Abstract
Mixed-lineage leukemia (MLL) fusions are potent oncogenes that initiate aggressive forms of acute leukemia. As aberrant transcriptional regulators, MLL-fusion proteins alter gene expression in hematopoietic cells through interactions with the histone H3 lysine 79 (H3K79) methyltransferase DOT1L. Notably, interference with MLL-fusion cofactors like DOT1L is an emerging therapeutic strategy in this disease. Here, we identify the histone H2B E3 ubiquitin ligase ring finger protein 20 (RNF20) as an additional chromatin regulator that is necessary for MLL-fusion–mediated leukemogenesis. Suppressing the expression of Rnf20 in diverse models of MLL-rearranged leukemia leads to inhibition of cell proliferation, under tissue culture conditions as well as in vivo. Rnf20 knockdown leads to reduced expression of MLL-fusion target genes, effects resembling Dot1l inhibition. Using ChIP-seq, we found that H2B ubiquitination is enriched in the body of MLL-fusion target genes, correlating with sites of H3K79 methylation and transcription elongation. Furthermore, Rnf20 is required to maintain local levels of H3K79 methylation by Dot1l at Hoxa9 and Meis1. These findings support a model whereby cotranscriptional recruitment of Rnf20 at MLL-fusion target genes leads to amplification of Dot1l-mediated H3K79 methylation, thereby rendering leukemia cells dependent on Rnf20 to maintain their oncogenic transcriptional program.
Keywords: epigenetics, cancer
The mixed-lineage leukemia (MLL) protooncogene (also called MLL1) was first cloned based on its involvement in chromosomal translocations found in leukemia, occurring at a frequency of 5–10% in acute myeloid leukemia (AML) and acute lymphoblastic leukemia (1). MLL translocations are especially common in infant acute leukemias and in secondary AMLs that arise following cancer treatment with topoisomerase II inhibitors (1). In both of these clinical settings, MLL rearrangements are associated with a poor outcome (1). MLL-rearranged leukemias are notable for (i) a paucity of additional gene mutations found in these cases (although NRAS mutations are among the most common cooperating lesion) (2), (ii) a unique transcriptional signature characterized by overexpression of homeobox A (HOXA) genes (1), and (iii) an absence of available targeted therapies. Regarding the latter, the current clinical management of MLL-rearranged leukemia with cytotoxic chemotherapy is similar to that of other genetic subtypes of leukemia that fall into the same risk category (1). Hence, a strong rationale exists to define unique therapeutic targets tailored to the molecular pathogenesis of this disease.
MLL encodes a chromatin regulator belonging to the SET1 family of histone H3K4 methyltransferases, which form multisubunit protein complexes that maintain active transcription (1, 3). MLL translocations found in leukemia generate fusions that encode an N-terminal fragment of MLL (which lacks the methyltransferase domain) linked to a C-terminal fragment of various partner proteins (1). This N-terminal fragment of MLL physically interacts with Menin, CpG-rich DNA, and the polymerase associated factor (PAF) complex, which collectively are sufficient for chromatin occupancy at specific genes such as HOXA9 (4–6). The translocation partners of MLL can be highly diverse; however, fusions with AF9 are the most common in AML (1). The aberrant recruitment of AF9-associated proteins to MLL-occupied genes (e.g., HOXA9) leads to transcriptional deregulation and, consequently, leukemia initiation. Several binding partners of AF9 are involved in MLL-AF9–mediated leukemogenesis, including the super elongation complex (SEC), the histone H3K79 methyltransferase DOT1L, and the chromodomain-containing protein CBX8 (7–10).
Targeting of MLL-fusion–associated factors has emerged as a promising therapeutic strategy in this subtype of leukemia. Genetic studies have validated that conditional inactivation Dot1l specifically inhibits progression of MLL-rearranged leukemias in vivo, in association with reduced expression of MLL-AF9 target genes (11, 12). Furthermore, a small-molecule inhibitor of DOT1L demonstrates antileukemia activity in MLL-rearranged disease models (13). Genetic or pharmacological disruption of the MLL:Menin interaction has also been validated as means of suppressing proliferation of MLL-fusion leukemias (5, 14). Targeting of DOT1L or Menin in cells that lack MLL rearrangements leads to remarkably little toxicity, suggesting a potential therapeutic window for this general approach (13, 14).
Ring finger protein 20 (Rnf20) (also called Bre1a) is the major H2B-specific ubiquitin ligase in mammalian cells that targets lysine 120 for monoubiquitination [H2B ubiquitination (H2Bub)] (15–18). Rnf20 can be recruited to chromatin via the PAF complex, resulting in the accumulation of H2Bub at genes in a transcription-dependent manner (19–22). Although found broadly at active genes, H2Bub is not strictly required for transcription elongation, but instead performs specialized roles in regulating nucleosome dynamics (22), the DNA damage response (23, 24), and the activity of other histone-modifying enzymes (19, 21, 22). Regarding the latter, it is known that the presence of H2Bub on nucleosomes can stimulate the activity of DOT1L in catalyzing H3K79 methylation in vitro and in vivo through apparent allosteric regulation (19, 25). H2Bub also promotes H3K4 methylation by the SET1 family of lysine methyltransferases (26). The role of H2Bub in supporting histone methylation in mammalian cells appears to be dependent on the specific cell type and/or on the specific genomic region examined (17, 27, 28). Although substantial evidence indicates cross talk between H2Bub and H3K79 methylation in various contexts, it has yet to be addressed whether mammalian Rnf20 supports the biological functions performed by Dot1l in vivo.
