Significance
Ribosomes synthesize all proteins in living cells. There are limits, however, to which sequences they can make. We identified short motifs within translating proteins that inhibit their own synthesis. We developed in vitro methods to determine the molecular mechanism of ribosome stalling by these motifs. Some act by blocking the formation of peptide bonds; in a few of these cases, a translation factor, elongation factor P, alleviates stalling. Other motifs block release of the protein at stop codons. Stalling motifs occur less often than expected in bacterial proteins, suggesting that proteins have evolved to be synthesized efficiently.
Keywords: EF-P, proline, ribosome stalling, tmRNA
Abstract
Although the ribosome is a very general catalyst, it cannot synthesize all protein sequences equally well. For example, ribosomes stall on the secretion monitor (SecM) leader peptide to regulate expression of a downstream gene. Using a genetic selection in Escherichia coli, we identified additional nascent peptide motifs that stall ribosomes. Kinetic studies show that some nascent peptides dramatically inhibit rates of peptide release by release factors. We find that residues upstream of the minimal stalling motif can either enhance or suppress this effect. In other stalling motifs, peptidyl transfer to certain aminoacyl-tRNAs is inhibited. In particular, three consecutive Pro codons pose a challenge for elongating ribosomes. The translation factor elongation factor P, which alleviates pausing at polyproline sequences, has little or no effect on other stalling peptides. The motifs that we identified are underrepresented in bacterial proteomes and show evidence of stalling on endogenous E. coli proteins.
We commonly think of the ribosome as capable of synthesizing any protein, regardless of its sequence. However, it turns out that some nascent peptides contain stalling motifs that inhibit core functions of the ribosome (1, 2). Why would a protein evolve to arrest its own synthesis? In one of the best-characterized examples, stalling in the secretion monitor (SecM) leader peptide up-regulates translation of SecA, a protein encoded downstream on the same mRNA (3). Known stalling motifs typically have been identified based on their function as genetic switches, regulating gene expression in response to levels of protein translocation factors (3, 4) or changes in the concentration of small-molecule metabolites (2, 5). Further understanding of the scope and mechanism of ribosome stalling may yield additional insight into programmed ribosome-stalling events that regulate gene expression in organisms from bacteria to humans (6, 7). In addition, by fine-tuning the rate of protein synthesis, stalling peptides may affect protein folding and function, as reported previously with rare codons (8, 9).
Analyses of natural motifs have identified three sites of interaction within the ribosome that lead to stalling. First, conserved residues at a motif’s N terminus often interact with the ribosome near a constriction in the exit tunnel between proteins L4 and L22. Ribosomal mutations near this site were isolated in genetic screens for reduced levels of stalling (3, 10). Second, conserved residues near a motif’s C terminus interact with nucleotides surrounding the ribosomal active site, the peptidyl-transferase center or PTC (11). Third, some motifs encode a specific aminoacyl-tRNA that acts as a poor peptidyl acceptor when bound in the A site (12, 13). The SecM consensus motif (FxxxxWIxxxxGIRAGP) showcases all three of these interactions, each of which contributes to stalling. A Trp side chain 11 residues upstream of the stall site binds near the constriction in the tunnel, the Arg residue three residues upstream is positioned close to the PTC (3), and Pro-tRNA binds in the A site but does not react (14). A recent cryoelectron microscopy structure of the stalled SecM complex visualized these interactions directly and begins to provide some molecular rationale for how stalling is induced (15).
A full understanding of the molecular mechanism underlying ribosome stalling has not been forthcoming because of the complexity of natural stalling motifs. In each case, stalling is reversible and is controlled by changes in the cellular environment. Stalling in TnaC, for example, is induced by high tryptophan concentrations through the binding of free tryptophan at an unknown site within the ribosome (5). Given that a single stalling motif interacts with the ribosome at multiple sites, deconvoluting the role of the conserved residues in the peptide is difficult enough without the added complexity of small-molecule binding. Moreover, even at well-validated sites of interaction, such as the L4/L22 constriction, different stalling motifs appear to work via different mechanisms. Ribosomal mutations that reduce stalling by one motif may have no effect or even increase stalling by another (13, 16). These complexities make it challenging to obtain general conclusions about the mechanism of ribosome stalling by natural stalling peptides.
To characterize better the scope and mechanism of ribosome stalling, we set out to find a series of artificial motifs that inhibit translation during their own synthesis. We reasoned that, by selecting directly for stalling peptides, we might identify motifs that are simpler than natural ones because they are not required to stall reversibly or to regulate downstream genes. The fact that only a few residues are essential for stalling by SecM, TnaC, and ErmCL (1) and the fact that these motifs share little or no sequence similarity led us to believe that more stalling motifs exist but have not yet been identified. To this end, we developed a powerful genetic selection in Escherichia coli that ties stalling on a reporter protein to cellular survival.
Here we report several stalling motifs, some that block peptide release during translational termination and others that block peptidyl transfer. Because these motifs are short, we are able to recapitulate the stalling phenomenon using pre–steady-state kinetic assays, an important step toward achieving mechanistic insights. Of particular interest, we show that polyproline sequences induce ribosome stalling. The translation factor elongation factor P (EF-P), which alleviates stalling at polyproline stretches, does not affect the other short motifs we identified, further defining its scope of action. Finally, our analysis of bacterial proteomes reveals that stalling motifs are underrepresented, implying that they have been selected against. Where they do occur in endogenous E. coli proteins, pausing is detectable by ribosome profiling. These findings argue that these short motifs have an impact on protein synthesis in bacteria.
Results
Genetic Selection for Stalling Motifs.
We previously reported the first systematic search for nascent polypeptide motifs that induce ribosome stalling (13). In that study, we identified stalling motifs from random libraries using a genetic selection based on tmRNA, part of the machinery that rescues stalled ribosomes in bacteria (17, 18). tmRNA recognizes stalled ribosomes and directs the addition of a short peptide tag to the nascent polypeptide; thus we were able to detect stalling events in living cells. We linked tmRNA tagging of the KanR protein to cellular survival and identified the stalling motif, FxxYxIWPPP (13). Because the selection depended on functional KanR, we suspect that many motifs were missed. To survive the selection, a motif within the KanR sequence had to induce ribosome stalling but not interfere with the enzyme’s folding and activity.
