja2048122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lincecum TL, Jr, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11(4):951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 27.Cusack S, Yaremchuk A, Tukalo M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19(10):2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat Struct Mol Biol. 2005;12(10):923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 29.Palencia A, et al. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat Struct Mol Biol. 2012;19(7):677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boniecki MT, Martinis SA. Coordination of tRNA synthetase active sites for chemical fidelity. J Biol Chem. 2012;287(14):11285–11289. doi: 10.1074/jbc.C111.325795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Splan KE, Musier-Forsyth K, Boniecki MT, Martinis SA. In vitro assays for the determination of aminoacyl-tRNA synthetase editing activity. Methods. 2008;44(2):119–128. doi: 10.1016/j.ymeth.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francklyn CS, First EA, Perona JJ, Hou YM. Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods. 2008;44(2):100–118. doi: 10.1016/j.ymeth.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese CR. On the evolution of cells. Proc Natl Acad Sci USA. 2002;99(13):8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaher HS, Green R. Fidelity at the molecular level: Lessons from protein synthesis. Cell. 2009;136(4):746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham Y, et al. Tryptophanyl-tRNA synthetase Urzyme: a model to recapitulate molecular evolution and investigate intramolecular complementation. J Biol Chem. 2010;285(49):38590–38601. doi: 10.1074/jbc.M110.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pham Y, et al. A minimal TrpRS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases. Mol Cell. 2007;25(6):851–862. doi: 10.1016/j.molcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 37.O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol Mol Biol Rev. 2003;67(4):550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribas de Pouplana L, Schimmel P. Aminoacyl-tRNA synthetases: Potential markers of genetic code development. Trends Biochem Sci. 2001;26(10):591–596. doi: 10.1016/s0968-0004(01)01932-6. [DOI] [PubMed] [Google Scholar]
- 39.Cusack S, Berthet-Colominas C, Härtlein M, Nassar N, Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature. 1990;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 40.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 41.Guth EC, Francklyn CS. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol Cell. 2007;25(4):531–542. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fersht AR. Editing mechanisms in protein synthesis: Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry. 1977;16(5):1025–1030. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gruić-Sovulj I, Uter N, Bullock T, Perona JJ. tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase. J Biol Chem. 2005;280(25):23978–23986. doi: 10.1074/jbc.M414260200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.