Here, we identify a role for Rnf20 in the pathogenesis of MLL-fusion leukemia. Suppression of Rnf20 leads to impaired leukemia progression in vivo associated with reduced expression of MLL-AF9 target genes, a finding we link to a defect in maintenance of local H3K79 methylation. Hence, our findings implicate Rnf20 as a key requirement for MLL-fusion leukemia through regulatory cross talk with Dot1l.
Results
Rnf20 Is Required for Proliferation of MLL-Fusion Leukemia Cells.
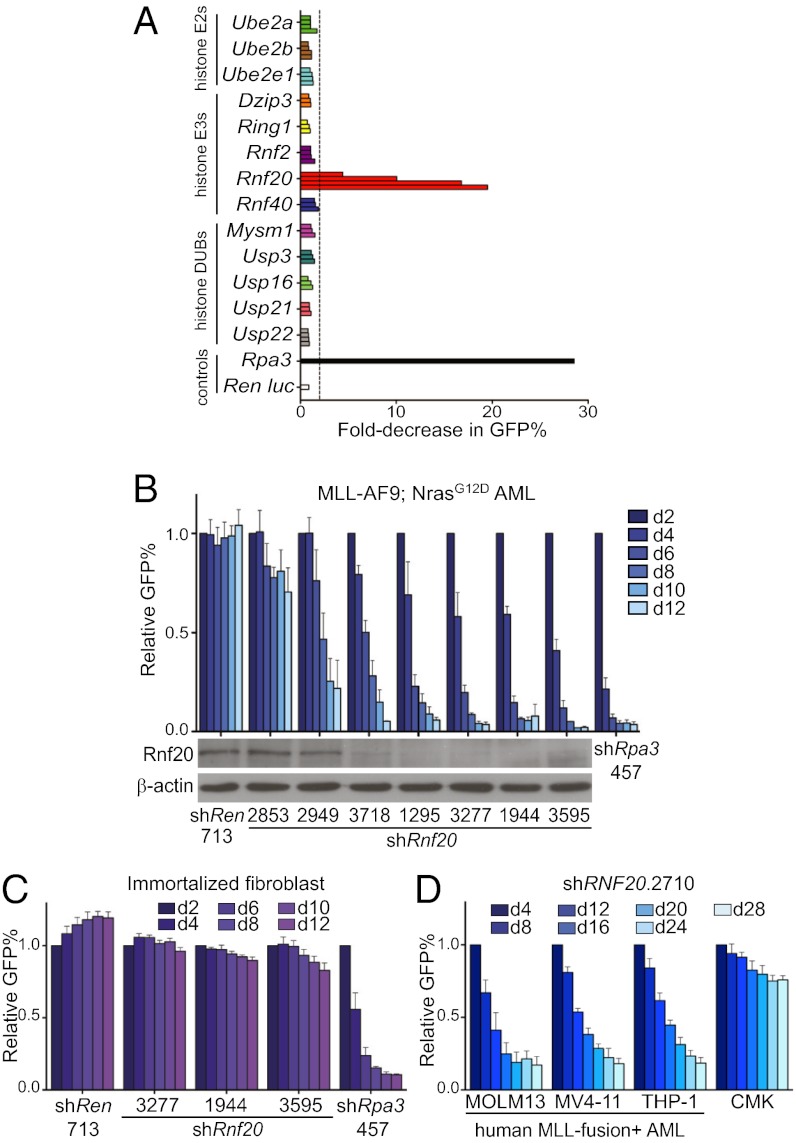
Based on the known role of H2Bub in stimulating H3K79 methylation in various systems (19, 25, 29), we hypothesized that Rnf20 might support the leukemogenic function of Dot1l in MLL-rearranged disease. We approached this question in a systematic fashion in which we suppressed expression of several histone monoubiquitination regulators and evaluated the impact on proliferation of MLL-fusion+ leukemia cells. We constructed a set of 45 shRNA vectors targeting the known E2 conjugating enzymes, E3 ligases, and deubiquitinating enzymes with specificity for histone H2A or H2B for monoubiquitination (13 genes in total) (30). Each shRNA (linked to a GFP reporter) was retrovirally introduced into cells derived from a genetically engineered mouse model of MLL-AF9/NrasG12D AML, shown previously to recapitulate an aggressive, chemotherapy-resistant disease subtype (31). The GFP positivity of partially infected cells was tracked by flow cytometry over 8 d to monitor the relative rate of accumulation of shRNA+/GFP+ cells relative to shRNA−/GFP− cells. Four shRNAs targeting Rnf20 were found to inhibit leukemia proliferation/viability compared with a negative control shRNA targeting Renilla luciferase and a positive control shRNA targeting the replication protein A3 (Rpa3) (Fig. 1A) (32). Notably, all of the other shRNAs targeting ubiquitination regulators led to negligible effects on leukemia proliferation (Fig. 1A). This includes shRNAs targeting Ube2a and Ube2b, which encode Rad6a and Rad6b, respectively, the E2-conjugating enzymes that work in conjunction with Rnf20 to catalyze H2Bub (21). Prior studies suggest that Rad6a and Rad6b function redundantly to maintain H2Bub in mammalian cells (21), which may explain why these shRNAs individually failed to evoke a phenotype in the screen. Based on these results, we focused our subsequent evaluation on Rnf20 as a unique chromatin regulator requirement in MLL-rearranged leukemia.
Fig. 1.
