Abstract
Long-lived corals, the foundation of modern reefs, often follow ecological gradients, so that populations or sister species segregate by habitat. Adaptive divergence maintains sympatric congeners after secondary contact or may even generate species by natural selection in the face of gene flow. Such ecological divergence, initially between alternative phenotypes within populations, may be aided by immigrant inviability, especially when a long period separates larval dispersal and the onset of reproduction, during which selection can sort lineages to match different habitats. Here, we evaluate the strength of one ecological factor (depth) to isolate populations by comparing the genes and morphologies of pairs of depth-segregated populations of the candelabrum coral Eunicea flexuosa across the Caribbean. Eunicea is endemic to the Caribbean and all sister species co-occur. Eunicea flexuosa is widespread both geographically and across reef habitats. Our genetic analysis revealed two depth-segregated lineages. Field survivorship data, combined with estimates of selection coefficients based on transplant experiments, suggest that selection is strong enough to segregate these two lineages. Genetic exchange between the Shallow and Deep lineages occurred either immediately after divergence or the two have diverged with gene flow. Migration occurs asymmetrically from the Shallow to Deep lineage. Limited recruitment to reproductive age, even under weak annual selection advantage, is sufficient to generate habitat segregation because of the cumulative prolonged prereproductive selection. Ecological factors associated with depth can act as filters generating strong barriers to gene flow, altering morphologies, and contributing to the potential for speciation in the sea.
Keywords: delayed reproduction, ecological speciation, developmental plasticity, conditional phenotypes, octocoral
Natural selection for adaptation to different ecological conditions has become increasingly recognized as an important factor in the formation and maintenance of species (1, 2). Among the ecological factors that may cause adaptive divergence to result in reproductive isolation is immigrant inviability (3). Immigrant inviability results when populations in different environments are locally adapted, so that immigration to the “wrong” habitats results in increased mortality. The effects of this screening process may depend on the life history traits of the species being studied. In species with fast generation times, such as insects and annual plants, immigrant inviability can contribute to reproductive isolation (3). Much less is known about this process and speciation in general in long-lived organisms, which often provide the foundation of species-rich ecosystems.
On coral reefs, the most diverse marine ecosystems, such long-lived foundation species include corals, sponges, and sea fans. Like many other reef-dwelling organisms, the life cycle of these animals includes a planktonic larval stage with great dispersal potential (4), and sister taxa and even more species-rich clades often share the same geographic range (5, 6). The co-occurrence of closely related species with high dispersal capabilities creates a challenge for evolutionary biologists: How can new marine species emerge without obvious geographic isolation? Caribbean coral reefs in particular provide several examples of young clades that have diverged more recently than their immediate cousins in the Eastern Pacific (6). Such species appear to have been generated by processes acting within the relatively small (∼3 million km2) confines of the Caribbean.
One explanation for within-Caribbean speciation is that geological processes (e.g., sea level fluctuations) may have transiently divided populations long enough for new species to form. Recently diverged sister species may have then expanded their ranges when conditions allowed and come into secondary contact. For example, the rise of the Isthmus of Panama altered ocean circulation, causing novel conditions in the Caribbean basin that generated both widespread extinction and speciation in Caribbean mollusks and corals (7), species that later expanded their distributions to the entire basin. Geographic isolation is also evident between some populations of widespread taxa at large (1,000 km) (8) and small (<100 km) spatial scales (9, 10). Thus, new species may result from historic geographic isolation within the Caribbean. However, ecological segregation is also widespread on Caribbean coral reefs. Genetic differences have been detected between ecomorphs with overlapping ranges (11–15), and thus habitat diversity may aid marine speciation. For example, the most common coral on Caribbean reefs was regarded as one species, but fertilization, genetic, and morphological data suggest it is a complex of three species segregated in part by depth (5, 16). In fact, depth segregates cryptic species on reefs more often than any other factor (17).
Candelabrum corals of the genus Eunicea constitute one such group where sister species are segregated by depth (13). Eunicea constitutes the most diverse genus of anthozoans (anemones, corals, and their kin) on Caribbean reefs, with 15 species plus some undescribed forms (18). Eunicea species share a mutualistic relationship with algae of the genus Symbiodinum clade B (19), which restricts them to the photic zone. All Eunicea are gonochoric and reproduce sexually by spawning gametes (18, 20).
Eunicea flexuosa is an especially suitable species in which to test the effects of immigrant inviability associated with adaptation to depth in a wide geographical context because (i) its geographic range spans the Caribbean (18), (ii) it is common [>5 colonies/m2 (21)] and present in virtually all habitats and depths on coral reefs and rocky walls (18), (iii) depth-segregated colonies show extensive internal and external morphological differences (13, 22), (iv) transplant experiments show colonies are adapted to different depths (13), and (v) first reproduction does not occur until a size of 30 cm, at least 15 y after larval dispersal (20). Such delayed reproduction during sedentary asexual growth allows selection to act for a long time before genes can be mixed [as in the strawberry-coral model of Williams (23)] and thus may increase the intensity of immigrant inviability between depth-segregated populations.
Here, we use field observations to estimate the fraction of E. flexuosa that survive until the onset of reproduction, and then measured survivorship advantage of native over foreign colonies at each depth by reciprocal transplants to show that native colonies have an advantage over foreigners, an advantage magnified by the cumulative effect of long prereproductive selection. We then evaluate morphological and genetic differentiation at two depths across the Caribbean range of E. flexuosa. If immigrant inviability operates between depths and is enhanced by delayed reproduction, then genetic divergence will be higher between sympatric populations separated by depth than among those at the same depth separated by geography. We found two depth-segregated lineages, each showing little evidence of subdivision among geographic populations across the Caribbean. Genetic lineages correlate well with morphology but some mismatched individuals are found at low frequency in both lineages. Previously observed adaptive phenotypic differences between depths (13) may thus provide both material for divergent selection and a mechanism for reproductive isolation in E. flexuosa. More generally, selection acting on habitat-dependent alternative phenotypes may be especially likely to enhance phenotypic and genetic divergence in long-lived sessile clonal animals with delayed postdispersal reproduction.
