Abstract
trans-acting small interfering RNAs (tasiRNAs) are plant-specific endogenous siRNAs produced via a unique pathway whose first step is the microRNA (miRNA)-programmed RNA-induced silencing complex (RISC)–mediated cleavage of tasiRNA gene (TAS) transcripts. One of the products is subsequently transformed into tasiRNAs by a pathway that requires several factors including SUPPRESSOR OF GENE SILENCING3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE6. Here, using in vitro assembled ARGONAUTE (AGO)1–RISCs, we show that SGS3 is recruited onto RISCs only when they bind target RNA. Following cleavage by miRNA173 (miR173)-programmed RISC, SGS3 was found in complexes containing cleaved TAS2 RNA and RISC. The 3′ cleavage fragment (the source of tasiRNAs) was protected from degradation in this complex. Depletion of SGS3 did not affect TAS2 RNA cleavage by miR173-programmed RISC, but did affect the stability of the 3′ cleavage fragment. When the 3′ nucleotide of 22-nt miR173 was deleted or the corresponding nucleotide in TAS2 RNA was mutated, the complex was not observed and the 3′ cleavage fragment was degraded. Importantly, these changes in miR173 or TAS2 RNA are known to lead to a loss of tasiRNA production in vivo. These results suggest that (i) SGS3 associates with AGO1–RISC via the double-stranded RNA formed by the 3′-terminal nucleotides of 22-nt miR173 and corresponding target RNA, which probably protrudes from the AGO1–RISC molecular surface, (ii) SGS3 protects the 3′ cleavage fragment of TAS2 RNA from degradation, and (iii) the observed SGS3-dependent stabilization of the 3′ fragment of TAS2 RNA is key to tasiRNA production.
Keywords: RNA silencing, secondary siRNA, transitivity
In RNA silencing, 20- to 30-nt small RNAs (sRNAs) mediate sequence-specific gene regulation. trans-acting siRNAs (tasiRNAs), a class of plant-specific endogenous small interfering RNAs (siRNAs), posttranscriptionally regulate mRNAs that have complementary sequences (1–4). tasiRNA biogenesis is initiated by the microRNA (miRNA)-directed cleavage of a transcript from the tasiRNA gene (TAS) loci (5). There are eight TAS loci in Arabidopsis thaliana (TAS1a–c, TAS2, TAS3a–c, and TAS4). The production of tasiRNAs from TAS1a–c and TAS2 is triggered by miRNA173 (miR173). miRNA390 (miR390) and miRNA828 (miR828) initiate tasiRNA production from TAS3a–c and TAS4 transcripts, respectively (3, 4, 6). The processing of the TAS3 transcript is distinct from that of other TAS transcripts, because it requires two miR390 target sites that are specifically recognized by an ARGONAUTE (AGO)7-containing RNA-induced silencing complex (RISC) (7, 8). In contrast, other TAS primary transcripts have single miRNA target sites, are cleaved by AGO1-containing RISCs, and produce tasiRNAs from 3′ cleavage fragments. Previous studies using engineered MIRNA genes showed that the primary miRNAs need to be 22 nt in length to trigger tasiRNA or secondary siRNA production, whereas the targets of these 22-nt miRNAs do not have any determinants beyond the presence of target sites (9–12). This implies that a 22-nt miRNA-guided RISC is fundamentally different from a 21-nt miRNA-guided RISC. However, a recent study demonstrated that an miRNA/miRNA* duplex (miRNA* or the star strand is the non-miRNA strand in the duplex) containing bulged bases is important for triggering secondary siRNA production, regardless of the size of the miRNA (13). The basis for the difference in the results of these studies remains to be established.
tasiRNA production is currently thought to proceed as follows (5). An RISC containing AGO1 that binds 22-nt forms of miR173 or miR828, or AGO7 that binds miR390, cleaves the primary TAS transcripts. SUPPRESSOR OF GENE SILENCING3 (SGS3) stabilizes the cleaved fragments, whereas RNA-DEPENDENT RNA POLYMERASE6 (RDR6) converts the 3′ cleavage fragments of TAS1a–c, TAS2, and TAS4 transcripts and the 5′ cleavage fragments of TAS3a–c transcripts to double-stranded RNAs (dsRNAs) in the SGS3/RDR6 body. Finally, DICER-LIKE (DCL)4 processes these long dsRNAs into 21-nt tasiRNAs (3, 4, 14–16). Although mutations in the genes encoding the components of TRanscription-EXport (TREX) complex and SILENSING DEFECTIVE5 also abolish tasiRNA production, the functions of these genes remain unclear (17–19).
Here we focus on SGS3, a plant-specific protein that is largely uncharacterized. SGS3 forms a homodimer and binds to a dsRNA with a 5′overhang (20, 21). The elimination of SGS3 not only leads to the loss of tasiRNA production but also impairs posttranscriptional gene silencing initiated by highly transcribed sense transgenes, and leads to enhanced susceptibility to Cucumber mosaic virus (22). Further, SGS3 colocalizes with AGO7 in the cytoplasm (23). These observations strongly suggest that SGS3 plays a pivotal role in RNA silencing. However, the molecular function of SGS3 in tasiRNA production remains unclear. In this paper, we show that SGS3 forms a complex with RISC and cleaved RNA in which the 3′ cleavage fragments (the source of tasiRNAs) are protected from degradation.
Results
SGS3 Associates with miR173-Cleaved TAS RNAs in A. thaliana.