To overcome these limitations, we developed a second-generation selection that allows more variability in the motif length and sequence than was possible in our earlier study. We established a variant of the bacterial two-hybrid system (19, 20) that links stalling with cellular survival (Fig. 1A). In this system, cells cannot synthesize histidine unless transcription of a HIS3 reporter gene is activated. Driven from a weak promoter, HIS3 expression is insufficient for cellular survival unless RNA polymerase is recruited to the transcriptional start site by a DNA-binding protein, in this case a modified form of lambda cI. We encoded stalling motifs at the C terminus of the full-length cI protein, where they have little or no effect on protein structure or function. When ribosomes stall during cI synthesis, the protein is tagged by tmRNA and therefore can recruit RNA polymerase fused to SspB. The SspB protein binds specifically to the peptide tag encoded by tmRNA (21); the resulting interaction between tagged cI and RNA polymerase alpha (RpoA)-SspB leads to recruitment of RNA polymerase to the HIS3 gene, transcriptional activation, and survival of cells on selective medium lacking histidine. Note that the natural function of SspB is to deliver tagged proteins to the ClpXP protease for destruction (21). By including only the tag-binding domain of SspB (residues 1–117), not the ClpXP-binding domain (22), and by changing the last two residues of the tmRNA tag to DD (23), we prevent degradation of tagged cI.
Fig. 1.
Two-hybrid selection for nascent peptides that stall ribosomes. (A) The lambda cI protein binds to DNA upstream of the HIS3 gene. When ribosomes stall during cI synthesis, a short tag is added to the protein by tmRNA, recruiting an SspB-RNA polymerase fusion protein. In this way, ribosome stalling in cI activates transcription of HIS3, restoring the cell’s ability to synthesize histidine and survive on minimal medium. (B) Four cI constructs were tested for their ability to recruit SspB-RpoA, activate HIS3, and grow on medium lacking histidine: cI translationally fused to the tmRNA tag (cI-tag); cI fused to two known inducers of stalling, Glu-Pro-stop (cI-EP-stop) and an mRNA lacking a stop codon (cI-nonstop); and cI with no stalling motif (cI only).
We validated the selection with four control cI constructs. First, expression of cI alone, with no stalling motif, does not support growth on minimal medium lacking histidine, because cI is not tagged by tmRNA and thus does not bind RpoA-SspB. When the modified tmRNA tag is translationally fused to the C terminus of cI in a second construct, the resulting protein recruits RpoA-SspB and activates HIS3 transcription, leading to robust survival on selective medium (cI-tag, Fig. 1B). Furthermore, we showed that known stalling motifs survive the selection by generating sufficient levels of tmRNA-tagged cI. We added Glu-Pro-stop to the C terminus of cI; this short sequence previously was shown to induce high levels of stalling and tagging in vivo (24). In another construct, we expressed cI on a nonstop mRNA, where ribosomes translate to the 3′ end of the message because there is no stop codon. These ribosomes are effectively stalled and are known to be rescued by tmRNA (25). Both these constructs support cellular survival on selective medium that is as robust as the cI-tag translational fusion (Fig. 1B). These data demonstrate that the selection successfully ties ribosome stalling during cI synthesis with cellular survival on selective medium.
To identify stalling motifs, we fused 20 random codons onto the C terminus of cI and subjected the resulting library to the two-hybrid selection. Although we could sample only a tiny sliver of sequence space (∼108 clones out of ∼1026 possibilities), we found that a significant fraction of the library survived the selection, roughly 1 in 104 colonies plated. This high rate of survival suggests that it is remarkably easy to find sequences that induce stalling and tagging by tmRNA. Because two-hybrid systems are notoriously rich in false positives, we performed a secondary screen in which lysates from 150 colonies were immunoblotted with antibodies against the tmRNA-DD tag (26). More than a quarter (41/150) showed significant levels of tmRNA-tagged cI protein, suggesting that they survived the selection because ribosome stalling occurred during cI synthesis. Known causes of stalling could be attributed to 21 of these 41 clones. The most common motif was Pro-stop at the C terminus of the cI protein (18 clones). Pro is known to induce stalling during termination (24). In contrast, clusters of rare codons, another known cause of stalling, were found in only three clones. For the remaining 20, no known cause of stalling was apparent, although tagged cI clearly was being produced. Presumably something in the variable region led to tagging, but the site of stalling and the residues responsible for stalling had to be identified for each clone by additional experiments.
Determination of the Site of Stalling and the Consensus Stalling Motifs.
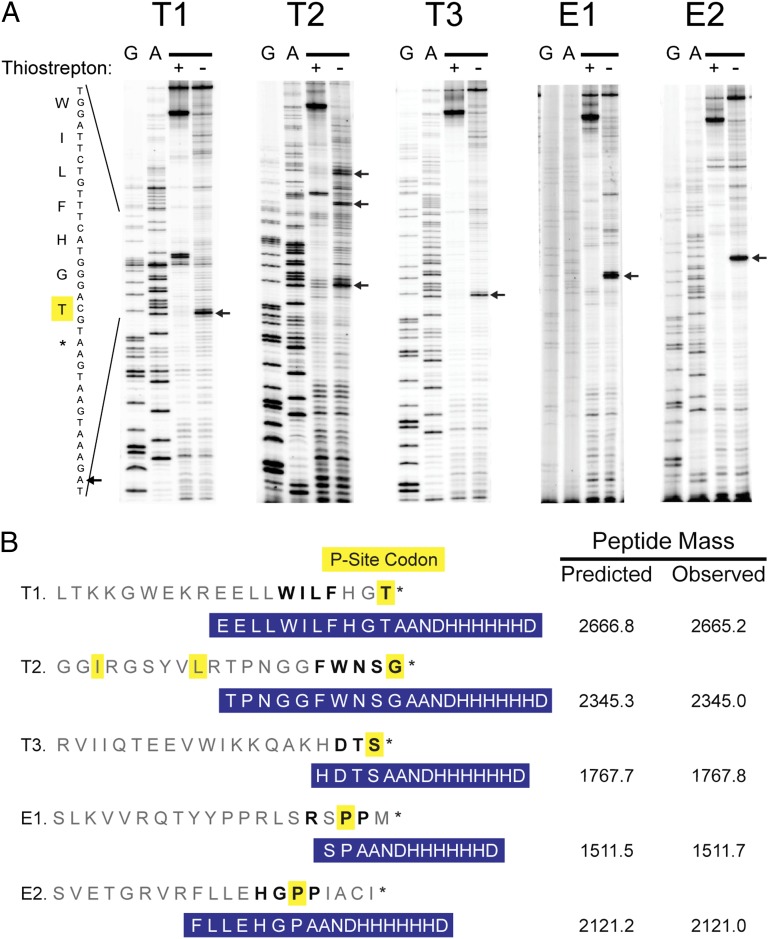
We used two different approaches to determine the site of stalling in clones that survived the two-hybrid selection. First, we selected 10 clones that showed the highest levels of tagging in the immunoblot assay described above and performed toeprinting assays to detect stalling in these clones directly during in vitro translation. A radiolabeled primer was annealed to the 3′ end of the cI transcript and was extended by reverse transcriptase. When it encounters a stalled ribosome, reverse transcriptase stops 15–16 nt downstream of the first nucleotide in the P site codon, thus revealing which codon is in the P site when stalling occurs. As a control, the antibiotic thiostrepton was added to trap ribosomes in the initiation stage. The disappearance of the toeprint band when thiostrepton is added demonstrates that the block in reverse transcription is caused by stalled ribosomes and is not an artifact of mRNA sequence or structure.