Rnf20 is required for proliferation of MLL-AF9/NrasG12D leukemia cells in vitro. (A) shRNA screen of histone monoubiquitination regulators evaluating for effects on expansion of MLL-AF9/NrasG12D acute myeloid leukemia cells. Each horizontal bar represents an individual LMN-shRNA targeting the indicated gene. The fold decrease in GFP-positive cells between day 2 and day 8 was used to infer the competitive disadvantage of shRNA+/GFP+ cells relative to untransduced cells in each culture. The dashed line indicates an arbitrary twofold cutoff. shRNAs targeting Rpa3 and Renilla luciferase are included as positive and negative controls, respectively. (B) Correlation between degree of Rnf20 knockdown and degree of proliferation inhibition for seven independent shRNAs. The percentage of shRNA/GFP-positive leukemia cells at the indicated time points is shown in the upper panel, normalized to day 2. Mean of three independent experiments is shown. In the lower panel, Western blotting of whole-cell extracts prepared from G418-selected immortalized mouse embryonic fibroblasts transduced with the indicated LMN shRNAs. β-Actin is included as a loading control. Representative experiment of three independent repeats is shown. (C) Impact of Rnf20 shRNAs on proliferation of immortalized mouse embryonic fibroblasts. The percentage of shRNA+/GFP+ cells at the indicated time points is shown, normalized to day 2. (D) Impact of a RNF20 shRNA on proliferation of human AML cell lines. The percentage of shRNA+/GFP+ cells at the indicated time points is shown, normalized to day 4. Mean of three independent experiments is shown. All error bars denote SEM.
To determine whether the observed phenotypes represent on-target consequences of Rnf20 knockdown, we cloned three additional shRNAs and analyzed the correlation between knockdown potency and growth inhibition (Fig. 1B). Among the seven Rnf20 shRNAs evaluated, the knockdown efficiency correlated closely with the degree of proliferation inhibition, consistent with on-target effects (Fig. 1B). Knockdown of Rnf20 led to G1 arrest of leukemia cells (using a BrdU incorporation assay), without evidence of myeloid maturation (Fig. S1). Notably, Rnf20 suppression led to only minimal antiproliferative effects in immortalized fibroblasts (Fig. 1C) and in nontransformed hematopoietic cell lines 32D, EML, and G1E (Fig. S2A). Rnf20 shRNAs also showed minimal effects on proliferation of various epithelial cancer cell lines (Fig. S2 B and C). Furthermore, RNF20 knockdown inhibited the proliferation of human AML lines harboring MLL translocations (MOLM-13, MV4-11, and THP-1), whereas the non–MLL-rearranged AML line CMK was affected to a lesser extent (Fig. 1D and Fig. S3). MOLM-13 and THP-1 cells harbor MLL-AF9, whereas MV4-11 harbors an MLL-AF4 translocation, suggesting that RNF20 is required for proliferation in the setting of different MLL-fusion partners. Together, these results suggest that Rnf20 is required for proliferation of MLL-fusion leukemias in vitro.
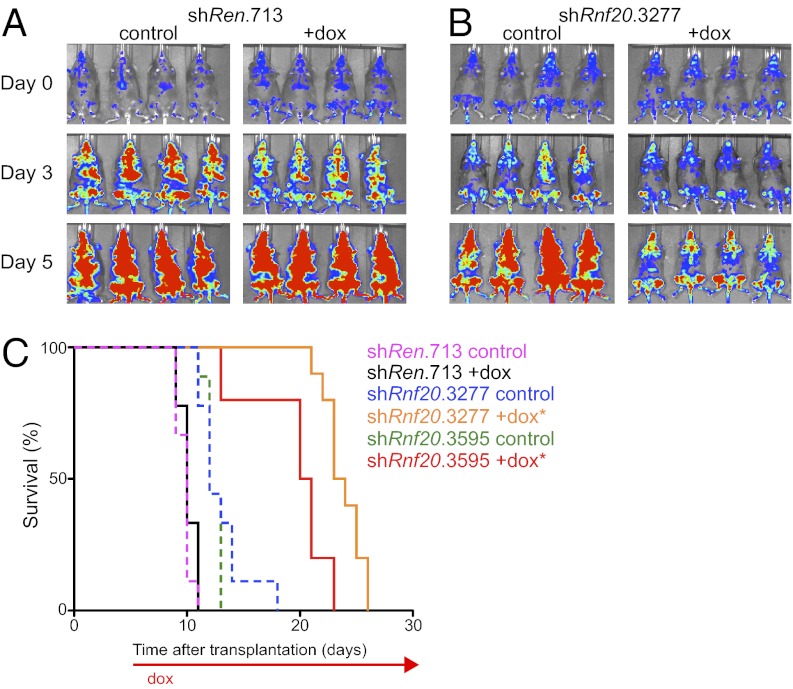
We next considered whether Rnf20 was required for leukemia proliferation in vivo. For this purpose, we used a Tet-On+/Luciferase+ MLL-AF9/NrasG12D leukemia line, called RN2 (33). RN2 cells were retrovirally transduced with Rnf20 or control shRNAs in the TRMPV-Neo vector, which links expression of a doxycycline (dox)-inducible shRNA to a dsRed reporter (33). Following neomycin selection, we derived clonal lines by limiting dilution (Fig. S4A). These lines were then transplanted into sublethally irradiated recipient mice followed by dox administration after 5–6 d, when the disease is engrafted and actively expanding. We found that two independent Rnf20 shRNAs inhibited leukemia progression, as indicated by a decreased rate of bioluminescent signal accumulation (Fig. 2 A and B, and Fig. S4 B and C) and an extension in overall survival compared with controls (Fig. 2C). The terminal disease of leukemias experiencing Rnf20 knockdown exhibited a reduced percentage of dsRed positivity compared with the pretransplanted clone (Fig. S4A). This suggests that subclones that have bypassed Rnf20 shRNA expression emerge in vivo under the negative-selection pressure imposed by Rnf20 suppression. These findings confirm that Rnf20 is necessary for rapid leukemia expansion under in vivo conditions.