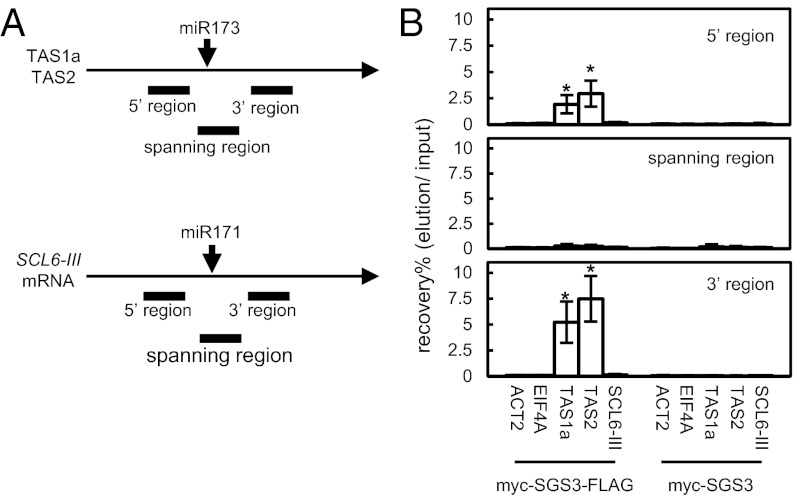
Previously, we proposed that SGS3 stabilizes the 5′ and 3′ fragments generated from TAS1a or TAS2 primary transcripts by miR173-directed cleavage, although these fragments lack poly(A) and a 5′ cap, respectively, and are otherwise unstable (4). To examine whether a ribonucleoprotein (RNP) complex containing the miR173-cleaved fragments and SGS3 is formed, we generated transgenic sgs3 rdr6 double mutant plants expressing myc and FLAG double epitope-tagged myc-SGS3-FLAG (myc epitope tag, Glu-Gln-Lys-Leu-Ile-Ser-Glu-GLu-Asp-Leu; FLAG epitope tag, Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Tyr) and used their floral tissue extracts for glycerol gradient centrifugation analysis. Importantly, the 5′ and 3′ fragments of TAS2 RNA overaccumulated in the transgenic plants, as in rdr6 single mutants, indicating that the introduced myc-SGS3-FLAG was functional. After fractionation and RNA isolation from each fraction, we analyzed the distribution of TAS2-derived RNAs. The result showed that the 5′ and 3′ fragments of TAS2 RNA are enriched in the fraction corresponding to the boundary between the 20% and 50% glycerol layers. The fraction also contained ribosomes (Fig. S1, fraction 8), suggesting that the 5′ and 3′ fragments of TAS2 RNA are contained in a large RNP complex. To determine whether SGS3 is in this RNP complex, the fractions that contained the RNP complexes (Fig. S1, fraction 8) were used for immunoprecipitation with anti-FLAG antibody. RNA copurified with FLAG-tagged SGS3 was analyzed by quantitative RT-PCR using primers for three distinct regions, namely the 5′, 3′, and the miRNA target site-spanning regions, of TAS1a, TAS2, and SCARECROW-LIKE TRANSCRIPTION FACTOR (SCL)6-III transcripts (Fig. 1A). ACTIN (ACT)2 and EUKARYOTIC TRANSLATION INITIATION FACTOR (EIF)4A mRNAs and myc-SGS3–expressing transgenic sgs3 rdr6 double mutant plants were used as controls. Both the 5′ and 3′ regions of the TAS1a and TAS2 transcripts were detected in the immunoprecipitates of FLAG-tagged SGS3 (Fig. 1B), but transcripts spanning the miR173 target site, as well as ACT2 and EIF4A mRNA, were absent from these immunoprecipitates (Fig. 1B). These data indicate that SGS3 specifically associates with the 5′ and 3′ fragments of the TAS1a and TAS2 transcripts, rather than with the uncleaved primary transcripts. However, these immunoprecipitates did not contain SCL6-III mRNA, which is cleaved by miR171 and does not produce tasiRNAs. These results suggest that SGS3 is a component of the RNP complex that contains miR173-cleaved RNAs.
Fig. 1.
Complex containing SGS3 and miR173-cleaved 5′ and 3′ fragments is formed during tasiRNA production. (A) Diagrams of TAS1a, TAS2, and SCL6-III transcripts. The 5′ regions, spanning regions, and 3′ regions correspond to upstream, spanning, and downstream of each miRNA cleavage site, respectively. (B) Analysis of SGS3-bound RNAs. Floral tissue extracts were prepared from transgenic plants from a self-pollinated primary transgenic plant expressing myc- and FLAG-double-tagged SGS3 (myc-SGS3-FLAG) or myc-tagged SGS3 (myc-SGS3) and subjected to stepwise glycerol gradient centrifugation. FLAG-tagged SGS3 was immunoprecipitated with anti-FLAG antibody from the TAS2 RNA-rich fractions (Fig. S1, fraction 8). RNA was purified from the fractions, and the amounts of the 5′ regions (Top), spanning regions (Middle), and 3′ regions (Bottom) of TAS1a, TAS2, and SCL6-III RNA were quantified by RT-PCR. Transcripts of ACT2 and EIF4A were used as controls. SDs were calculated from three biological repeats. Asterisks indicate that values are significantly different from those of tag minus controls (*P < 0.05 by Tukey–Kramer test). Similar results were obtained in another experiment using an independent transgenic line for each tag.
SGS3 Is Recruited onto RISC in Vitro When It Binds a Target.