The toeprinting results show that the 10 clones fall into two classes. Some stall with a stop codon in the A site, indicating that termination is inhibited; toeprinting data for three examples of this class, T1–T3, are shown in Fig. 2A. Others stall with a sense codon in the A site, indicating that elongation is inhibited; toeprinting data are shown for two such motifs, E1 and E2. All 10 sequences and their stalling sites are given in Table S1. The fact that stalling was recapitulated in a reconstituted translation system [the protein synthesis using recombinant elements (PURE) system] rules out alternate explanations such as mRNA cleavage or degradation that might have led to tagging by tmRNA and survival in the two-hybrid selection.
Fig. 2.
Stalling motifs that block termination (T1–T3) or elongation (E1 and E2). (A) Stalling sites for five motifs were determined by toeprinting assays. Blocks in cDNA synthesis are marked with arrows. The antibiotic thiostrepton traps ribosomes in initiation complexes; bands seen in both treated and untreated lanes are reverse transcriptase artifacts. (B) Key amino acids in each motif are written in bold. The codon labeled in yellow is positioned in the P site of the stalled ribosome. The site of stalling in vivo was confirmed by purifying tagged cI, digesting the protein with trypsin, and determining the mass of the C-terminal peptide (highlighted in blue) by MALDI-MS.
We performed a second series of experiments to detect where stalling and tmRNA tagging occurred in vivo. We did so by analyzing tagged cI protein by MS; the residue upstream of the tag corresponds to the codon positioned in the P site in the stalling event. We analyzed the five clones depicted in Fig. 2A because they gave strong toeprints. We expressed each cI clone with a modified tmRNA that encodes six His residues in its tag sequence (27). Tagged cI was purified over Ni-NTA resin and digested with trypsin. The peptide fragment corresponding to the C terminus of the tagged cI protein contains the altered tmRNA tag and a few upstream residues, depending on where trypsin cleavage occurs. This peptide was enriched on an Ni-NTA resin, and its mass was determined by MALDI-MS. The amino acid sequence of the peptide also was confirmed by MS/MS. The five peptides and their masses are shown in Fig. 2B. In each case, the residue immediately before the tag corresponds to the codon in the P site in the stalled complex in the toeprinting assays, confirming that stalling occurs at the same site in vitro and in vivo.
We determined which residues in these five stalling motifs are required for stalling through a process of mutagenesis and reselection. We introduced mutations into the 20 codons of a given motif at a frequency of 30% per nucleotide. The resulting library was subjected to the two-hybrid selection, and a consensus sequence of surviving clones was determined. Taken together, the toeprinting, MS, and reselection experiments define five stalling motifs, their stalling sites, and consensus sequences. Data from the motifs that inhibit termination (T1–T3) will be discussed first, followed by the data for the elongation motifs (E1 and E2).
Stalling Motifs That Inhibit Termination.
The T1 motif contains the consensus residues WILFxxT-stop, where “x” is any amino acid. When the T1 sequence was subjected to mutagenesis and reselection, the highest enrichment was seen at the C-terminal residue, Thr, with P < 10−5 (Fig. 3A). We found that mutation of this Thr to Ala abolishes stalling in the toeprinting assay (Fig. 3B). The Trp residue seven codons back from the stop codon also is selected for (P < 10−4); it lies in a stretch of hydrophobic residues. Ala substitutions have little or no effect on stalling by this motif, with the exception of the Phe residue at the −4 position. In contrast, Glu substitutions often reduce stalling efficiency, presumably by blocking hydrophobic interactions between these residues and the ribosomal exit tunnel. It appears that stalling in the T1 motif is induced by the C-terminal Thr residue and this stretch of upstream hydrophobic and aromatic residues.
Fig. 3.
Key residues in motifs that block termination. (A) The consensus sequence of each motif was determined by randomizing all 20 codons at 30% per nucleotide and subjecting the resulting library to the two-hybrid selection. (B) Toeprinting analyses of a series of mutants for each motif. The original residues are shown above the line, and the substitutions are given below. A thiostrepton-treated control reaction (TS) is shown in the left lane.
The T2 motif is FWNSG-stop. Although two additional sites of stalling are detectable in the toeprinting assay (Fig. 2A), only trace amounts of the corresponding peptides were detected in the MS analysis, suggesting that stalling during termination was the predominant cause of tmRNA tagging in vivo. The mutagenesis and reselection experiments show conservation of the final four codons WNSG (each with P < 0.01), although some flexibility is evident. Asn and Asp occur at the −3 position, and Ser and Thr occur at the −2 position (Fig. 3A). We found that the Asn to Asp mutation and the Ser to Thr mutation both retain significant stalling activity, whereas replacing any residue in the FWNSG motif with Ala abolishes stalling in the toeprinting assay (Fig. 3B). These mutations also can occur in combination: Changing the final three amino acids from NSG to DTG had little or no effect on stalling efficiency.