Results
Survivorship to Reproductive Size is Low in Shallow Areas.
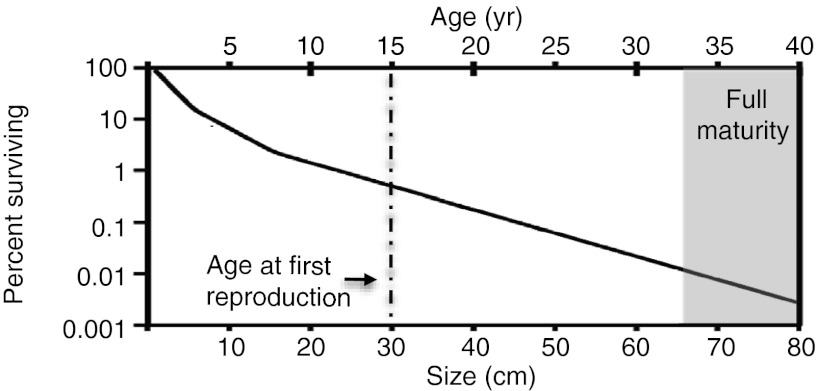
We estimated mean annual survivorship rates (averaged over 4 y) for colonies divided in three size categories (<10, 11–30, and >31 cm). Survivorship rates range from 69.1% for the smallest to 90.1% for the largest category (Table S1). Based on these values, the proportion of individuals (Fig. 1) that would survive to first reproduction (30 cm) is 1% and only 0.01% would reach full maturity (70 cm).
Fig. 1.
Age/size survivorship curve for Eunicea flexuosa (log scale). Age at first reproduction (20) is shown by the dotted line and full reproductive maturity (20) with the gray bar. Age calculated based on a 2-cm/y growth rate (13).
Selection Is Strong Between Habitats.
We selected 40 adult colonies from shallow and 40 from deep habitats, divided them, and then transplanted the resulting clones (>25 cm) to both depths. We estimated annual survivorship rates based on those observed for the 2 y following transplantation and by pooling our data with previous estimates (13). Mean annual survivorship was highest (Table S2) for colonies transplanted to the same depth (87.1% and 94.0% for shallow and deep, respectively) and lowest for reciprocally transplanted colonies (78.5% and 63.0% for colonies transplanted to shallow and to deep, respectively). We estimated an annual increase in survivorship of native colonies over foreign ones of 9.9% in shallow areas and 33.0% in deep areas. Over the span of a generation (about 40 y), the survivorship advantage would be 98.3% for shallow areas and 99.9% for deep. Assuming a 1:1 ratio of settlers and Hardy–Weinberg proportions, we expect 96% of the mating events in shallow habitats to be between Shallow × Shallow, with only 4% between Shallow × Deep (hybrid crosses). In deep habitats, even fewer hybrid crosses are expected, one in a million.
To compare our selection estimates with other studies, we translated them into difference in fitness between native and foreign colonies as in ref. 24. There are increases of 180% and 190% in fitness of native over foreigners for shallow and deep habitats, respectively. These estimates are based on a growth rate (1.8 cm/y) at the high end of those seen for E. flexuosa (13), the size (70 cm) at which colonies significantly contribute to reproduction (20), and adults showing the highest survivorship rates. Estimates based on faster growth rates, reproduction at younger ages, and survivorships from younger colonies result in greater asymmetry in the selective advantage between habitats.
Two Genetic Lineages Are Segregated by Depth.
We sampled populations from four locations: Bahamas, Panama, Puerto Rico, and Curaçao (Table S3). At each location, we sampled adult colonies (>50 cm) at two depths, as earlier work in Puerto Rico suggested deep and shallow populations might be genetically differentiated (13).
All our analyses of genetic variation indicated unambiguously that there are two lineages within E. flexuosa that segregate between deep and shallow depths. Patterns of genetic subdivision revealed by STRUCTURE showed strong support for two clusters (Fig. 2Ab, ∆L > 95%) that correlated closely with depth (Fig. 2A). The probability of membership to either cluster was high (>90%) for most colonies, although it dropped to 89–60% for 20 individuals (11%), perhaps as a result of hybridization between lineages. Similarly, analysis of molecular variance showed that depth explains most of the variation for both mitochondrial and nuclear markers (61% and 71%, respectively). Less variation was explained by geography (7% and 1% for mtDNA and nuclear markers, respectively) and within site variation (31% and 27% for mtDNA and nuclear markers, respectively). All fixation indices were significant (10,000 permutations) at the 0.01 level for both types of markers. Although haplotype networks for each gene did not sort completely (Fig. S1), they revealed two distinct lineages that largely correlate with depth (at least for MSH and I3P). For EF1A and CT2, haplotypes sampled from different depths mixed, but clinal variation was still evident.
Fig. 2.
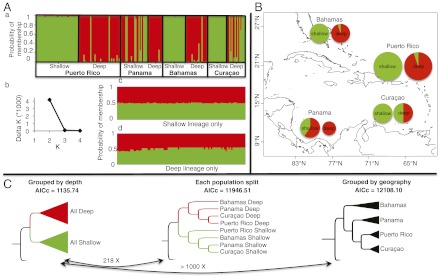
(A) Graphical summary of Bayesian clustering (a) STRUCTURE results for all samples combined. (b) Likelihood score differences (ΔK) as a function of K (number of clusters), with highest difference between K = 2 and K = 3. Separate runs for the (c) Shallow and (d) Deep lineages revealed in a show no further subdivision. (B) Proportion of individuals from shallow and deep habitats falling into the Shallow (green) and Deep (red) clusters revealed by STRUCTURE. Two individuals with ancestry probability <70% were excluded. The size of the circle is proportional to the number of sampled individuals. (C) STEM results for each of three data arrangements: by depth, by depth and geography, or by geography. Corrected Akaike information criteria scores are indicated. The black arrows denote the evidence ratios between alternative topologies from information theory.