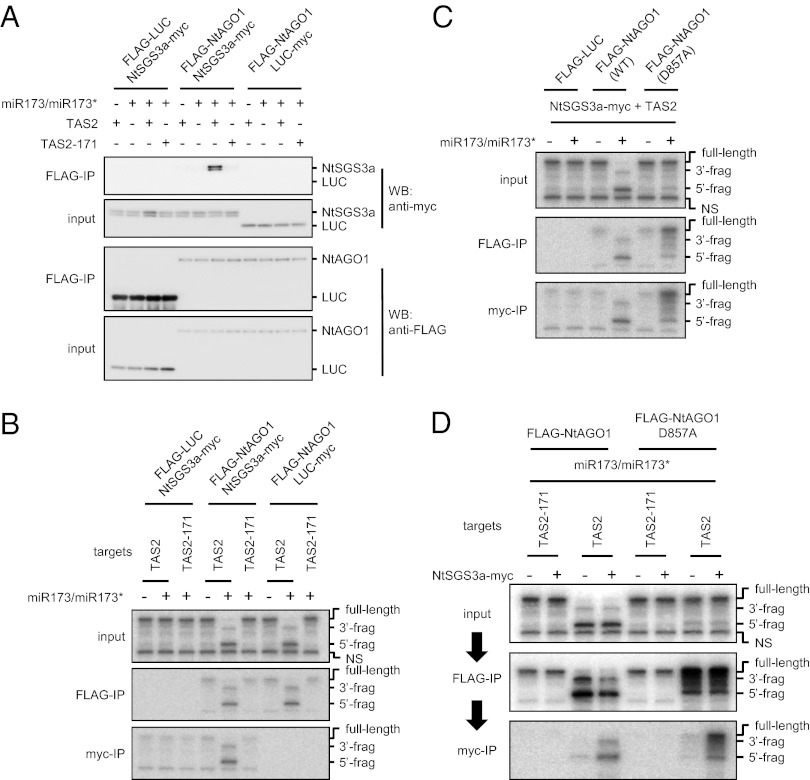
To explore the molecular function of SGS3 in more detail, we took advantage of an in vitro RISC assembly system based on extracts of evacuolated tobacco protoplasts (BYL) (24, 25). We cloned an SGS3 ortholog from Nicotiana tabacum (NtSGS3a; Fig. S2), tagged it with the myc epitope, and then determined whether the myc-tagged NtSGS3a (NtSGS3a-myc) is capable of associating with FLAG-tagged N. tabacum AGO1 (FLAG-NtAGO1). The proteins were produced from these mRNAs using BYL extracts and then mixed. The mixture was incubated with miR173/miR173* to form miR173-programmed RISC (miR173–RISC), and further incubated with a TAS2 target RNA containing a 5′ cap and 70 nt of 3′ poly(A). FLAG-NtAGO1 was then immunoprecipitated using anti-FLAG antibody. As negative controls, a mutant form of TAS2 RNA, in which the miR173 target site was replaced by the miR171 target site (TAS2-171) (Fig. S3A), myc-tagged luciferase, and FLAG-tagged luciferase (LUC-myc and FLAG-LUC, respectively), were used. We found that NtSGS3a-myc copurified with FLAG-NtAGO1 in the presence of both miR173 and TAS2 RNA, but not in the absence of either RNA or in the presence of miR173 and TAS2-171 RNA (Fig. 2A, Upper). NtSGS3a-myc was not detected when FLAG-LUC was precipitated in the presence of miR173 and TAS2 RNA. These results suggest that miR173–RISC and the target RNA are needed for the association of SGS3 with AGO1.
Fig. 2.
SGS3 specifically associates with miR173–RISC when RISC binds to TAS2 target RNA in vitro. (A) Association of NtSGS3a-myc with FLAG-NtAGO1 in the presence of miR173/miR173* and TAS2 RNA. FLAG-NtAGO1 was immunoprecipitated from the BYL-based reaction mixtures containing FLAG-NtAGO1, NtSGS3a-myc, 50 nM miR173/miR173*, and 50 nM target RNAs with anti-FLAG antibody. The immunoprecipitates were analyzed by Western blotting (WB) with anti-myc or anti-FLAG antibodies. The input samples were analyzed in parallel. (B) Association of FLAG-NtAGO1 and NtSGS3a-myc with cleaved fragments of target RNA. FLAG-NtAGO1 and NtSGS3a-myc were immunoprecipitated with anti-FLAG and anti-myc antibodies, respectively, from BYL-based reaction mixtures containing FLAG-NtAGO1, NtSGS3a-myc, 50 nM miR173/miR173*, and 5 nM 32P-labeled target RNAs. RNA was extracted from the input samples and immunoprecipitates and analyzed by denaturing 5% PAGE and autoradiography. As controls, FLAG-LUC, LUC-myc, and TAS2-171 RNA were used in place of FLAG-NtAGO1, NtSGS3a-myc, and TAS2 RNA, respectively. (C) Association of NtSGS3a-myc with slicer-defective FLAG-NtAGO1. Experiments were performed as described for B, except that FLAG-NtAGO1D857A defective in slicer activity was used. (D) Formation of a complex containing RISC, target RNA, and SGS3. Experiments were performed as described for B, except that immunoprecipitation was performed first with anti-FLAG antibody, followed by a second immunoprecipitation with anti-myc antibody. The band marked “NS” shows an in vitro transcription by-product that does not have the miR173 target site. The structures of the target RNAs and sRNAs are shown in Fig. S3.
We next examined whether SGS3 can bind to miR173–RISC–cleaved RNAs in vitro as observed in planta (Fig. 1B). As expected, miR173–RISC could cleave TAS2 RNA but not nontarget TAS2-171 RNA (Fig. 2B, Upper). The 5′ and 3′ fragments of TAS2 RNA copurified with FLAG-NtAGO1 in these preparations (Fig. 2B, Middle). Both fragments also copurified with NtSGS3a-myc (Fig. 2B, Lower), which is consistent with the in vivo data.
To determine whether miR173-directed cleavage is necessary for the association between SGS3 and AGO1, we used a mutant NtAGO1 protein whose slicer activity was disrupted by the substitution of 857th aspartic acid for alanine (FLAG-NtAGO1D857A). This mutant protein can load miRNA/miRNA* and release the miRNA* to form an miRISC, but cannot form an siRISC due to its inability to remove the passenger strand from the siRNA duplex (24). We found that when FLAG-NtAGO1D857A is incubated with NtSGS3a-myc, miR173/miR173*, and TAS2 RNA, the uncleaved TAS2 RNA copurified with FLAG-NtAGO1D857A and NtSGS3a-myc (Fig. 2C). Thus, RISC-directed cleavage is not essential for the association of SGS3 with a target RNA and AGO1.
The above results prompted us to examine formation of a complex that contains RISC, SGS3, and RNA fragments that were cleaved by RISC in these mixtures. miR173–RISCs containing FLAG-NtAGO1 or FLAG-NtAGO1D857A were formed in the presence or absence of NtSGS3a-myc. Then, the mixture was further incubated with TAS2 or TAS2-171 RNAs. After sequential immunoprecipitation of FLAG-NtAGO1 and NtSGS3a-myc with anti-FLAG antibody and anti-myc antibody, respectively, the copurified RNAs were analyzed. We found that cleaved TAS2 RNA fragments coprecipitate with a complex containing NtAGO1 and NtSGS3a, whereas uncleaved full-length TAS2 RNA coprecipitates with a complex containing NtAGO1D857A and NtSGS3a (Fig. 2D).