The T3 motif, DTS-stop, is sharply defined by the consensus sequence from the mutagenesis and reselection data. Each codon is conserved with a P value < 0.001 (Fig. 3A). Although the Asp and Thr codons are invariant, in a few surviving clones the final Ser codon is replaced with Gly. Toeprinting analyses confirm that the Ser to Gly mutation has little or no effect, whereas mutating any residue in the DTS sequence to Ala abolishes stalling (Fig. 3B). Because the T2 motif was able to accommodate either Asp or Asn at the first codon and Thr or Ser at the second, we reasoned that T3 also might exhibit some sequence plasticity. We found, however, that both the Asp to Asn and the Thr to Ser mutations strongly inhibit stalling. We conclude that DTS-stop or DTG-stop is sufficient for stalling ribosomes but that NSG-stop stalls poorly without the upstream FW residues found in T2. In support of this conclusion, we found that replacing the last three residues of T1 with DTS yields robust stalling, but replacing them with NSG does not (Fig. 3B, Top). Finally, we note that DTS stalls only during the termination step, not during elongation. Although stalling occurs at both UAG [recognized by release factor 1 (RF1)] and UGA [recognized by release factor 2 (RF2)] stop codons, mutating the stop codon to an Ala sense codon abolishes stalling (Fig. 3B). Taken together, our analysis of the T1–T3 motifs show that short, polar peptides can induce ribosome stalling at stop codons.
Stalling Motifs That Inhibit Elongation.
Characterization of the two elongation motifs with the strongest toeprints, E1 and E2, revealed that the amino acid Pro plays a dual role in both motifs. The toeprinting data indicate that stalling occurs with the first Pro codon in the P site and the second Pro codon in the A site (Fig. 2A). Analysis of tmRNA-tagged cI protein by MS confirmed that ribosomes stall in this same position in vivo (Fig. 2B). Randomization of the E1 motif and reselection define the key residues as RxPP, where “x” can be Ser, Ala, Gly, or Pro (Fig. 4A). Similarly, the consensus E2 motif is HGPP. In both cases, the two Pro codons are invariant, and mutation of either to Ala completely abolishes stalling in toeprinting assays (Fig. 4B). An analysis of the nucleotide sequence of surviving clones in the E2 library shows that there is strong selection for the first two nucleotides of each Pro codon but not for the third (Fig. 4A). This observation suggests that the amino acid is critical, not the nucleotide sequence, or even a particular tRNA.
Fig. 4.
Key residues in motifs that block elongation. (A) The consensus sequence of the E1 and E2 motifs was determined through random mutagenesis and reselection. In addition, the nucleotide consensus for the HGPP codons is shown below E2. (B) Toeprinting assays reveal the effects of substitutions in the E1 motif (RxPP) (Left) and the E2 motif (HGPP) (Right). (C) Toeprinting assays of E1 and E2, including the minimal motif alone (4), the minimal motif plus seven additional upstream codons (11), and the full-length construct (FL). Ch1 is a chimera containing the minimal motif of E1 (RSPP) with seven upstream codons taken from E2. Ch2 corresponds to the minimal motif of E2 (HGPP) with seven upstream codons from E1.
Although these two motifs have clear similarities, a subtle context dependence arises from residues upstream of the consensus motif. At first glance, it seems that E1 and E2 are examples of the same family, R/HxPP, where “x” is a small amino acid. The first residue (R or H) is critical for both motifs: Stalling is lost when it is mutated to Ala. However, replacing the Arg residue in E1 with His significantly reduced stalling levels, as did replacing the His residue of E2 with Arg (Fig. 4B). We thought that the requirement for a particular residue (Arg or His) might arise from the peptide context; Perhaps R/HxPP is sufficient for stalling, but the upstream sequence imposes a specific requirement for either Arg or His. In support of this idea, we found that the minimal motifs of E1 (RSPP) and E2 (HGPP) induce robust stalling, as strong as the original 20-codon sequence (compare the lanes labeled “4” and “FL” in Fig. 4C). This result demonstrates that no upstream residues are necessary. We then created chimeras containing elements of both motifs. In chimera 1 (Ch1), the seven residues upstream of the E2 motif were put upstream of the minimal E1 motif, RxPP. As shown in Fig. 4C, the toeprint was reduced dramatically, indicating that these additional seven residues block stalling at the minimal E1 motif. Likewise, stalling was inhibited in chimera 2 (Ch2), where the seven residues upstream of the E1 motif were added to the N terminus of the minimal E2 motif, HGPP. These findings show that secondary elements in the extra seven residues somehow restrict which residues work in the minimal motif, making the sequence specific for either Arg or His.
Not only is stalling by the E1 motif sensitive to the upstream peptide sequence, it also is sensitive to the length of the nascent polypeptide. Although ribosomes stall robustly on the minimal E1 sequence RSPP, stalling is reduced dramatically when a longer truncated form of the E1 motif is translated, YYPPRLSRSPP (compare lanes labeled “4” and “11” in Fig. 4C). Note that we have not altered the motif sequence; this truncated form is part of the full motif. The explanation for this paradox may be that the RSPP residues cannot achieve the right conformation in the context of the longer YYPPRLSRSPP sequence. In the context of the full motif, however, the peptide conformation permits productive stalling; in effect, the stalling suppressor is suppressed.
Nascent Peptides Dramatically Influence Rates of Peptide Release and Peptidyl Transfer.
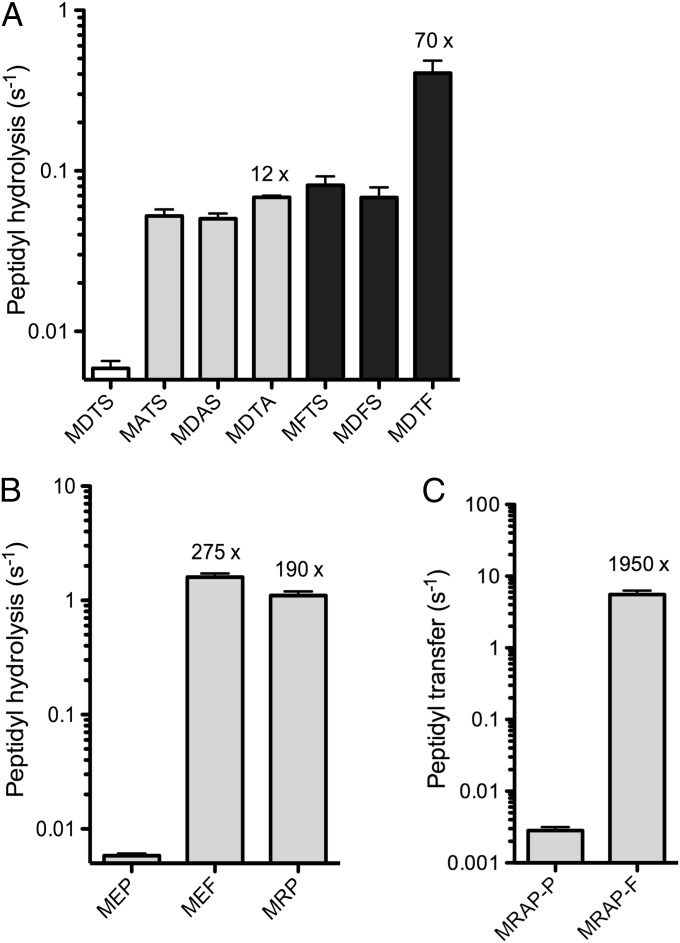
One limitation in the study of ribosome stalling has been the lack of biochemical tools to probe the mechanism. Because they are short, simple, and robust, the motifs defined above enabled us to recapitulate stalling in pre–steady-state kinetic assays (28–30). For example, we observed inhibition of peptide release on the T3 motif, DTS-stop. From purified components, we assembled ribosome complexes with MDTS peptidyl-tRNA bound in the P site and the UAA stop codon poised in the A site. We reacted this complex with saturating concentrations of RF1 and measured the rate of release of the peptide from its tRNA. We found that this rate was quite slow, 0.006 s−1.