To test the extent to which depth, geography, and their interaction have played roles in lineage splitting and to recover the evolutionary history of all populations, we used a combined gene trees/species tree (STEM) (25) and model-based inference approach. The Shallow and Deep lineages were well supported by STEM. The tree with the best corrected Akaike information criterion (AICc) score showed two lineages: one composed of all Shallow samples and one of all Deep (Fig. 2C). When we further split populations by depth and geography, the best tree still joins populations by depth, but was 218 times less likely than the all Shallow and all Deep tree. Conversely, if we grouped populations by geography (i.e., combine Panama Shallow and Panama Deep), the tree with the highest likelihood score was >1,000 times worse than the best tree.
Depth segregation was significant (Fisher’s exact test, P < 0.0001), but the degree of segregation varied among sites (Fig. 2B). In Puerto Rico and Bahamas, it was high, with >80% of colonies of each lineage segregated into either shallow or deep habitats. In Curaçao, the Shallow lineage dominated (100%) shallow environments, but was also common (50%) in deep areas. Thus, although the two lineages segregate over a depth gradient, the absolute depths defining the gradient vary among localities. In Puerto Rico, our sampling captured the ends, but in Curaçao and Bahamas the corresponding deep habitat may be at greater depths.
Panama was an extreme case in this regard: the Deep lineage dominated both deep and shallow environments, with the Shallow lineage present at only modest levels in shallow environments (even after adding an extra shallow collecting site). This Panamanian oddity was not unexpected: particulate matter around Bocas del Toro is high, which increases food for suspension feeders and attenuates light, allowing deep-water suspension feeders such as black corals to occur at depths as shallow as 5 m (26), whereas in other locations they are not visible until 25 m (26).
Little Geographical Differentiation Within Lineages.
In contrast to the striking pattern of depth segregation between lineages, there was little evidence of geographical differentiation within lineages. Fig. 2 Ac and Ad shows STRUCTURE plots for each lineage after samples were assigned to either cluster from the global analysis. No clear clusters were visible in either. A within-lineage analysis of molecular variance showed that geography significantly explains just 3% and 11% of the variation for the Shallow and Deep lineage, respectively. A pairwise FST analysis found that some within-lineage geographical comparisons were significant (Table S4), but the magnitude of these differences was not large.
No Strict Allopatry and Asymmetric Migration from Shallow to Deep.
The rank of all Isolation with Migration models by our information theory approach suggested that migration has occurred during the divergence of the Shallow and Deep lineages (Fig. 3). The best model had three parameters for population size and two for migration, regardless of whether we classified shallow and deep individuals by their lineage, morphology, or habitat (Table S5). Models without migration parameters (strict isolation) were thousands of times less likely than the best model. Migration from Shallow to Deep is higher overall and in each locality, except in Panama when samples are classified by habitat or morphology (Fig. S2 and Table S6). This is not unexpected as in Panama morphologies are less distinct (Fig. 4) and deep water species tend to come up shallower than elsewhere in the Caribbean (27).
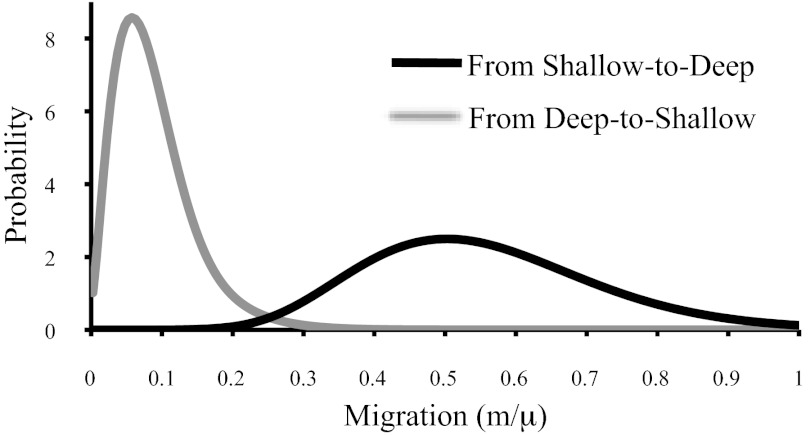
Fig. 3.
Migration rate estimates between Shallow and Deep obtained by fitting the IMa model to all four loci. Estimates are scaled by the neutral mutation rate. Samples were partitioned by morphology (excluding Panama). The number of migrants between lineages per generation (Nm) is 0.4 [90% highest posterior density (HPD) interval, 0.147–0.756] from Shallow to Deep and 0.078 (90% HPD interval, 0.02–0.189) for Deep to Shallow.
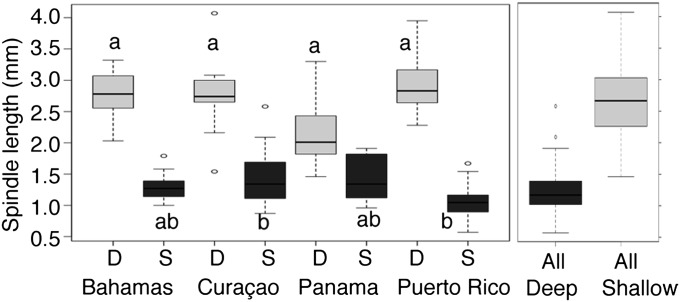
Fig. 4.