In keeping with the in vitro results, AGO1 was coimmunoprecipitated with FLAG-tagged SGS3 from the extract of the transgenic Arabidopsis plants used in Fig. 1 (Fig. S4).
Length of miR173 Affects the Stability of Cleaved RNA–RISC Complexes.
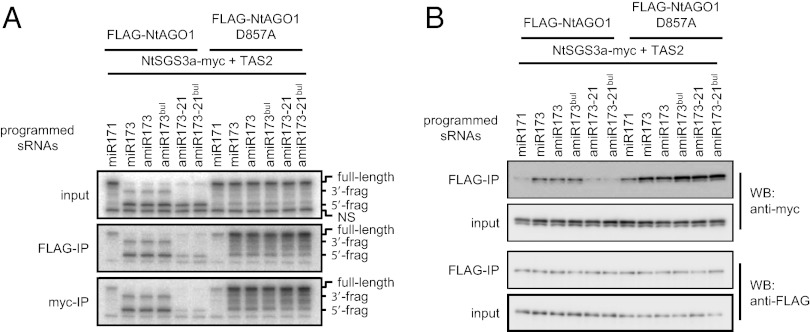
In vivo analyses of tasiRNA production triggered by miR173 showed that 22-nt forms of artificial miR173 (amiR173) can trigger tasiRNA production but 21-nt forms of amiR173 (amiR173-21) cannot (11, 12). In addition, a recent study showed that bulged forms of miRNA/miRNA* duplexes are important for miRNA-triggered siRNA production (13). Thus, we prepared 22-nt and 21-nt forms of amiR173 generated from nonbulged or bulged forms of amiR173/amiR173* duplexes (amiR173, amiR173bul, amiR173-21, or amiR173-21bul) (Fig. S3B) (12, 13). These amiR173s have an identical sequence to miR173, except that amiR173-21 is 1 nt shorter than miR173. We incubated the four forms of amiR173/amiR173* duplexes with FLAG-NtAGO1 or FLAG-NtAGO1D857A, followed by further incubation with TAS2 RNA. With FLAG-NtAGO1, the 3′ cleavage fragments that were generated by miR173–RISC and amiR173–RISC accumulated to higher levels than fragments generated by amiR173-21–RISC (Fig. 3A, Upper). After FLAG-NtAGO1 or NtSGS3a-myc was immunoprecipitated, the 5′ and 3′ cleavage fragments were copurified when miR173 or amiR173 was used, but not when amiR173-21–RISCs were used (Fig. 3A, Middle and Lower). When this miR173-related sRNA-programmed FLAG-NtAGO1D857A or NtSGS3a-myc was immunoprecipitated, TAS2 RNA copurified at similar levels (Fig. 3A, Middle and Lower). The introduction of the bulge in miR173 duplexes produced no significant differences. These results suggest that 3′ TAS2 RNA fragments generated by 22-nt forms of miR173 form stable complexes with AGO1 and SGS3 and are thereby protected from degradation, whereas the 3′ fragments produced by 21-nt forms of miR173 are degraded. A similar amount of the 5′ cleavage product was detected in Fig. 3A, Upper, irrespective of whether the TAS2-derived RNAs associated with AGO1 and SGS3 or not. This is probably because the capped RNA is stable in BYL even when it is not in the complex.
Fig. 3.
Length of miR173 affects the stability of cleaved fragment–RISC–SGS3 complexes. (A) Copurification of TAS2 RNAs with FLAG-NtAGO1 (Middle) programmed with modified miR173/miR173* duplexes or with NtSGS3a-myc (Lower). Experiments were performed as described in the legend for Fig. 2B, except that 50 nM miR173/miR173*–related duplexes were used. (B) Copurification of NtSGS3a-myc with FLAG-NtAGO1 in the presence of TAS2 RNA and miR173/miR173*–related duplexes (Upper two panels). Experiments were performed as described in the legend for Fig. 2A, except that 50 nM miR173/miR173*–related duplexes were used. The structures of miR173/miR173*–related duplexes are shown in Fig. S3B.
When FLAG-NtAGO1 programmed by these sRNAs was immunoprecipitated after incubation with TAS2 RNA and NtSGS3a-myc, larger amounts of NtSGS3a-myc were copurified with FLAG-NtAGO1 in the presence of 22-nt forms of amiR173 than in the presence of 21-nt forms (Fig. 3B, Upper). In contrast, when a similar experiment was performed using FLAG-NtAGO1D857A, the amount of NtSGS3a-myc was similar, irrespective of the forms of miR173 and amiR173 (Fig. 3B, Upper). Thus, the length of miR173 is important for stable association of SGS3 with RISCs and cleaved target RNAs and stabilization of 3′ cleavage products.
Mismatches at the 3′ End of miR173 Affect the Stability of Cleavage Fragment–RISC Complexes.