Peptides containing single Ala substitutions (MATS, MDAS, MDTA) were released about 10-fold faster than the original MDTS peptide (Fig. 5A). This result corroborates the consensus sequence and toeprinting data, indicating that each of these three residues is important for stalling by the T3 motif. In addition, we found that although Phe substitutions at the first two positions yielded a similar 10-fold decrease in stalling, changing the final residue to Phe decreased stalling by 70-fold. We speculate that the larger Phe side chain prevents Asp and Thr from making necessary contacts because of steric effects or conformational changes in the peptide.
Fig. 5.
Pre–steady-state kinetic analysis of ribosome stalling. (A) A ribosome complex containing MDTS-peptidyl-tRNA in the P site and the UAA stop codon in the A site was reacted with RF1 to determine the rate of peptidyl-tRNA hydrolysis. Ala (gray) and Phe (black) mutants were characterized to test the contribution of each residue in the DTS motif. (B) A ribosome complex containing MEP-peptidyl-tRNA or derivatives was reacted with RF1. (C) Peptidyl-transfer rates for the E1 motif were obtained by reacting ribosome nascent chain complexes with excess ternary complex composed of Pro- or Phe-tRNA, EF-Tu, and GTP.
In considering the importance of the size and geometry of the final residue, we wondered about the effect of Pro on termination rates. In this selection and in previous studies (13, 24), proteins ending in Glu-Pro, Asp-Pro, and Pro-Pro were found to induce high levels of stalling and tagging by tmRNA in vivo. We measured the rate of release of the MEP peptide by RF1 and found it to be 0.006 s−1, about the same as MDTS. Changing the last residue from Pro to Phe increased the rate of release 275-fold (MEF; Fig. 5B), highlighting the importance of Pro at the final position. The −2 position also is critical: Replacing Glu with Arg led to a 190-fold rate increase (MRP; Fig. 5B). These data are consistent with earlier findings that when a protein ends in Pro, tagging by tmRNA is lowest when Arg is found in the −2 position (24). Taken together, these data show that the peptide sequence, and in particular the final three amino acids of the protein, can have a profound effect on the rate of release by RF1. The effect is primarily on catalysis, not binding, because saturating concentrations of RF1 were used (representative rate data are shown in Fig. S1).
In addition to these studies on peptide release, we also recapitulated stalling during peptidyl transfer in a pre–steady-state kinetic assay (28). To characterize the E1 motif, RxPP, further, we assembled ribosome complexes on an mRNA encoding MRAPP. A ribosome complex containing MRA peptidyl-tRNA was formed, purified, and reacted with a ternary complex of Pro-tRNA, elongation factor Tu (EF-Tu), and GTP. Products were resolved by electrophoretic TLC, and, importantly, the appearance of MRAP and MRAPP could be distinguished. The rate of peptidyl transfer of MRAP to Pro-tRNA was determined to be 0.003 s−1. In contrast, we found that the rate of peptidyl transfer of MRAP to Phe-tRNA was 5.5 s−1, nearly 2,000-fold faster (Fig. 5C). These data show that the incoming aminoacyl-tRNA in the A site can have dramatic effects on peptidyl transfer rates. Taken together, these studies of the kinetics of peptide release and peptidyl transfer demonstrate the value of these motifs as tools for future mechanistic studies.
Stalling Occurs at Various PPX Motifs in Vitro.
It is striking that peptidyl transfer is blocked in the E1 and E2 motifs by such short peptide sequences. We therefore asked if a motif we had identified previously, FxxYxIWPPP, might also contain a shorter minimal motif capable of inducing stalling (13). To test this possibility, we monitored stalling using toeprinting assays with various derivatives of the original FxxYxIWPPP motif. As reported previously, stalling in the full-length sequence occurs with the second Pro codon in the P site and the third Pro codon in the A site. Mutation of the second Pro to Ala abolishes stalling (Fig. 6A).
Fig. 6.
EF-P alleviates stalling at polyproline stretches but not other stalling motifs. (A) Toeprinting analyses of Ala substitutions in the FxxYxIWPPP motif and a truncated derivative, WPPP. (B) Analysis of pausing in the 20 PPX motifs. The arrow points to the toeprint with the second Pro codon in the P site and the X codon in the A site. (C) Toeprinting analyses of three endogenous E. coli genes with polyproline stretches: lepA, ligT, and amiB. (D) Various motifs were translated in the presence or absence of purified EF-P; the relevant toeprint is labeled with an arrow. (E) MWPPP and MFQKYGIWPPP were translated in vitro with [35S]-methionine in the presence or absence of EF-P. Peptidyl-tRNA accumulates in stalled ribosomes; it was visualized by SDS/PAGE and autoradiography after 3- or 7-min reactions.
Additional analysis of the FxxYxIWPPP motif established that three Pro codons are sufficient to induce a high level of stalling in toeprinting assays. Two strong pauses are detected during translation of WPPP and APPP; these correspond to either the first or second Pro codon positioned in the P site (Fig. 6A). Pausing at these two sites is interdependent: Mutation of any of the three Pro codons to Ala reduces stalling at both sites dramatically. Although a low level of stalling is detectable with two consecutive Pro codons in the WAPP and WPPA mutants, these data argue that three Pro codons are necessary and sufficient for robust stalling.