Fig. 2.
Rnf20 is required for expansion of MLL-AF9/NrasG12D leukemia cells in vivo. (A and B) Bioluminescent imaging. Mice were transplanted with clonal RN2 leukemia lines (MLL-AF9+; NrasG12D+; rtTA3+, firefly luciferase+) transduced with indicated TRMPV-Neo shRNA constructs. Dox was administered upon disease onset, 5–6 d after transplant. Day 0 refers to the start of dox administration. (C) Kaplan–Meier survival curves of recipient mice transplanted with the indicated TRMPV-Neo-shRNA RN2 lines. The interval of dox treatment is indicated by the arrow. Each shRNA group contained 8–10 mice. Statistical significance compared with shRen +dox was calculated using a log-rank test; *P < 0.0001.
Rnf20 Is Required to Maintain Expression of MLL-AF9 Target Genes.
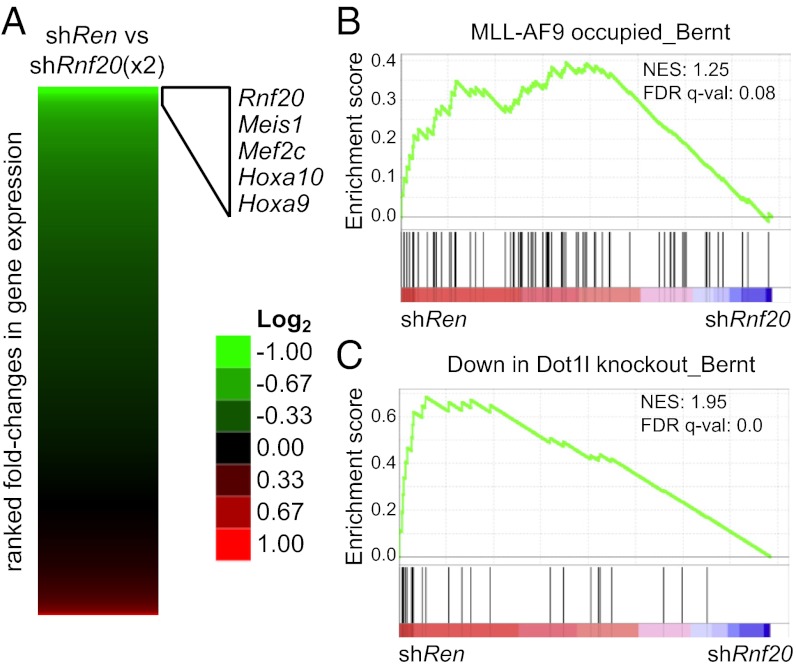
To gain mechanistic insight into the role of Rnf20 in MLL-fusion leukemia, we performed global gene expression profiling. Two independent shRNAs targeting Rnf20 were induced in RN2 cells with dox for 4 d followed by RNA sequencing analysis. We ranked all expressed genes based on their fold change in mRNA level following Rnf20 suppression, which, importantly, validated Rnf20 itself as among the most down-regulated genes (Fig. 3A). This further supports the on-target effects of the shRNAs used here. Inspection of the other affected genes revealed that many of the known direct MLL-AF9 target genes [homeobox A9 (Hoxa9), homeobox A10 (Hoxa10), Meis homeobox 1 (Meis1), and myocyte enhancer factor 2C (Mef2c)] were also among the most down-regulated (Fig. 3A). Interestingly, these changes in expression were unique to Rnf20 suppression, as inhibiting other chromatin regulators (BRD4 or PRC2) in this same leukemia model fails to influence expression of this set of genes (Fig. S5) (34). Using gene set enrichment analysis (GSEA), we further verified a systematic suppression of MLL-AF9 target gene expression upon Rnf20 knockdown (35). We also noted a pronounced decrease in the expression of Dot1l-dependent genes (Fig. 3C), using a gene set defined previously in a Dot1l−/−; MLL-AF9 leukemia model (11). Together, these data highlight a significant overlap between the genes up-regulated by Rnf20 and those up-regulated by MLL-AF9/Dot1l.
Fig. 3.
Knockdown of Rnf20 leads to decreased expression of MLL-AF9/Dot1l target genes. (A) RNA-seq data heat map indicating fold change in gene expression comparing shRen.713 to two independent Rnf20 shRNAs (3,595 and 3,277) after 4 d of dox induction. An expression cutoff (RPKM > 20) was implemented. (B and C) GSEA plots evaluating changes in MLL-AF9 targets and Dot1l-dependent gene signatures upon Rnf20 knockdown. FDR q-val, false discovery rate q-value (the probability that a gene set with a given normalized enrichment score represents a false-positive finding); NES, normalized enrichment score.