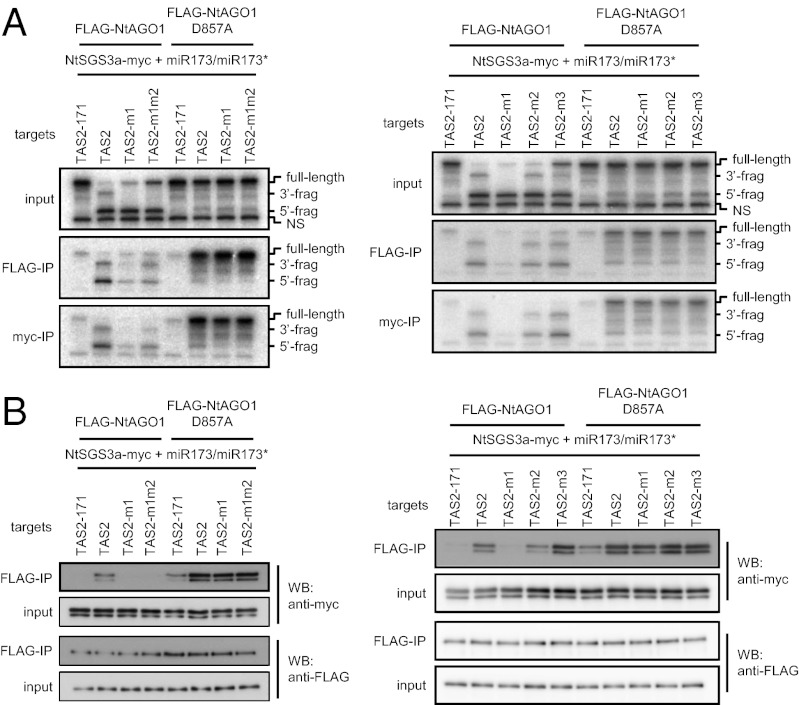
A previous study showed that a 2-nt mismatch between the 3′ end of miR173 and the miR173 target site in TAS1c RNA blocks tasiRNA production (26). To examine the effect of this mismatch on the association of RISC with a target RNA and SGS3, we introduced 2-nt or 1-nt mutations at the 5′ end of the miR173 target site in TAS2 RNA (TAS2-m1m2 or TAS2-m1, respectively) (Fig. S3A). TAS2-m1m2 mimics the 173mTd mutant used by Zhang et al. (26). miR173–FLAG–NtAGO1–RISC and NtSGS3a-myc were produced as described above, mixed, and further incubated with native or mutated TAS2 RNAs. Whereas the 5′ fragment accumulated to similar levels in all mixtures, the 3′ fragment was only detectable when the native TAS2 RNA was used as a target (Fig. 4A, Left Upper). The loss of the 3′ cleavage fragment derived from the target RNA with a 2-nt mismatch between the 3′ end of miR173 and its target site is consistent with the in vivo data reported by Zhang et al. (26) and indicates that the introduction of 1-nt or 2-nt mismatches at the 5′ end of the miR173 target site (i.e., the 3′ end of miR173) enhances the subsequent degradation of the 3′ cleavage fragment. The 5′ and 3′ cleavage fragments of the native TAS2 RNA were detected in immunoprecipitates of FLAG-NtAGO1, and considerable amounts of the 5′ and 3′ fragments of TAS2-m1m2 and smaller amounts of the 5′ and 3′ fragments of TAS2-m1 were also detected in these immunoprecipitates (Fig. 4A, Left Middle). Because the 3′ fragments of the mutated TAS2 RNAs were not detected in the input samples (Fig. 4A, Left Upper), we interpret these results to indicate that uncleaved RNAs associated with FLAG-NtAGO1, and that the 5′ and 3′ fragments were generated after immunoprecipitation. After immunoprecipitation of NtSGS3a-myc, the 5′ and 3′ fragments from the native TAS2 RNA were copurified, whereas for TAS2-m1 and TAS2-m1m2 the 5′ and 3′ fragments were copurified with lower efficiency (Fig. 4A, Left Lower). These findings are reminiscent of previous results that SGS3 expressed in Escherichia coli binds dsRNAs with a 5′ overhang (21), and suggest that SGS3 binds to the dsRNA formed between the 3′-terminal nucleotides of miR173 and TAS2 RNA and somehow facilitates the association with RISC and stabilization of the 3′ cleavage fragment of TAS2 RNA.
Fig. 4.
Mismatches between the 3′ end of miR173 and TAS2 target RNAs affect stability of cleaved fragment–RISC–SGS3 complexes. (A) Copurification of TAS2-derived RNAs with FLAG-NtAGO1 (Middle) or NtSGS3a-myc (Lower). Experiments were performed as described in the legend for Fig. 2B. (B) Copurification of NtSGS3a-myc with miR173-programmed FLAG-NtAGO1 in the presence of TAS2-related RNAs (Upper two panels). Experiments were performed as described in the legend for Fig. 2A. The structures of the mutated TAS2 RNAs are shown in Fig. S3A.
Use of FLAG-NtAGO1D857A resulted in copurification of TAS2-m1 and TAS2-m1m2 RNAs with FLAG-NtAGO1D857A or NtSGS3a-myc as efficiently as the native TAS2 RNA (Fig. 4A, Left Middle and Left Lower). We next examined the amount of NtSGS3a-myc copurified with miR173-bound FLAG-NtAGO1 in the presence of TAS2-m1 and TAS2-m1m2 RNAs and found that it was negligible, whereas NtSGS3a-myc was readily detectable in the presence of native TAS2 RNA. NtSGS3a-myc copurified with FLAG-NtAGO1D857A in the presence of TAS2-m1 and TAS2-m1m2 RNA as efficiently as in the presence of TAS2 RNA (Fig. 4B, Left Upper). These results indicate that the mismatches at the 5′ end of the miR173 target site seriously affect the association of cleaved target RNAs with miR173–RISC and SGS3, as well as the stability of the 3′ fragments of cleaved TAS2 RNAs, whereas the same mismatches do not affect the association of SGS3 with uncleaved TAS2 RNA–miR173–RISC complexes.
The effect of internal mismatches between the miRNA and target RNA was then examined. We prepared two additional TAS2 RNA derivatives with mutations at the second or third nucleotide from the 5′ end of the miR173 target site (TAS2-m2 and TAS2-m3, respectively) (Fig. S3A), mixed these with miR173–FLAG–NtAGO1–RISC and NtSGS3a-myc, and immunoprecipitated with FLAG-NtAGO1 or NtSGS3a-myc. These mutations (m2 and m3) did not drastically affect the accumulation of the 3′ fragments, the association of cleaved RNA with RISC or SGS3, or the association of SGS3 with RISC (Fig. 4 A and B, Right). When similar experiments were performed using FLAG-NtAGO1D857A, the amount of uncleaved TAS2-m2 and TAS2-m3 RNAs that copurified with FLAG-NtAGO1D857A or NtSGS3a-myc, and the amount of NtSGS3a-myc that copurified with FLAG-NtAGO1D857A, were similar to that for native TAS2 RNA.