Our previous in vivo studies on the FxxYxIWPPP motif indicated that, in addition to Pro-tRNA, other aminoacyl-tRNAs also can act as poor peptidyl acceptors when bound in the A site, prohibiting peptidyl transfer (13). When the third Pro codon is mutated to Trp or Asp (e.g., FxxYxIWPPW), robust ribosome stalling and tmRNA tagging occur in vivo. We revisited this phenomenon in the context of the shorter, core PPP motif, testing all 20 amino acids as peptidyl acceptors in toeprinting assays. We found that robust stalling by PP(X) occurs when X is an Asn, Asp, Glu, Gly, Pro, or Trp codon (Fig. 6B). These aminoacyl-tRNAs act as poor peptidyl acceptors for peptides ending in Pro-Pro, further defining the scope of this phenomenon.
Stalling at Polyproline Stretches in Endogenous E. coli Proteins.
Because of the simple nature of these motifs, they occur often in endogenous proteins. We next asked how these proteins can be translated in vivo, given the strong pauses that occur in vitro. The E. coli MG1655 genome encodes ∼100 proteins containing three or more consecutive Pro codons, and even more if Asn, Asp, Glu, Gly, or Trp is allowed at the third position. Perhaps the context of the motif within the proteins prohibits stalling, as we have seen with the E1 and E2 motifs when upstream sequences are swapped. To address this question, we performed in vitro translation of LepA and LigT, both of which contain PPP motifs, and AmiB, a protein with eight consecutive Pro codons. As shown in Fig. 6C, toeprinting analyses reveal that protein synthesis of LepA and LigT does stall dramatically at the PPP motif with the second Pro codon in the P site. These are the clear pause sites in these genes. In AmiB, where there are eight Pro codons in a row, the stalling occurs primarily with the second Pro codon in the P site but also to a lesser extent at downstream Pro codons. These data show that endogenous E. coli proteins stall at PPP motifs in vitro; their sequence context does not suppress stalling effectively.
EF-P Alleviates Stalling at Polyproline Stretches but Not Other Stalling Motifs.
During the course of these studies, it was discovered that the translation factor EF-P relieves translational stalling at polyproline stretches, thus explaining how proteins with polyproline sequences are translated efficiently in living cells (31, 32). EF-P has a tRNA-like structure (33) and binds between the E and P sites on the ribosome (34). EF-P is posttranslationally modified (35–37), and the modified lysine residue is likely positioned close to the peptidyl-transferase center. As expected, addition of EF-P to the translation reactions abolishes stalling at the polyproline stretches present in LepA, LigT, and AmiB as seen in the toeprinting assays (Fig. 6D). Furthermore, although a peptidyl-tRNA intermediate is the primary product in the absence of EF-P, synthesis of the full-length protein is strongly enhanced by addition of EF-P (Fig. S2).
We next asked whether EF-P functions universally to alleviate stalling. Toeprinting assays revealed that EF-P has little or no effect on stalling by the E1 (RxPP), T3 (DTS-stop), and Glu-Pro-stop motifs (Fig. 6D). Likewise, EF-P had no effect on stalling by the full-length FxxYxIWPPP motif, although it abolished stalling by the minimal WPPP motif (Fig. 6D). Analysis of the products of in vitro translation further confirmed the ability of EF-P to prevent stalling at WPPP but not at the full-length FxxYxIWPPP motif (Fig. 6E). Translation of the MWPPP peptide led to the synthesis of an ∼20-kDa intermediate, consistent with the size of the tRNA with a tetrapeptide attached. This intermediate does not accumulate, however, when EF-P is added to the translation reaction (Fig. 6E). In contrast, during MFQKYGIWPPP synthesis, peptidyl-tRNA accumulates regardless of whether EF-P is added. Taken together, these data show that our motifs identified by genetic selection are resistant to EF-P, further defining its scope of action.
Stalling Motifs Have Been Selected Against.
The motifs we identified from random libraries induce ribosome stalling both in vivo and in vitro. What effect would these motifs have on the synthesis of endogenous proteins in vivo? Are they found in endogenous bacterial proteomes? Or have they been selected against?
Analysis of bacterial proteomes supports the idea that, in fact, some of these motifs have been selected against. We searched for stalling motifs in ∼13.7 million bacterial proteins in the NCBI RefSeq database from 4,277 organisms in the bacterial kingdom. We found that the DTS motif occurred sixfold less at the C terminus of proteins than one would expect based on amino acid frequencies in this dataset. Glu-Pro-stop and Pro-Pro-stop were underrepresented by 1.7- and 1.8-fold, respectively, and Asp-Pro-stop was underrepresented by 3.6-fold. Elongation motifs RSPP and HGPP also were underrepresented, by 3.3- and 2.8-fold, respectively. All these changes were significant with P < 10−6. These findings suggest that stalling motifs may have been broadly selected against during the course of evolution. In contrast, the six PP(X) motifs that stall in vitro occur at expected levels in bacterial proteomes; when analyzed together, they were slightly enriched at <0.1% above the expected frequency (P > 0.35). We conclude that there is no evidence of selection against the six stalling PP(X) motifs.
Stalling Occurs at Several Motifs in Endogenous E. coli Proteins.
Ribosome-profiling data published by Weissman and coworkers (38) allow us to quantify ribosome occupancy at potential stalling sites in endogenous E. coli proteins. We define a pause score as the number of reads at the pause site divided by the median number of reads for the entire ORF. Sites where ribosomes are enriched have higher pause scores than sites of low ribosome occupancy. We calculated pause scores for all 8,000 tripeptide combinations. The highest reliable pause score was 5.9 ± 0.3 for the tripeptide GGT. Indeed, many of the top 100 tripeptide motifs are rich in Gly residues (Table S2). In contrast, PPP has a pause score of 4.4 ± 0.4, and other PPX stalling motifs have lower scores (PPD = 4.3 ± 0.5; PPG = 3.9 ± 0.4; PPE = 3.1 ± 0.3).
Stalling motifs identified in our genetic selection have higher pause scores in the ribosome-profiling data. We first looked at motifs that block termination. No proteins in the E. coli MG1655 genome end in Pro-Pro-stop. At the Asp-Pro-stop sequence in the SgrR gene, the ribosome pauses robustly with a score of 84, meaning that a ribosome is 84 times more likely to be found at this site than a typical position in the SgrR ORF. The Glu-Pro-stop motif in TreF has a pause score of 28. None of our termination motifs (T1–T3) are found in this strain, but the RxPP elongation motif in the RecG protein has a pause score of 14, and the HGPP motif in the YaaX protein has a pause score of 10. Ribosome profiles for these four genes are shown in Fig. 7. These data argue that these motifs indeed delay ribosomes in vivo, even in endogenous proteins.