Upon Rnf20 knockdown, we also noted a paradoxical up-regulation of Myc expression and associated leukemia stem cell gene signatures (Fig. S6) (36). This unexpected result may explain the lack of myeloid maturation observed following Rnf20 suppression, as Myc levels are known to play a role in regulating the differentiation program in this disease (37). Prior studies also found that RNF20 can repress MYC expression in HeLa cells (27) but, conversely, can also promote MYC expression in LNCaP cells (38). These data would suggest that Rnf20 influences Myc expression in nonleukemia cellular contexts, albeit in either a positive or a negative manner depending on cell type. Nevertheless, Rnf20 inhibition leads to a unique situation in leukemia where G1 arrest occurs despite increased levels of Myc expression.
RNF20 has been found previously to play a role in the DNA damage response, with RNF20-deficient HeLa cells being more sensitive to DNA-damaging agents (23, 24). Interestingly, we failed to observe increased sensitivity of RNF20-deficient leukemia cells to cytotoxic agents etoposide or daunorubicin (Fig. S7). In contrast, we found that RNF20-deficient leukemia cells are hypersensitive to the BRD4 inhibitor JQ1, which exerts its effects in part through lowering of MYC expression in AML (37). The observed antileukemia effects when combining BRD4 and RNF20 inhibition might be related to the paradoxical MYC up-regulation that occurs in RNF20-deficient leukemia cells as described above (Fig. S6). These results further highlight a unique, context-specific role for RNF20 in AML.
H2B Ubiquitination Is Enriched in the Transcribed Region of MLL-AF9 Target Genes.
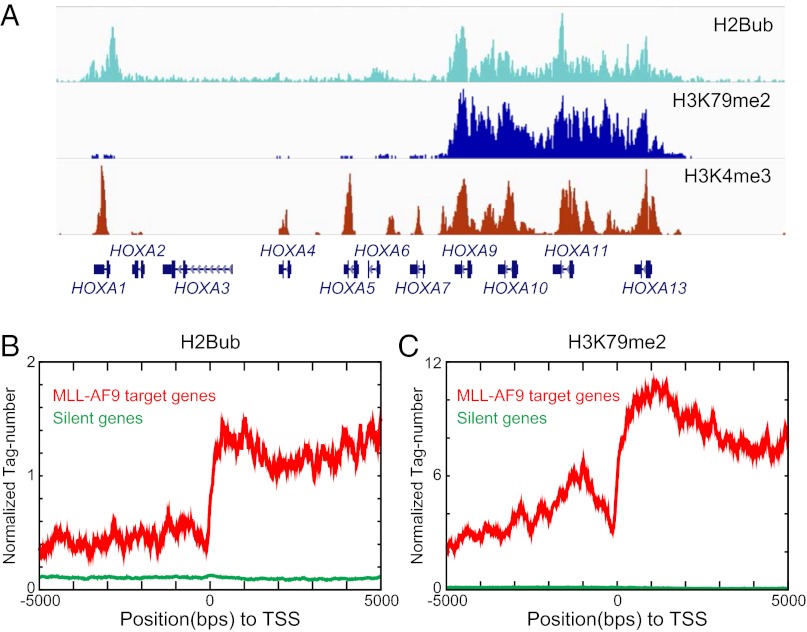
We next used ChIP-seq to evaluate whether Rnf20 and H2Bub might play direct roles in regulating transcription of MLL-AF9 target genes. Our attempts at immunoprecipitating Rnf20 were unsuccessful, so as an alternative we used an H2Bub-specific antibody since Rnf20 is the major E3 ligase catalyzing this modification in mammalian cells (21, 27). For these ChIP-seq experiments, we evaluated H2Bub, H3K79me2, and H3K4me3 in the human AML cell line THP-1, which harbors an endogenous MLL-AF9 translocation. Using this approach, we identified a broad enrichment of H2Bub at the HOXA cluster that reached its highest levels within the gene bodies, a localization pattern that resembled that of H3K79me2 (Fig. 4A). We also found H3K4me3 to be enriched in this region, albeit in a more focal pattern with prominent peaks near the transcription start site (TSS) of each gene (Fig. 4A and Fig. S8A). The correlation between H2Bub and H3K79me2 enrichment within coding regions was further verified using a metaanalysis of known MLL-AF9 target genes (Fig. 4 B and C). Interestingly, knocking down expression of MLL-AF9 led to reduced levels of H3K79me2 and H2Bub at Hoxa9 without an effect on H3K4me3 (Fig. S9). These results together suggest that H2Bub and H3K79me2 are coupled to one another at MLL-AF9 targets.
Fig. 4.
H2Bub and H3K79me2 are enriched in the transcribed portion of MLL-AF9 target genes. (A) ChIP-seq data of indicated histone modifications over the HOXA locus in THP-1 cells. Cumulative reads were visualized using the Integrated Genomics Viewer (IGV). (B and C) A metaprofile of histone modification levels at 139 MLL-AF9 target genes (gene list obtained from ref. 11) compared with 6,221 silent genes. The x-axis shows the relative distance to each TSS, whereas the y-axis shows the normalized read count of the histone modification.
H2Bub and H3K79me2 are coupled generically to transcription elongation by RNA polymerase II (39, 40). Hence, it should be noted that we found MLL-AF9 target genes to be profoundly H3K79 hypermethylated, likely as a result of the MLL-AF9:DOT1L interaction (Fig. S8), consistent with prior observations (11). In contrast, we found that H2Bub was present at only moderate levels in the body of MLL-AF9 target genes, in relative proportion to the expression level of this set of genes (Fig. S8). This result suggests that Rnf20 recruitment to MLL-AF9 target genes might be mediated through an association with the elongating form of RNA polymerase II, rather than via direct interactions with MLL-AF9.