We obtained similar results when these experiments were performed using the myc-tagged SGS3 from A. thaliana (AtSGS3-myc) in place of NtSGS3a-myc, except when AtSGS3-myc was combined with miR173-programmed FLAG-NtAGO1D857A in the presence of either TAS2m1 or TAS2m1m2 RNA (Fig. S5). The amount of uncleaved target RNA that copurified with AtSGS3-myc, and the amount of AtSGS3-myc that copurified with FLAG-NtAGO1D857A for TAS2-m1 and TAS2-m1m2 RNAs, were lower than that for native TAS2 RNA (and TAS2-m2 and m3 RNAs) but higher than that for TAS2-171 RNA (Fig. S5). These results suggest that in addition to the ability to bind the dsRNA formed by miR173 and the target RNA in RISC, SGS3 can interact with the protein moiety of AGO1–RISC and that the interaction of tobacco AGO1–RISC with AtSGS3 is weaker than that with NtSGS3a.
Depletion of Endogenous NtSGS3 in BYL Causes Instability of Cleavage Fragment–miR173–RISC Complexes.
The addition of in vitro translated NtSGS3a-myc did not have any significant effect on TAS2 RNA cleavage or stability (Fig. 2). We speculated that endogenous NtSGS3 in the BYL extract may mask the effect of the added SGS3 on TAS2 RNAs. To test this idea, we immunoprecipitated FLAG-tagged and miR173-programmed NtAGO1 or NtAGO1D857A in the presence of native or mutated TAS2 RNAs. Endogenous NtSGS3 was then detected using antibodies raised against peptides common to NtSGS3a and NtSGS3b (Fig S2, red underline). The results showed that endogenous NtSGS3 behaves similarly to in vitro translated NtSGS3a-myc (Fig. S6A).
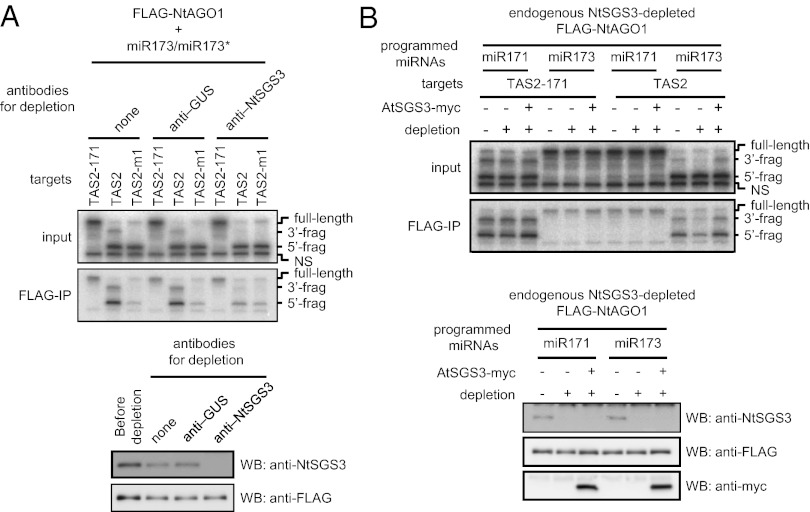
To examine the contribution of SGS3 to the stabilization of cleaved RNA–RISC complexes, we depleted endogenous NtSGS3 using anti-NtSGS3 antibody after the formation of miR173–RISC. We used anti-β-glucuronidase (GUS) antibody as a negative control. The depletion step successfully reduced the amount of endogenous NtSGS3 (Fig. 5A and Fig. S6B). After the depletion, TAS2 RNA (or TAS2-171 or TAS2-m1 RNAs as controls) was added to the mixtures containing miR173–RISC. We found that depletion of endogenous NtSGS3 reduced the accumulation of the 3′ fragment of cleaved TAS2 RNA and the amount of the 3′ fragment that copurified with miR173–RISC to the levels found for TAS2-m1 RNA (Fig. 5A and Fig. S6B). In contrast, depletion of endogenous NtSGS3 did not affect copurification of target RNA with miR173-bound FLAG-NtAGO1D857A (Fig. S6C).
Fig. 5.
Depletion of endogenous NtSGS3 in BYL destabilizes cleavage fragment–miR173–RISC complexes. (A) Copurification of TAS2-derived RNAs with FLAG-NtAGO1 after depletion of endogenous NtSGS3. The BYL-based reaction mixtures containing FLAG-NtAGO1 and 50 nM miR173/miR173* were incubated with affinity-purified anti-GUS or anti-NtSGS3 antibodies (or mock-conjugated), treated with protein A Sepharose, and then mixed with 5 nM 32P-labeled target RNAs. From these mixtures, FLAG-NtAGO1 was immunoprecipitated with anti-FLAG antibody. RNA was extracted from the immunoprecipitates and analyzed by denaturing 5% PAGE and autoradiography. RNA from input samples was analyzed in parallel (Upper two panels). Endogenous NtSGS3 or FLAG-NtAGO1 in each reaction mixture was detected with anti-NtSGS3 or anti-FLAG antibodies, respectively (Lower two panels). (B) In vitro complementation of endogenous NtSGS3 depletion by AtSGS3-myc with regard to stabilization of RISC-cleaved fragments and cleaved RNA–RISC complexes. BYL-based reaction mixtures containing miR171- or miR173-bound FLAG-NtAGO1 were incubated with anti-NtSGS3 antibodies, treated with protein A Sepharose, and then combined with mock-translated and mock-depleted lysate, mock-translated and NtSGS3-depleted lysate, or AtSGS3-myc mRNA-translated and NtSGS3-depleted lysate. 32P-labeled target RNAs (5 nM) were added to each reaction mixture, and FLAG-NtAGO1 was immunoprecipitated with anti-FLAG antibody. RNA was extracted from the immunoprecipitates and analyzed by denaturing 5% PAGE and autoradiography. RNA from input samples was analyzed in parallel (Upper). Endogenous NtSGS3, FLAG-NtAGO1, and AtSGS3-myc in the mixtures were detected with anti-NtSGS3, anti-FLAG, and anti-myc antibodies, respectively (Lower).