Mean spindle length between Deep (black) and Shallow (light gray) lineages in different populations. The letters indicate significant differences for within lineage comparisons. The different letters indicate significant differences in an honestly significant difference (HSD) Tukey test after Bonferroni correction. D, Deep; S, Shallow.
The difference between the asymmetry in selection (strongest in the deep habitat) and between-lineage migration (more from Shallow to Deep) may be explained by interlineage differences in gamete production. In E. flexuosa, large colonies produce on average two orders of magnitude more gametes than smaller colonies (20). Deep colonies are on average less than one-half the size of Shallow colonies and have one-half their polyp density (13, 22). Deep lineage colonies are also less abundant than Shallow across the Caribbean; in Puerto Rico their densities are one-third those of Shallows (21). Thus, by the time reproduction occurs the amount of Shallow lineage gametes may greatly outnumber those from the Deep lineage. This asymmetry in gamete production may result in asymmetrical introgression, as previously shown in oaks (28).
Lineages Match Phenotypes at Local Scales.
To assess morphological divergence among populations, we measured the length of spindles, the calcium carbonate particles that provide structure to the colony. Spindle length correlates well with other morphological characters (e.g., calice size, intercalice distance, branch thickness) and is the most divergent morphological character between deep and shallow colonies of E. flexuosa in Puerto Rico (13).
Patterns of morphological differentiation mirrored those seen genetically; lineage was a better predictor of spindle length than either geography or habitat (Table 1). All of the four best models had lineage as a variable, along with its interaction with either geography or habitat. When we excluded lineage or its interactions as variables, the remaining models were 100 times less likely than the best model with lineage included. Lineage fit the data better than depth, despite the two being highly correlated (Table 1). Although morphology and genetics match well, mismatched individuals occur at low frequency (2 of 97 and 7 of 92 for Shallow and Deep, respectively).
Table 1.
Model ranking for spindle length analysis using a general linear model
| Model | AIC | ∆i | ML | wi | Evidence ratio |
| GDL L*G | 165.56 | 1.00 | 0.61 | ||
| GDL D*L G*L | 167.31 | 1.75 | 0.42 | 0.25 | 2 |
| GDL D*L G*D | 170.47 | 4.91 | 0.09 | 0.05 | 12 |
| GDL G*L G*D | 171.08 | 5.52 | 0.06 | 0.04 | 16 |
| Full model | 171.08 | 5.52 | 0.06 | 0.04 | 16 |
| GDL D*G | 175.15 | 9.59 | 0.01 | 0.01 | 121 |
| GL G*L | 185.25 | 19.69 | 0 | 0 | 18,864 |
| All other models | >188 | >20 | 0 | 0 | >105 |
G, geography; D, depth; L, lineage assignment from STRUCTURE clustering for K = 2; ML, maximum likelihood; AIC, Akaike information criterion inferred from the generalized linear model fitting; ∆i, difference in AIC score with respect to the best model; model likelihoods, relative likelihood of the model given the data; wi, model probabilities; evidence ratio, fold difference in model probabilities against the best model.
Once lineage is accounted for, some differences between locations remained (Fig. 4). For the Shallow lineage, colonies from Curaçao had larger spindles than colonies in Puerto Rico. For the Deep lineage, colonies in Panama had smaller spindles than for all other locations. This difference in spindle length in Panama made the morphological boundary between Shallow and Deep narrower and shifted shallower than in other locations. Thus, morphologies were diagnosable, correlated well with lineage, and segregated into shallow and deep habitats at local scales, but lineage by depth by locale interactions generated morphological overlap between taxa when populations were considered together.
Discussion
Our study found that (i) selection is strong in both habitats for native over foreign colonies, (ii) colonies of the candelabrum coral Eunicea flexuosa occurring in shallow and deep habitat are segregated into two genetically and morphologically divergent lineages, (iii) genetic divergence is weak among geographic samples within each lineage, (iv) the Shallow and Deep lineages have exchanged genes since their initial divergence, and (v) migration between them has been higher from Shallow to Deep. Reciprocal transplantation suggests divergence between colonies found at different depths is adaptive and selection is strong enough to maintain the separation of the two lineages at the ends of the depth gradient. This divergent selection may by itself result in reproductive isolation via immigrant inviability (poor performance and viability of larvae settling at the wrong depth) and temporal isolation (spawning at different times) (29). We suggest divergence between Shallow and Deep lineages of E. flexuosa is driven by adaptation to different conditions at different depths (1), facilitated by developmental plasticity (30, 31) and prolonged opportunity for depth-specific selection before reproduction.
Ecological Gradients, Adaptive Divergence, and Speciation.
Divergent selection along depth gradients is generated by associated factors such as the quantity and quality of light, the force of waves and currents, sediment load, the distribution of predators and mutualists, and the availability and composition of food. In response to this variation in environmental conditions, closely related species or populations found at different depths have been found to differ in their morphologies (5, 13), feeding strategies (32, 33), metabolic rates (33), symbiotic relationships (34), resistance to predators (35), and spawning times (16, 29). In E. flexuosa's close relative Plexaura homomalla (Eunicea and Plexaura are paraphyletic), depth-segregated sister species increase heterotrophy and decrease photosynthesis as depth increases (32).
Such depth-related divergence seems to be correlated with the differentiation of closely related species, as in bryozoans (36). Depth gradients in light are hypothesized as a cause of speciation by sensory drive in cichlids (37) and factors associated with depth appear to have driven speciation in rockfish (38). Among marine sibling species reported by Knowlton (17), over one-half involved depth as a dividing factor, even though not all comparisons included depth. Thus, adaptive divergence by depth is consistent with a role for depth in maintaining and generating species differences in marine and aquatic settings.