Fig. 7.
Ribosome-profiling data highlight pauses at stalling motifs in vivo. Ribosome density is shown across four genes that contain stalling motifs highlighted in red. Read densities are reported in units of reads per million mapped reads. These analyses are of published datasets of E. coli MG1655 ribosome-profiling experiments (38).
Discussion
Our selection identified nascent peptide motifs that induce ribosome stalling and tmRNA tagging. Several lines of evidence support the claim that the peptide is the primary cause of stalling. First, stalling occurs in a reconstituted in vitro translation system, where cleavage or degradation of the mRNA is ruled out. Second, the consensus sequences show a high degree of conservation of the first two nucleotides in a codon but considerable variation at the wobble position. Third, the nascent peptide and incoming amino acid seem to play a key role in slowing reaction rates in kinetic assays. We cannot rule out a role for associated tRNAs, however, because many aspects of tRNA structure and function are common among the isoacceptors for a given amino acid.
Several of the peptides we identified inhibit translational termination with small, polar residues at the C terminus. For example, the sequence DTS-stop is sufficient for stalling. This motif expands to D/N, T/S, and S/G if additional aromatic residues are present upstream. With an even longer stretch of hydrophobic residues, termination is slowed by a single Thr residue at the C terminus of the WILFxxT-stop motif. Perhaps the potential of Thr to block peptide release explains why Thr is the most underrepresented residue at the C terminus of bacterial proteins (2.1-fold). These findings define this class of stalling peptides and argue that release factors are sensitive to the sequence of the polypeptide being released.
In these examples, upstream aromatic or hydrophobic residues probably enhance stalling by increasing the binding of the peptide to the ribosomal exit tunnel. Conserved upstream residues in natural stalling motifs such as SecM and TnaC work in the same way (15, 39). However, in some of the motifs we identified, the upstream sequence has the opposite effect, suppressing stalling by a motif that otherwise stalls effectively. In the R/HxPP motif, for example, upstream residues impose sequence specificity for either Arg or His. In addition, although the minimal motif R/HxPP is sufficient to block peptidyl transfer, stalling is suppressed if the peptide is of intermediate length (∼11 amino acids), perhaps because specific upstream residues induce a peptide conformation that disrupts interactions between the ribosome and the minimal motif. We speculate that longer versions of this motif (∼20 amino acids) stall efficiently because the N terminus of the peptide moves past the L4/L22 constriction, restricting which conformations are available to the nascent peptide.
Pro poses a particular challenge for the ribosome, serving as both a poor peptidyl donor and a poor peptidyl acceptor (40, 41). These roles are combined in the PPP motif, where the ribosome stalls with the second Pro codon in the P site and the third in the A site, so that transfer of peptidyl-tRNA to Pro-tRNA is blocked. We found that peptidyl transfer to Asn-, Asp-, Glu-, Gly-, or Trp-tRNA also is inhibited after two Pro residues. Two of these tripeptide motifs, PPD and PPE, were shown recently to pause ribosomes in mammalian cells (42), suggesting that stalling at Pro-rich motifs may be a general phenomenon, not limited to bacteria.
Stalling by the PPP motif is abolished by EF-P, explaining how ∼100 E. coli proteins containing this motif are translated in vivo (31, 32). Proteins with PPD, PPE, PPG, PPN, or PPW probably also require EF-P for their synthesis. The fact that the PP(X) motifs are not underrepresented in bacterial proteomes suggests that EF-P alleviates stalling by these motifs efficiently. In contrast, EF-P has no effect on our newly identified motifs, even those including Pro residues, such as FxxYxIWPPP and RxPP. This observation makes sense, because these motifs were selected for their ability to induce tmRNA tagging in vivo, where EF-P is present. We speculate that the aromatic residues in FxxYxIWPPP interact with the ribosomal exit tunnel to stabilize the stalled conformation, blocking the activity of EF-P, which otherwise could resolve stalling by the PPP motif. In addition, it seems that EF-P primarily affects peptidyl transfer and not peptide release, given that it does not alleviate stalling at Glu-Pro-stop, even though stalling occurs with a Pro codon in the P site. These findings begin to define the scope of EF-P’s ability to relieve translational pauses.
The requirement in the PP(X) motifs for specific aminoacyl-tRNAs bound in the A site is intriguing. In a similar manner, Asp-, Glu-, Gly-, and Trp-tRNA were also found to act as poor peptidyl acceptors in the analysis of the ErmAL1 stalling peptide by Ramu et al. (12). In contrast, the strongest stalling in the ErmAL1 study was with Lys-, Arg-, and His-tRNA, but these did not stall robustly after two Pro codons. It appears that different nascent peptides modulate the reactivity of different subsets of aminoacyl-tRNAs.
Insight into the mechanism of ribosome stalling has been limited by the lack of biochemical assays to measure directly the individual steps in the synthesis of stalling peptides. In many studies, stalling is inferred from changes in gene expression or tagging of the protein by tmRNA. Although stalling can be detected directly in toeprinting assays, this method cannot always pinpoint the precise step that is blocked or determine reaction rates. In theory, many steps in the translational cycle could be inhibited during ribosome stalling, including tRNA or factor binding, translocation, peptide-bond formation, and peptide release. We were able to recapitulate stalling in pre–steady-state kinetic assays using the motifs we identified, showing that peptide release and peptidyl transfer are inhibited by roughly 100- and 1,000-fold, respectively. The fact that our motifs are short makes interpretation of their interactions with the ribosome far simpler than interpreting the function of natural motifs such as SecM. Our stalling peptides cannot contact the L4/L22 constriction; we speculate that they interact directly with rRNA nucleotides near the peptidyl-transferase center to induce conformational changes that inhibit chemistry at the active site. Going forward, these assays will allow us to determine the role of the peptide, tRNA, and rRNA sequence in the stalling mechanism.
What do our data tell us about the likelihood of finding more stalling motifs in the future? On one hand, well-characterized motifs containing Pro residues appeared often in the two-hybrid selection. The Pro-stop motif, for example, was found in half of the clones that induce tmRNA tagging. On the other hand, several additional motifs were discovered that block termination with polar residues, and the cause of stalling remains unknown for several elongation motifs. Given the tiny fraction of sequence space we accessed in our library and the high rate of survival in the selection, it seems likely that more motifs remain undiscovered.