Rnf20 Is Required to Maintain Local H3K79 Hypermethylation at MLL-AF9 Target Genes.
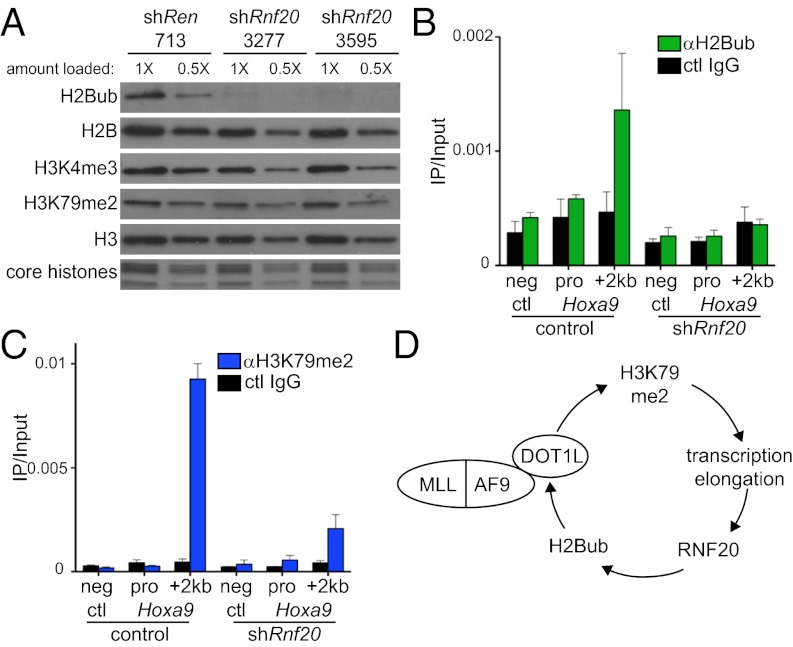
Because the presence of H2Bub on nucleosomes is known to increase the methyltransferase activity of DOT1L (25), we hypothesized that many of the phenotypes described above could be explained by Rnf20 acting to maintain H3K79 methylation in leukemia cells. We first evaluated this on a global level by Western blotting with histone modification-specific antibodies (Fig. 5A). Dox-induced Rnf20 knockdown led to a global reduction of H2Bub; however, we found only minimal reductions in global H3K79me2 under these conditions (Fig. 5A). Global H3K4me3 was also similarly unaffected by Rnf20 knockdown (Fig. 5A). We next considered whether Rnf20 might instead influence the local accumulation of H3K79 methylation in the vicinity of genes regulated by MLL-AF9. Using ChIP-quantitative PCR (qPCR), we found that levels of H3K79me2 were decreased at the Hoxa9 and Meis1 transcribed regions upon Rnf20 knockdown (Fig. 5C and Fig. S10A), whereas levels of this mark were unaffected at the highly expressed splicing factor arginine/serine rich 10 (Sfrs10) gene, which is not regulated by MLL-AF9 (Fig. S10B). Consistent with the Western blotting results, Rnf20 knockdown led to a decrease in H2Bub levels at all sites examined (Fig. 5B and Fig. S10B). H3K4me3 levels were unaffected at Hoxa9 and Sfrs10 upon Rnf20 knockdown (Figs. S10B and S10C). These findings suggest that Rnf20 is necessary for local Dot1l-mediated H3K79 methylation at MLL-AF9 target genes.
Fig. 5.
Rnf20 is required for local H3K79 methylation at Hoxa9. (A) Western blotting of acid-extracted histones prepared from RN2 cells after 4 d of dox-induced shRNA expression using the TRMPV-Neo vector. The bottom panel shows a Coomassie-stained SDS/PAGE gel indicating levels of the four core histones. A twofold dilution of each sample was included to highlight the minimal effects of Rnf20 shRNAs on H3K4me3 and H3K79me2. Representative experiment of three independent repeats is shown. (B and C) ChIP-qPCR evaluation with indicated antibodies following conditional Rnf20 knockdown (shRnf20.3595 was used). Different PCR amplicons are labeled along the x-axis. Neg ctl refers to negative control primers that amplify a nontranscribed region downstream of the Hoxa locus. Pro, Hoxa9 promoter. “+2 kb” is relative to the Hoxa9 TSS. Control cells were not treated with dox. Mean of three independent experiments is shown. All error bars denote SEM. (D) Model depicting the role of Rnf20 in amplifying MLL-AF9–mediated transcriptional activation through cross talk with Dot1l.
Discussion
The findings presented here implicate Rnf20 as a participant in the mechanism of leukemic transformation by the MLL-AF9 oncoprotein. This is primarily supported by (i) the RNF20 requirement for rapid proliferation of MLL-rearranged human AML cell lines and for disease progression in a genetically engineered mouse model and (ii) the similarity between gene expression changes that occur upon RNF20 knockdown and the known target genes of MLL-AF9 and its cofactor DOT1L. These results lead us to propose the following model, depicted in Fig. 5D. MLL-AF9 is known to up-regulate Hoxa9 transcription, in part, via the physical interaction between AF9 and DOT1L (9, 11). Through mechanisms that are not fully understood, DOT1L-dependent H3K79 methylation in the vicinity of Hoxa9 leads to increased rates of transcription by RNA polymerase II (9, 11, 13). RNF20 is likely to be recruited to Hoxa9 by transcription elongation complexes, resulting in H2Bub accumulation in the gene body in a manner dependent on MLL-AF9–mediated transcriptional activation. Prior studies have demonstrated that RNF20 directly binds the PAF elongation complex, which could account for the observed H2Bub enrichment in the gene body of MLL-AF9 targets (19, 21). Interestingly, MLL-fusion proteins can also bind directly to PAF (4, 41), an interaction that may also stabilize RNF20 recruitment to Hoxa9. As nucleosomes containing H2Bub are better substrates for DOT1L (25), the presence of RNF20 at Hoxa9 would trigger increased H3K79 methylation at this region. As such, the net consequence of RNF20 recruitment would be to strengthen MLL-AF9/DOT1L–mediated transcriptional activation through a feedforward loop (Fig. 5D). In this model, RNF20 acts as an amplifier of MLL-AF9 function, enabling more robust H3K79 hypermethylation and consequent increases in transcriptional activation. As a result, leukemia cells rely on RNF20 to maintain their leukemogenic gene expression program in a form of “nononcogene addiction” (42).