These patterns associated with depth are analogous to those seen for other environmental gradients, such as altitude, salinity, or temperature. Such gradients generate adaptive divergence in vertebrates (39), invertebrates (40), and plants (41). Because such abiotic factors generate adaptive divergence across multiple species, they also generate gradients in biotic interactions (such as predator–prey interactions) (42), creating additional axes of complexity.
High Gene Flow Within Species.
Divergence between depths at small scales (as little 200 m separate deep and shallow populations we sampled) in E. flexuosa contrasts with the high connectivity seen among populations within lineages across thousands of kilometers. Such high connectivity is not uncommon to marine species with planktonic larval dispersal (4), ranging to extremes in which gene flow spans the broadest expanses of the Pacific Ocean (43). Thus, adaptive divergence occurs here despite the potential for high levels of gene flow (39). Such adaptive divergence under high gene flow generates a pattern where neutral genetic divergence is primarily partitioned by habitats, with little geographic structure. Ecotypes are connected across vast geographical spans, thus producing a pattern of single ecological segregation as seen in E. flexuosa and shared by other broadcast spawners such as the star coral Montastraea annularis (5), some coral reef fishes (14), and the Hawaiian limpets Cellana (44).
In contrast, under low gene flow, genetic divergence first occurs geographically and then ecologically. The pattern has been detailed in the marine snail Littorina saxatillis (45) and is similar to those in other marine brooders where ecology has been proposed to drive speciation, such as the corals Favia fragum (15) and Seriatopora hystrix (46). Clonal brooders also tend to reproduce at younger ages (smaller sizes) (47), increasing the role of geography in local adaptation. This difference between brooders and broadcasters has parallels in winged and wingless phytophagous insects. In pea aphids, segregation by host plant occurs at large geographical scales with little geographical differentiation (48). Conversely, in walking sticks, pairs of habitat-segregated ecotypes have occurred multiple times (3). Thus, the partition of genetic diversity during ecological speciation depends on the dispersal biology of the studied species.
Divergence Between Habitat-Segregated Populations with Prolonged Prereproductive Selection.
Ecological filtering can help maintain species differences and contribute to reproductive isolation of populations occurring in different habitats (3). In E. flexuosa, adult colonies native to their depth survive far better than do foreign colonies, an advantage enhanced by the cumulative effect of many years of prereproductive selection. For a settling larva to reach adulthood, it must pass a series of ecological challenges until it reaches a safe size of at least 30 cm (>15 y) and begins reproductive activity. Significant genetic contributions to the population likely take far longer, however, not until attaining a colony size of 70 cm (>35 y), because such large colonies contribute 98% of all eggs in E. flexuosa (20).
The difference in fitness between native and foreign colonies that we found here is three times higher than any reported for similar transplant experiments (24). It also exceeds values used in mathematical models of ecological speciation in which adaptive divergence is sufficient to generate reproductive isolation in discrete habitats (49, 50) or along clines (51, 52). The long period between when a larva settles and when it becomes a colony that contributes genetically to the next generation thus decreases gene flow between depth-adapted lineages and enhances the opportunity for habitat-specific selection and divergent adaptation. Such immigrant inviability may be common in slowly maturing sessile species with broad dispersal [like some scrubs and trees (53, 54)]. In contrast, when adults are the primary dispersers, immigrant inviability would have to be exceptionally strong to have such an effect, as migrants can reproduce as soon as they settle, allowing locally adapted genotypes to mix with maladapted foreign ones.
Strong divergent selection between habitats could also stifle the early stages of divergence if dispersal is limited and a narrow range of phenotypes limit opportunities to colonize new habitats (31). In E. flexuosa, however, larval dispersal is high (Fig. 2) and colonies quite phenotypically plastic (13).
Temporal isolation may also emerge from adaptation to different depths and serve to further isolate depth-segregated populations. As for flowering plants segregated by altitude (41, 55), marine broadcasters segregated by depth often differ in their time of spawning, decreasing the probability of cross-fertilization between depth-segregated colonies (16, 29). Before spawning events, colonies in deep areas of E. flexuosa lack mature eggs compared with shallow ones (22). One-hour difference in the timing of spawning is sufficient for sperm to dilute to the point where crossbreeding is unlikely in other corals (16, 29). Although the exact traits that affect ecological fitness are unknown, population segregation by depth thus has the potential to increase reproductive isolation by pleiotropic interactions (31, 56) between fitness traits and those that control timing of reproduction, increasing the chance of genetic divergence in depth-segregated populations adapted to different ecological conditions.
Speciation by Prolonged Selection on Adaptively Plastic Habitat-Specific Contrasting Forms: Divergence in the Sea.
Our data on E. flexuosa show how a developmentally plastic organism with ecologically variable phenotypes may diverge genetically to produce different species without geographic isolation, even in groups with widely dispersing propagules.
Habitat-specific alternative phenotypes, whether discrete or at the ends of a cline, represent different habitat-mediated sets of expressed genes (57). To the degree that they are independently expressed, they are independently exposed to habitat-specific selection and can, given genetic variation in their form (57, 58), diverge genetically without reproductive isolation (30). Then, due to genetic accommodation or assimilation of the contrasting phenotypes (30, 58), they may diverge genetically such that migrants between habitats become effectively inviable, as suggested by our data and as hypothesized for speciation between alternative phenotypes (30, 59) and in clines (51, 52). A broader analysis of the evolution of plasticity in the genus Eunicea would provide a framework to further test this hypothesis.