Ribosome profiling has potential for shedding light on variation in translational rates and for identifying additional stalling motifs. A recent ribosome-profiling study showed that most strong pauses could be explained by Shine–Dalgarno-like sequences 6–12 nt upstream of the pause site (38). In the E5 motif (Table S1), stalling occurs with the nucleotides GGAGGA within 6–12 nucleotides of the A site codon, consistent with this report. Our analysis of the same profiling dataset revealed that Gly-rich tripeptide motifs have the highest pause scores in E. coli. Because Gly is encoded by GGN, it may be tempting to attribute this result to SD-like sequences. However, the middle codon of the tripeptide is positioned in the P site in our analysis, making it unlikely that these codons bind strongly to the anti-SD sequence in 16S rRNA during pausing. Consistent with the high pause score for the GGG tripeptide (Table S2), we note that the E6 motif stalls on three Gly codons with the second Gly in the P site (Table S1).
Our data argue that bacterial proteomes have been shaped by the demand for translational efficiency. The motifs that we identified were three- to sixfold underrepresented in a dataset including ∼13.7 million bacterial proteins. For those stalling motifs that are retained in the E. coli genome, we find substantial accumulation of ribosome density at the motif in ribosome-profiling datasets. These data show that short motifs stall on endogenous proteins in vivo and have the potential to modulate protein synthesis rates in a biologically relevant way.
Experimental Procedures
Two-Hybrid Selection.
The selection for stalling motifs is based on Bacteriomatch II (Agilent). Residues 1–117 of SspB were fused to the C terminus of RpoA. To generate the library, 20 random codons were added to the 3′ end of the cI coding sequence. A modified tmRNA encoding the ANDENYALDD tag was expressed in the reporter strain, which lacks hisB and expresses HIS3 from a weak promoter downstream of cI binding sites. A library of 3 × 108 cI mutants was introduced into the reporter strain, and transformants were plated at 30 °C on M9 minimal medium with His dropout supplement, 10 μM isopropylthio-β-galactoside, and 5 mM 3-aminotriazole. The cI gene was PCR amplified from the pool of surviving clones, inserted into fresh expression vector, and passaged through the selection again. About 10% of clones survived in this second round, and 150 from this enriched pool were sequenced. Additional details on plasmid construction and the cI controls are given in SI Experimental Procedures.
In Vitro Translation Assays.
In vitro translation and toeprinting assays were performed in the PURExpress translation system (New England BioLabs) as described previously (13). Each set of experiments was repeated at least twice. The DNA constructs are described in SI Experimental Procedures. Where indicated, 1 μM EF-P was added to the translation reaction. Modified EF-P was purified as described previously (36).
MS.
cI clones were expressed in pET-15b in BL21 (DE3) cells together with a modified tmRNA encoding the ANDHHHHHHD tag. Tagged cI was purified over Ni-NTA resin and digested with trypsin, and the C-terminal–tagged peptide was purified and analyzed as previously described (13).
Kinetics.
Ribosome nascent chain complexes were assembled and reacted with excess release factor 1 or excess ternary complex containing EF-Tu, GTP, and aminoacyl-tRNA at 23 °C in polymix buffer. Reported rates are the average of three independent experiments, and the SE is given. Details of the reaction conditions and materials are discussed in SI Experimental Procedures.
Statistical Analysis.
We developed a likelihood ratio test to detect conserved motifs in sequences of surviving clones from the two-hybrid selection. Under the null hypothesis, we assume that the mutation rate is uniform across all the bases in the sequence. Enriched motifs therefore are the locations that deviate from the overall frequencies across the entire 20-codon sequence. Details are provided in SI Experimental Procedures.
Pause scores for specific stalling motifs were obtained from E. coli MG1655 ribosome-profiling data from the Weissman laboratory (38). The pause score for known motifs was computed by averaging the number of reads per base over the P-site codon and two codons downstream, normalized to the median number of reads per base for the given ORF. This approach was used because it effectively captured the increased read density in the SecM motif. Pause scores for tripeptides (Table S2) were computed with the average of density at all three codons and were calculated using only well-translated genes (>10 reads per million kilobases). Motifs with fewer than 20 occurrences in the genome were discarded, as were motifs with higher than 25% error in the pause score. The error in the distribution in the pause scores was computed by bootstrapping.
Supplementary Material
Acknowledgments
We thank Hiroji Aiba for sharing antibodies against the tmRNA-DD tag; Levi Lowry for constructing plasmids; Bruce Jackson, Aman Makaju, and Ryan Taylor for assistance with MS; Megan McDonald and Hani Zaher for help in developing the kinetic assays; and Rachel Green for comments on the manuscript. This work was supported by National Institutes of Health Grant GM77633 (to A.R.B.). N.R.G. was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219536110/-/DCSupplemental.
References
- 1.Ito K, Chiba S, Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun. 2010;393(1):1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71(4):811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 3.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108(5):629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 4.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28(22):3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297(5588):1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 6.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331(6017):586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 7.Lovett PS, Rogers EJ. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60(2):366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16(3):274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 9.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34(1):16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell. 2005;19(3):333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30(2):190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 12.Ramu H, et al. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41(3):321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem. 2009;284(50):34809–34818. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell. 2006;22(4):545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Bhushan S, et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9(1):e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vázquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29(18):3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 18.Janssen BD, Hayes CS. The tmRNA ribosome-rescue system. Adv Protein Chem Struct Biol. 2012;86:151–191. doi: 10.1016/B978-0-12-386497-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386(6625):627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 20.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci USA. 2000;97(13):7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289(5488):2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 22.Wah DA, et al. Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol Cell. 2003;12(2):355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- 23.Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18(16):4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277(37):33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 25.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271(5251):990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 26.Sunohara T, Abo T, Inada T, Aiba H. The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA. 2002;8(11):1416–1427. doi: 10.1017/s1355838202020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche ED, Sauer RT. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem. 2001;276(30):28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- 28.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117(5):589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 29.Shaw JJ, Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell. 2007;28(3):458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol. 2007;14(1):30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- 31.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 32.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339(6115):85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 33.Hanawa-Suetsugu K, et al. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci USA. 2004;101(26):9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: The structure of EF-P bound to the 70S ribosome. Science. 2009;325(5943):966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarre WW, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39(2):209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peil L, et al. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol. 2012;8(8):695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- 37.Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol. 2010;17(9):1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- 38.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484(7395):538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seidelt B, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326(5958):1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muto H, Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem Biophys Res Commun. 2008;366(4):1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- 41.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283(47):32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 42.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.