A remarkable feature of Rnf20 and Dot1l function in this system is that inhibiting either enzyme leads to relatively selective effects on transcription of MLL-AF9 target genes, despite H2Bub and H3K79me2 being enriched in the transcribed portion of most active mammalian genes (11, 27, 40). One possible explanation for this finding is that H2Bub and H3K79me2 only promote transcription elongation in specific chromatin environments. It is interesting to note that the HOXA cluster possesses a unique chromatin architecture: a dense arrangement of genes in identical orientation, broad domains of histone modifications (43), extensive higher order chromatin interactions (44), and several interspersed long noncoding RNAs (45). We speculate that this scenario, together with the aberrant activation stimulus provided by the MLL-fusion protein, creates a situation in which cross talk between Dot1l and Rnf20 becomes critically required for productive transcription.
Our findings also implicate RNF20 as a possible therapeutic target in the MLL-rearranged subtype of leukemia. Targeting of other E3 ligase proteins with known drugs provides proof-of-principle that potent small-molecule inhibitors can be developed against this class of enzymes (46, 47). However, because E3s rely on extensive protein:protein interactions to promote ubiquitin ligation to substrates, the ideal mode of RNF20 inhibition for therapeutic purposes is nonobvious at present and will require additional genetic and biochemical experiments to elucidate. Interestingly, overexpression of RNF20 might be a means through which leukemia cells acquire resistance to drugs that inhibit MLL-fusion cofactors (e.g., Dot1l and Menin inhibitors). In such a circumstance, an elevation of H2Bub levels could amplify the function of residual MLL-fusion complexes. Hence, targeting of RNF20 may have clinical utility by undermining MLL-fusion protein function and/or by overcoming acquired resistance to other agents.
Materials and Methods
MLL-AF9;NrasG12D AML Mouse Model.
All murine leukemia models used here have been described previously (31, 33). In brief, leukemias were derived by retroviral cotransduction of embryonic day 13.5 fetal liver hematopoietic stem and progenitor cells (HSPCs) with MSCV-MLL-AF9 and MSCV-Luciferase-IRES-NrasG12D, followed by transplantation of infected cells into sublethally irradiated recipient mice. The Tet-on-competent leukemia cells (RN2) were generated by cotransduction of HSPCs with MSCV-rtTA3-IRES-MLL-AF9 and MSCV-Luciferase-IRES-NrasG12D. The MLL-AF9-Tet-off model was generated by cotransduction of HSPCs with pSIN-TREtight-dsRed-IRES-MLL-AF9-5XFlag and MSCV-NrasG12D-IRES-tTA. All murine leukemia cultures were derived from bone marrow and spleen of terminally diseased mice. All mouse experiments included in this work were approved by The Cold Spring Harbor Animal Care and Use Committee.
Competition Assay to Score Effects of shRNAs on Cell Proliferation/Viability.
For evaluating the impact of shRNAs on leukemia expansion, cell cultures were retrovirally transduced with individual LMN-shRNA vectors (MSCV-miR30-shRNA-PGKp-NeoR-IRES-GFP), followed by measurement of the GFP percentage at various days postinfection using a Guava Easycyte (Millipore). The rate of GFP percentage decline over time is used to infer a defect in cell accumulation conferred by a given shRNAs relative to the uninfected cells in the same culture. For human leukemia cell line experiments, we used the MLS vector (MSCV-miR30-shRNA-SV40p-GFP), which allows a higher retroviral transduction efficiency in these lines. This competition assay is sensitive to an effect on either cell cycle progression or on cell survival.
In Vivo shRNA Studies.
For dox-inducible shRNA experiments in vivo, RN2 leukemia cultures were retrovirally transduced with TRPMV-Neo constructs (pSIN-TRE-dsRed-miR30-shRNA-PGK-Venus-IRES-NeoR) followed by G418 selection (1 mg/mL for 6 d). Clonal lines (obtained by limiting dilution) were then transplanted into secondary recipient animals, followed by initiation of dox administration (in drinking water and food) after 5–6 d. Mice were monitored thereafter by bioluminescent imaging, quantified using Living Image Software 4.0 (IVIS Spectrum system; Caliper LifeSciences). For bioluminescent imaging, mice were injected intraperitoneally with D-luciferin (Gold Biotechnology).
RNA-Seq.