As we discuss here, habitat-related divergence may apply especially in organisms with a long period of prereproductive selection, which affords an increased “opportunity for selection” (60), and in organisms that are resident for a prolonged period in a single place. In sessile clonal animals, independent selection on condition-sensitive (depth-sensitive) phenotypes is enhanced because immotile colonies cannot escape the selective factors of a given stable habitat. Selection is intense in juveniles and decreases exponentially with colony size, so that clonal organisms delay (for a couple of decades in E. flexuosa) sexual reproduction and allocate all resources to vegetative growth (23). Even once these younger colonies begin to reproduce, most gametes in a population will still come from the large, old (over one-half a century), well-adapted colonies that dominate sexual reproduction. These conditions characterize E. flexuosa and other sessile clonal organisms, such as anthozoans, sponges, long-lived trees (e.g., oaks and pines), and shrubs (e.g., Ceanothus) (23, 61), which invest heavily in vegetative growth before reproduction (23, 61). This process of habitat-related divergence without reproductive isolation may therefore be more common than usually appreciated. It can occur whether the contrasting phenotypes are initially genetically cued (51, 52) or environmentally induced (13, 30, 62), for it is the contrast between the different phenotypes and the underlying sets of expressed genes that, under selection, drives divergence—not the cues (genetic or environmental) that govern their expression.
Taken together, the combination of habitat-responsive plasticity, long prereproductive selection, sessile residence in distinctive habitats, and the potential for temporal separation of reproduction in different habitats, exemplified in E. flexuosa, may help solve the long-standing question of how speciation can occur in marine populations with widely dispersing larval forms.
Materials and Methods
We estimated annual survivorship per size class in 1-m2 quadrats. We monitored 161 quadrats spread across eight reefs in Puerto Rico over 4 y (SI Materials and Methods). We sampled survivorship advantage of native over foreign by reciprocally transplanting 40 adult (>25 cm) colonies from each depth to both shallow and deep areas (80 colonies per depth). Survivors were recorded annually for 2 y.
We sampled populations in Bahamas, Panama, Puerto Rico, and Curaçao for genetic analysis. At each location, we sampled 17–38 adult colonies (>50 cm) from each of two depths (Table S3). We developed three new nuclear sequence markers (Table S7) using sequences generated from a partial 454 run. Sequences from these three and a previously described mtDNA marker were generated using standard PCR procedures (SI Materials and Methods). We edited and assembled all sequences using standard procedures. We resolved haplotypes probabilistically (SI Materials and Methods) and corroborated inferred haplotypes by directly cloning 10% of our sequences. We recoded sequences as frequency data and used them to infer population subdivision using Bayesian clustering in STRUCTURE (63). We ran STRUCTURE without information of the origin of each individual, thereby reducing potential biases. We used the STRUCTURE cluster assignment for two subsequent analyses: STEM and IM.
We inferred gene genealogies and estimated likelihood scores for species trees using STEM 2.0 (25). We manipulated the settings file to estimate the likelihood score for the best tree for each of three arrangements: (i) populations divided by geography (four lineages), (ii) populations divided by depth (two lineages), and (iii) each population as an independent lineage (eight lineages). We then used the average likelihood scores across replicates to infer information theory statistics and rank all possible topologies (64).
We used IMa, the isolation with migration model (65), and model-based inference (64) to evaluate all possible scenarios of divergence. Our aim was to compare the support for strict isolation (zero migration) of the Shallow and Deep lineages (as delineated by STRUCTURE) to that for a model of divergence with gene flow. We also evaluated the IMa model when individuals were partitioned by habitat and morphology. We then used model-based selection to calculate evidence ratios and rank all possible models (64).
We took images of each colony through a Diagnostic Instrument SPOT RT Slider CCD camera attached to a Leica MZ7 stereomicroscope. All morphological measurements were carried out at the Louisiana State University Socolofsky Microscopy Center. We fit the spindle length to a generalized linear model. We used a Gaussian distribution and geography, depth, and lineage as factors. We constructed all 17 possible models and used model based approaches to rank each model (64). All statistical tests were carried out in R.
Supplementary Material
Acknowledgments
Comments from D. Beltrán, M. DeBiasse, J. Niegel, A. Whitehead, two anonymous reviewers, the editor, and P. Yoshioka improved earlier versions of this manuscript and allowed use of the Caribbean Coral Reef Institute data. We thank the Department of Marine Sciences of the University of Puerto Rico (R. Appeldoorn and N. Schizas), The Smithsonian Tropical Research Institute, Caribbean Research and Management of Biodiversity (CARMABI), and H. Lasker. This work was supported by the Rosemary Grant Research Award from the Society for the Study of Evolution, the Society of Systematic Biologists, Louisiana State University Department of Biological Sciences, National Science Foundation Grant OCE-0550270 (to M.E.H. and I. Baums), and National Oceanic and Atmospheric Administration through Caribbean Coral Reef Institute (Grant NA06NOS4780190). C.P. is supported by a Huel Perkins fellowship from Louisiana State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC310499–KC310687 and KC333998–KC335131).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208931110/-/DCSupplemental.
See Commentary on page 3713.