To confirm that the instability of the 3′ fragment of TAS2 RNA and the inability to form cleaved RNA–miR173–RISC complexes was indeed due to the decrease in NtSGS3 concentration, we used AtSGS3-myc, which is not affected by the anti-NtSGS3 antibody. We combined NtSGS3-depleted miR171– or miR173–RISC mixtures with mock-translated and mock-depleted (anti-GUS antibody) lysate, mock-translated and NtSGS3-depleted lysate, or AtSGS3-myc mRNA-translated and NtSGS3-depleted lysate. After incubation of the mixtures with TAS2 or TAS2-171 RNAs, the accumulation of their 3′ cleavage fragments and the copurification of TAS2 RNA fragments with FLAG-NtAGO1 were evaluated. When miR173–RISC was mixed with TAS2 RNA, the accumulation of the 3′ fragment of cleaved TAS2 RNA in the mixture and the amount of cleaved TAS2 RNA copurified with miR173–RISC were reduced by depletion of endogenous NtSGS3 and recovered by supplementation of AtSGS3-myc (Fig. 5B). These results indicate that SGS3 is required for the maintenance of cleaved TAS2 RNA–miR173–RISC complexes and protection of the 3′ fragment from degradation, and that the 22-nt forms of miR173 alone are not sufficient for these functions.
SGS3 Is Not Necessary for Stabilization of the Cleavage Fragment Directed by Non-tasiRNA-Producing sRNA-Programmed RISC.
To know whether the revealed SGS3 function is relevant to tasiRNA biosynthesis, the association of SGS3 with RISCs directed by miR171, which does not trigger tasiRNA production, was examined. This analysis showed that NtSGS3a-myc associates with miR171-programmed FLAG-NtAGO1 and or FLAG-NtAGO1D857A and target RNAs in the presence of TAS2-171 (Fig. S7), suggesting that SGS3 associates with AGO1 even in the presence of a non-tasiRNA-producing miRNA and its target RNA. This was in contrast to the in vivo results obtained using plant tissue extracts, which indicated that miR171 target RNA (SCL6-III transcript) was not copurified with SGS3 (Fig. 1B).
Finally, we determined the effect of depletion of endogenous NtSGS3 on the stability of the cleavage fragments directed by miR171-programmed RISCs. In contrast to the results obtained with miR173–TAS2 RNA, the stability of the 3′ fragments generated by miR171 was not affected by the depletion of NtSGS3 (Fig. 5B). We also used 20-, 21-, and 22-nt forms of siRNAs (gf698-20, gf698-21, and gf698-22, respectively) complementary to a target RNA that has a partial sequence of GFP mRNA (GF-s) (figure 1a in ref. 24). The gf698-20 and -21 siRNAs are identical to gf698-22 except for 2- and 1-nt deletions, respectively, at the 3′ termini. The stability of the 3′ fragments generated by these siRNA-programmed RISCs was also not affected by the depletion of NtSGS3 (Fig. S8).
Discussion
Our data indicate that miR173-programmed AGO1–RISC remains associated with cleaved TAS2 RNA fragments and SGS3, and that depletion of SGS3 affects the stability of the 3′ fragment of cleaved TAS2 RNA. These observations suggest that the 3′ fragment of cleaved TAS2 RNA, which lacks the 5′ cap and so is otherwise unstable, is in a complex in which SGS3 is an essential component and thereby protected from degradation. Previous in vivo studies have shown that tasiRNA biogenesis is inhibited when the 3′ nucleotide of miR173 is deleted or when the corresponding nucleotide in the target RNA is mutated (11, 12, 26). In keeping with this fact, when a similar deletion in miR173 or mutations in the TAS2 RNA were introduced, the association of SGS3 with miR173-programmed RISC was not observed and the 3′ cleavage products were degraded in a plant cell extract. These findings suggest that the formation of the cleaved TAS2 RNA–miR173–RISC–SGS3–containing complex is key to tasiRNA biogenesis. We propose that to serve as the template for cRNA synthesis by RDR6 toward tasiRNA production, the 3′ TAS RNA fragments need to be in this complex.
A previous study demonstrated that SGS3 protein expressed in E. coli binds specifically to dsRNA with a 5′ overhang (21). A structural analysis of the Thermus thermophilus Ago (TtAgo) protein guide DNA–target RNA ternary complex indicated that the 3′-terminal region of the guide DNA, which is bound to the Piwi-Argonaute-Zwille (PAZ) domain when target RNA is not present, hybridizes to the target RNA to form a duplex and detaches from the PAZ domain. As a result, a dsRNA with a 5′ overhang protrudes from the TtAgo protein surface (27). Based on these observations, we propose that SGS3 binds to the dsRNA with a 5′ overhang formed between miR173 (22 nt) and the 5′ cleavage fragment, which protrudes from the AGO1 protein surface.
This possibility is consistent with the observed negative effect of mismatches at the 3′ end of the miRNA on complex formation and 3′ fragment stabilization and the previous results by Fukunaga and Doudna that E. coli-expressed SGS3 protein has a lower affinity for 5′ overhang-containing dsRNAs with terminal mismatches than for those with a perfect match (21). Moreover, the failure of the complex formation and 3′ fragment stabilization observed with shorter versions of miR173 (<21 nt) is also consistent with this hypothesis because the double-stranded region may be hidden within AGO1 (or RISC) and inaccessible to SGS3 when the miRNA is too short. Consistent with this scenario, it was reported that 22-nt forms of amiRNAs that were modified from 21-nt forms of originally non-tasiRNA-producing miRNAs can trigger secondary siRNA production in vivo (11, 12). Thus, this dsRNA-mediated mode of SGS3 binding to cleaved RNA–RISC complexes should be important for tasiRNA or secondary siRNA production.