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. One microgram of total RNA was subjected to “not-so-random” (NSR) primer-based RNA-seq library preparation according to protocols described previously (48). The prepared DNA was sequenced using an Illumina HiSEq 2000. The obtained reads were trimmed into 28 base reads corresponding to 9th to 36th position from the 5′ ends of the reads. These reads were mapped to the mouse genome (mm9) using Bowtie allowing no mismatch. Gene expression was analyzed by using Cufflinks software (http://cufflinks.cbcb.umd.edu/). Heat maps were generated with Java Tree View using a 20 reads per million per kilobase (RPKM) expression cutoff (49). Gene set enrichment analyses were performed according to the instructions described at www.broadinstitute.org/gsea/index.jsp (35).
ChIP-Seq.
ChIP assays were performed as previously described (50). Briefly, cells were fixed with 0.4% formaldehyde at room temperature for 10 min, and chromatin was sonicated in RIPA buffer with 0.3 M NaCl to the size range of 500–100 bp. For each ChIP-seq library, around 10 ng of immunoprecipitated DNA was used for library construction. After end repair and addition of “A” base to 3′ ends, barcoded adaptors were ligated to DNA fragments. Following a 17-cycle PCR, libraries were purified by two rounds of Ampure XP beads (Beckman Coulter; A63881) purification and one step of agarose gel purification. ChIP-seq libraries were sequenced on HiSEq 2000 (50 bp, single end). Reads were mapped to human genome (hg19) using BWA (http://bio-bwa.sourceforge.net/). Data were visualized at representative loci using the Integrative Genomics Viewer (www.broadinstitute.org/igv/). To examine the levels of histone modifications in specific gene groups, we separated the genes into four groups according to their expression. We ranked the genes according to their RPKM values from low to high, using RNA-seq data obtained from THP-1 cells. Genes with 0 RPKM were defined as the silent genes (6,221 genes). The remaining genes were then divided into three roughly equal-sized groups of 5,000 genes: low, medium, and high. See SI Materials and Methods for details of experimental methods for tissue culture and for sequences of all primers and shRNAs used here.
Histone Extraction and Western Blotting.
Cells were washed with PBS and lysed with buffer A solution (10 mM Hepes⋅KOH, pH 7.9, 15 mM MgCl2, 10 mM KCl). Cell pellets were resuspended in 0.2 N HCl. Supernatant was isolated followed by the addition of trichloroacetic acid (Sigma; T6399). Precipitate was carefully washed with acetone and resuspended in deionized water and run on an SDS/PAGE gel. For Rnf20 Western blot, ∼50,000 cells were lysed directly into Laemmli buffer and resolved using SDS/PAGE electrophoresis, followed by transfer to nitrocellulose for blotting.
Antibodies.
Anti–β-actin HRP (Sigma; A3854), anti-Rnf20 antibody (Novus Biological; NB100-2242), anti-H3K4me3 (Millipore; 07-473), anti-H3K79me2 (Abcam; ab3594), anti-H2Bub (Millipore; 05-1312), and anti-H3 (Abcam; ab1791).
Supplementary Material
Acknowledgments
We thank M. Taylor, E. Earl, and L. Bianco for support with mouse work; F. Karginov for assistance with RNA-seq analysis; and G. Blobel and C. Hammell for comments on the manuscript. This work was supported by Starr Cancer Foundation Grant I4-A430 (to C.R.V. and R.G.R), and additional funding was provided by the Edward P. Evans Foundation, the Martin Sass Foundation, and the F. M. Kirby Foundation. R.G.R. was also supported by Leukemia and Lymphoma Society Specialized Center of Research (SCOR) Grant 7132-08. C.R.V. is also supported by a Burroughs-Wellcome Career Awards for Medical Scientists award and National Cancer Institute Cancer Center Support Grant Development Funds (Grant CA45508).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE43725).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301045110/-/DCSupplemental.
References
- 1.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 2.Radtke I, et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci USA. 2009;106(31):12944–12949. doi: 10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shilatifard A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne TA, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama A, et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24(23):10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37(3):429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16(1):92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 9.Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 10.Tan J, et al. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20(5):563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grembecka J, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8(3):277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang WW, et al. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11(1):261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 16.Wood A, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11(1):267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20(5):759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, et al. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20(4):601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278(37):34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 20.Xiao T, et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25(2):637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137(3):459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavri R, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 23.Moyal L, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41(5):529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura K, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41(5):515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 25.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453(7196):812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20(3):341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shema E, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22(19):2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vethantham V, et al. Dynamic loss of H2B ubiquitylation without corresponding changes in H3K4 trimethylation during myogenic differentiation. Mol Cell Biol. 2012;32(6):1044–1055. doi: 10.1128/MCB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chernikova SB, et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012;72(8):2111–2119. doi: 10.1158/0008-5472.CAN-11-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weake VM, Workman JL. Histone ubiquitination: Triggering gene activity. Mol Cell. 2008;29(6):653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23(7):877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McJunkin K, et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci USA. 2011;108(17):7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29(1):79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2012 doi: 10.1038/onc.2012.110. 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jääskeläinen T, et al. Histone H2B ubiquitin ligases RNF20 and RNF40 in androgen signaling and prostate cancer cell growth. Mol Cell Endocrinol. 2012;350(1):87–98. doi: 10.1016/j.mce.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 39.Jung I, et al. H2B monoubiquitylation is a 5′-enriched active transcription mark and correlates with exon-intron structure in human cells. Genome Res. 2012;22(6):1026–1035. doi: 10.1101/gr.120634.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steger DJ, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muntean AG, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17(6):609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Ferraiuolo MA, et al. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010;38(21):7472–7484. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327(5971):1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 47.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 48.Armour CD, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6(9):647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- 49.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 50.Yu M, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45(3):330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.