References
- 1.Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323(5915):737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- 2.Darwin C. On the Origin Of Species. London: John Murray; 1859. [Google Scholar]
- 3.Nosil P, Vines TH, Funk DJ. Perspective: Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution. 2005;59(4):705–719. [PubMed] [Google Scholar]
- 4.Hellberg M. Gene flow and isolation among populations of marine animals. Annu Rev Ecol Evol Syst. 2009;40:291–310. [Google Scholar]
- 5.Fukami H, et al. Geographic differences in species boundaries among members of the Montastraea annularis complex based on molecular and morphological markers. Evolution. 2004;58(2):324–337. [PubMed] [Google Scholar]
- 6.Taylor MS, Hellberg ME. Marine radiations at small geographic scales: Speciation in neotropical reef gobies (Elacatinus) Evolution. 2005;59(2):374–385. [PubMed] [Google Scholar]
- 7.Jackson JBC, Jung P, Coates AG, Collins LS. Diversity and extinction of tropical american mollusks and emergence of the isthmus of panama. Science. 1993;260(5114):1624–1626. doi: 10.1126/science.260.5114.1624. [DOI] [PubMed] [Google Scholar]
- 8.Baums IB, Miller MW, Hellberg ME. Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol Ecol. 2005;14(5):1377–1390. doi: 10.1111/j.1365-294X.2005.02489.x. [DOI] [PubMed] [Google Scholar]
- 9.DeBiasse MB, Richards VP, Shivji MS. Genetic assessment of connectivity in the common reef sponge, Callyspongia vaginalis (Demospongiae: Haplosclerida) reveals high population structure along the Florida reef tract. Coral Reefs. 2010;29(1):47–55. [Google Scholar]
- 10.Taylor MS, Hellberg ME. Genetic evidence for local retention of pelagic larvae in a Caribbean reef fish. Science. 2003;299(5603):107–109. doi: 10.1126/science.1079365. [DOI] [PubMed] [Google Scholar]
- 11.Duran S, Rützler K. Ecological speciation in a Caribbean marine sponge. Mol Phylogenet Evol. 2006;40(1):292–297. doi: 10.1016/j.ympev.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Brazeau DA, Harvell CD. Genetic structure of local populations and divergence between growth forms in a clonal invertebrate, the Caribbean octocoral Briareum asbestinum. Mar Biol. 1994;119(1):53–60. [Google Scholar]
- 13.Prada C, Schizas NV, Yoshioka PM. Phenotypic plasticity or speciation? A case from a clonal marine organism. BMC Evol Biol. 2008;8:47. doi: 10.1186/1471-2148-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocha LA, Robertson DR, Roman J, Bowen BW. Ecological speciation in tropical reef fishes. Proc Biol Sci. 2005;272(1563):573–579. doi: 10.1098/2004.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlon DB, Budd AF. Incipient speciation across a depth gradient in a scleractinian coral? Evolution. 2002;56(11):2227–2242. doi: 10.1111/j.0014-3820.2002.tb00147.x. [DOI] [PubMed] [Google Scholar]
- 16.Knowlton N, Mate JL, Guzman HM, Rowan R, Jara J. Direct evidence for reproductive isolation among the three species of the Montastraea annularis complex in Central America (Panama and Honduras) Mar Biol. 1997;127(4):705–711. [Google Scholar]
- 17.Knowlton N. Sibling species in the sea. Annu Rev Ecol Evol Syst. 1993;24:189–216. [Google Scholar]
- 18.Sanchez J. Systematics of the candelabrum gorgonian corals (Eunicea Lamouroux; Plexauridae; Octocorallia; Cnidaria) Zool J Linn Soc. 2009;157(2):237–263. [Google Scholar]
- 19.Santos SR, Taylor DJ, Coffroth MA. Genetic comparisons of freshly isolated vs. cultured zooxanthellae: Implications for extrapolating to the intact symbiosis. J Phycol. 2001;37(5):900–912. [Google Scholar]
- 20.Beiring EA, Lasker HR. Egg production by colonies of a gorgonian coral. Mar Ecol Prog Ser. 2000;196:169–177. [Google Scholar]
- 21.Yoshioka P. Sediment transport and the distribution of shallow-water gorgonians. Caribb J Sci. 2009;45(2):254–259. [Google Scholar]
- 22.Kim E, Lasker HR, Coffroth MA, Kim K. Morphological and genetic variation across reef habitats in a broadcast-spawning octocoral. Hydrobiologia. 2004;530/531:423–432. [Google Scholar]
- 23.Williams G. Sex and Evolution. Princeton: Princeton Univ Press; 1975. [Google Scholar]
- 24.Hereford J. A quantitative survey of local adaptation and fitness trade-offs. Am Nat. 2009;173(5):579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- 25.Kubatko LS, Carstens BC, Knowles LL. STEM: Species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics. 2009;25(7):971–973. doi: 10.1093/bioinformatics/btp079. [DOI] [PubMed] [Google Scholar]
- 26.Opresko DM, Sanchez JA. Caribbean shallow-water black corals (Cnidaria: Anthozoa: Antipatharia) Caribb J Sci. 2005;41(3):492–507. [Google Scholar]
- 27.Guzmán HM, Guevara CA. Coral reefs of Bocas del Toro, Panama: III. Distribution, structure, diversity and conservation status of reefs in Pastores, Cristobal, Popa and Cayo Agua islands. Rev Biol Trop. 1999;47(2):659–676. [Google Scholar]
- 28.Lepais O, et al. Species relative abundance and direction of introgression in oaks. Mol Ecol. 2009;18(10):2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- 29.Levitan DR, et al. Mechanisms of reproductive isolation among sympatric broadcast-spawning corals of the Montastraea annularis species complex. Evolution. 2004;58(2):308–323. [PubMed] [Google Scholar]
- 30.West-Eberhard MJ. Developmental Plasticity and Evolution. New York: Oxford Univ Press; 2003. [Google Scholar]
- 31.Fitzpatrick BM. Underappreciated consequences of phenotypic plasticity for ecological speciation. Int J Ecol. 2012;2012:256017. [Google Scholar]
- 32.Lasker HR, Gottfried MD, Coffroth MA. Effects of depth on the feeding capabilities of two octocorals. Mar Biol. 1983;73(1):73–78. [Google Scholar]