On the other hand, we showed that NtSGS3 associates with slicer-defective RISCs that bind uncleaved target RNAs, but not with free RISCs. In this case, however, neither deletion of the 3′ nucleotide of miR173 nor a mutation in the target RNA at the nucleotide that corresponds to the 3′ terminus of miR173 affected SGS3 binding to RISC and the target RNA. This suggests that SGS3 binding to slicer-defective RISC-uncleaved target RNA complexes is not via a dsRNA with a 5′ overhang. The crystal structure of TtAgo protein bound to a 21-nt guide DNA shows that the 5′-terminal phosphate group of the guide DNA is bound to the middle domain of AGO, termed the MID domain, whereas the 3′-terminal nucleotide is anchored in the PAZ domain pocket before TtAgo binds to a target. Binding of this complex to a target RNA causes pivot-like domain movements within the Ago scaffolds and release of the 3′ end of the guide DNA from the PAZ domain pocket to form a duplex with the target RNAs (27, 28). The AGO1 molecular surface, which is exposed after similar conformational changes, may be involved in the binding of SGS3 to uncleaved target RNA-bound slicer-defective (and perhaps slicer-active) AGO1 RISCs.
Last, we would like to comment on the discrepancy between the in vivo and in vitro results with regard to miR171- or gf698-programmed AGO1–RISCs. First, in plant tissue extracts (in vivo), miR171-cleaved SCL6-III RNA did not copurify with SGS3. However, in the in vitro system, both uncleaved and cleaved TAS2-171 RNA stably associated with miR171–AGO1–RISC and SGS3, although depletion of SGS3 did not affect RISC–target RNA association. Second, it was reported that DCL2-produced 22-nt, but not DCL4-produced 21-nt, siRNAs trigger RDR6-dependent secondary siRNA production in vivo (29). However, in vitro, both uncleaved and cleaved GF-s RNA stably associated with gf698-22–AGO1–RISC, and 3′ nucleotide deletions in gf698-22 siRNA did not affect the association. Depletion of SGS3 also did not affect this association. In animals, autoantigen La has been shown to facilitate dissociation of cleaved RNAs from RISCs (30). Although the factors that facilitate dissociation of cleaved RNAs from RISCs have not been identified in plants, we speculate that plant cells are equipped with such factors (hereafter “putative dissociation factors”), which are missing or present at low levels in the BYL extract. Unlike miR173–TAS2 RNA, miR171 and gf698 are perfectly complementary to TAS2-171 and GF-s RNAs, respectively. Due to the increased thermostability of the sRNA-cleaved target RNA duplexes, the lack of the putative dissociation factors in the BYL extract may have resulted in the inability of in vitro cleaved target RNAs to dissociate from miR171– or gf698–RISCs. In plant cells, RISCs programmed by gf698-22 (DCL2-produced 22-nt siRNAs), but not gf698-21 or -20 (DCL4-produced 21-nt siRNAs), may be able to protect cleaved target RNAs from degradation by forming a complex involving SGS3–dsRNA contact and lead to the pathway of secondary siRNA production.
Materials and Methods
In Vitro RISC Assembly and Immunoprecipitation.
In vitro RISC assembly and analyses of RISCs were performed as described previously (24). To analyze the association of NtSGS3a-myc with FLAG-NtAGO1, EZview Red Anti-FLAG M2 affinity gel (Sigma) was used. To analyze the association of target RNAs with NtSGS3a-myc or FLAG-NtAGO1, anti-myc (clone 4A6; Millipore) or anti-FLAG antibody (clone M2; Sigma) was used, respectively.
Additional Methods.
Full materials and methods are described in SI Materials and Methods. The oligonucleotides used in this study are listed in Table S1 (primers), and Table S2 (oligoRNAs).
Supplementary Material
Acknowledgments
We thank Yoriko Fujibayashi for technical assistance. This work was supported by a grant from Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency (to M.Y.) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to M.Y. and M.I.). Work in R.S.P.’s laboratory is supported by a grant from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cDNA sequences reported in this paper have been deposited in the GenBank database [accession nos. AB690269 (NtSGS3a) and AB690270 (NtSGS3b)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217050110/-/DCSupplemental.
References
- 1.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18(19):2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vazquez F, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16(1):69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 3.Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19(18):2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20(24):3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127(3):565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 8.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133(1):128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 9.Montgomery TA, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA. 2008;105(51):20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 2009;10(3):264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HM, et al. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 2010;107(34):15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuperus JT, et al. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol. 2010;17(8):997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA. 2012;109(7):2461–2466. doi: 10.1073/pnas.1200169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102(36):12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15(16):1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Kumakura N, et al. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009;583(8):1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 17.Jauvion V, Elmayan T, Vaucheret H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell. 2010;22(8):2697–2709. doi: 10.1105/tpc.110.076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yelina NE, et al. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci USA. 2010;107(31):13948–13953. doi: 10.1073/pnas.0911341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez-Pinzon I, et al. SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J. 2007;50(1):140–148. doi: 10.1111/j.1365-313X.2007.03043.x. [DOI] [PubMed] [Google Scholar]
- 20.Elmayan T, et al. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J. 2009;276(3):835–844. doi: 10.1111/j.1742-4658.2008.06828.x. [DOI] [PubMed] [Google Scholar]
- 21.Fukunaga R, Doudna JA. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28(5):545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101(5):533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 23.Jouannet V, et al. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 2012;31(7):1704–1713. doi: 10.1038/emboj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iki T, et al. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell. 2010;39(2):282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Iki T, Yoshikawa M, Meshi T, Ishikawa M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2011;31(2):267–278. doi: 10.1038/emboj.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Ng DW, Lu J, Chen ZJ. Roles of target site location and sequence complementarity in trans-acting siRNA formation in Arabidopsis. Plant J. 2012;69(2):217–226. doi: 10.1111/j.1365-313X.2011.04783.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461(7265):754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456(7224):921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mlotshwa S, et al. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One. 2008;3(3):e1755. doi: 10.1371/journal.pone.0001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Autoantigen La promotes efficient RNAi, antiviral response, and transposon silencing by facilitating multiple-turnover RISC catalysis. Mol Cell. 2011;44(3):502–508. doi: 10.1016/j.molcel.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.