- 33.Lesser MP, et al. Photoacclimatization by the coral Montastraea cavernosa in the mesophotic zone: Light, food, and genetics. Ecology. 2010;91(4):990–1003. doi: 10.1890/09-0313.1. [DOI] [PubMed] [Google Scholar]
- 34.Kirk NL, Andras JP, Harvell CD, Santos SR, Coffroth MA. Population structure of Symbiodinium sp. associated with the common sea fan, Gorgonia ventalina, in the Florida Keys across distance, depth, and time. Mar Biol. 2009;156(8):1609–1623. [Google Scholar]
- 35.West JM, Harvell CD, Walls AM. Morphological plasticity in a gorgonian coral (Briareum asbestinum) over a depth cline. Mar Ecol Prog Ser. 1993;94:61–69. [Google Scholar]
- 36.Jackson J, McKinney F. In: Ecological Processes and Progressive Macroevolution of Marine Clonal Benthos. Causes of Evolution. Ross R, Allmon W, editors. Chicago: Univ of Chicago Press; 1990. pp. 173–209. [Google Scholar]
- 37.Seehausen O, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455(7213):620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- 38.Ingram T. Speciation along a depth gradient in a marine adaptive radiation. Proc Biol Sci. 2011;278(1705):613–618. doi: 10.1098/rspb.2010.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehead A, Roach JL, Zhang S, Galvez F. Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proc Natl Acad Sci USA. 2011;108(15):6193–6198. doi: 10.1073/pnas.1017542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanford E, Kelly MW. Local adaptation in marine invertebrates. Annu Rev Mar Sci. 2011;3:509–535. doi: 10.1146/annurev-marine-120709-142756. [DOI] [PubMed] [Google Scholar]
- 41.Hodges SA, Arnold ML. Floral and ecological isolation between Aquilegia formosa and Aquilegia pubescens. Proc Natl Acad Sci USA. 1994;91(7):2493–2496. doi: 10.1073/pnas.91.7.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freeman AS, Byers JE. Divergent induced responses to an invasive predator in marine mussel populations. Science. 2006;313(5788):831–833. doi: 10.1126/science.1125485. [DOI] [PubMed] [Google Scholar]
- 43.Lessios HA, Robertson DR. Crossing the impassable: Genetic connections in 20 reef fishes across the eastern Pacific barrier. Proc Biol Sci. 2006;273(1598):2201–2208. doi: 10.1098/rspb.2006.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bird CE, Holland BS, Bowen BW, Toonen RJ. Diversification of sympatric broadcast-spawning limpets (Cellana spp.) within the Hawaiian archipelago. Mol Ecol. 2011;20(10):2128–2141. doi: 10.1111/j.1365-294X.2011.05081.x. [DOI] [PubMed] [Google Scholar]
- 45.Johannesson K. Parallel speciation: A key to sympatric divergence. Trends Ecol Evol. 2001;16(3):148–153. doi: 10.1016/s0169-5347(00)02078-4. [DOI] [PubMed] [Google Scholar]
- 46.van Oppen MJ, Bongaerts P, Underwood JN, Peplow LM, Cooper TF. The role of deep reefs in shallow reef recovery: An assessment of vertical connectivity in a brooding coral from west and east Australia. Mol Ecol. 2011;20(8):1647–1660. doi: 10.1111/j.1365-294X.2011.05050.x. [DOI] [PubMed] [Google Scholar]
- 47.Szmant AM. Reproductive ecology of Caribbean reef corals. Coral Reefs. 1986;5:43–54. [Google Scholar]
- 48.Ferrari J, West JA, Via S, Godfray HC. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution. 2012;66(2):375–390. doi: 10.1111/j.1558-5646.2011.01436.x. [DOI] [PubMed] [Google Scholar]
- 49.Thibert-Plante X, Hendry AP. The consequences of phenotypic plasticity for ecological speciation. J Evol Biol. 2011;24(2):326–342. doi: 10.1111/j.1420-9101.2010.02169.x. [DOI] [PubMed] [Google Scholar]
- 50.Giraud T, Gladieux P, Gavrilets S. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol. 2010;25(7):387–395. doi: 10.1016/j.tree.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Endler JA. Geographic Variation, Speciation, and Clines. Princeton: Princeton Univ Press; 1977. [PubMed] [Google Scholar]
- 52.Clarke BC. The evolution of morpho-ratio clines. Am Nat. 1966;100(914):389–402. [Google Scholar]
- 53.Savolainen V, et al. Sympatric speciation in palms on an oceanic island. Nature. 2006;441(7090):210–213. doi: 10.1038/nature04566. [DOI] [PubMed] [Google Scholar]
- 54.Alberto F, et al. Population differentiation of sessile oak at the altitudinal front of migration in the French Pyrenees. Mol Ecol. 2010;19(13):2626–2639. doi: 10.1111/j.1365-294X.2010.04631.x. [DOI] [PubMed] [Google Scholar]
- 55.Hall MC, Willis JH. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;60(12):2466–2477. [PubMed] [Google Scholar]
- 56.Servedio MR, Van Doorn GS, Kopp M, Frame AM, Nosil P. Magic traits in speciation: “Magic” but not rare? Trends Ecol Evol. 2011;26(8):389–397. doi: 10.1016/j.tree.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Moczek AP, et al. The role of developmental plasticity in evolutionary innovation. Proc Biol Sci. 2011;278(1719):2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150(3811):563–565. [Google Scholar]
- 59.Pfennig DW, et al. Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol Evol. 2010;25(8):459–467. doi: 10.1016/j.tree.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Crow J. Mechanisms and trends in human evolution. Daedalus. 1961;90:416–431. [Google Scholar]
- 61.Connell J. Population ecology of reef- building corals. In: Jones A, Endeam R, editors. Biology and Geology of Coral Reefs. London: Academic; 1973. pp. 205–245. [Google Scholar]
- 62.Scoville AG, Pfrender ME. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proc Natl Acad Sci USA. 2010;107(9):4260–4263. doi: 10.1073/pnas.0912748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson DR. Model Based Inference in the Life Sciences. New York: Springer; 2008. [Google Scholar]
- 65.Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167(2):747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.