Abstract
The N-end rule pathway is a proteolytic system in which destabilizing N-terminal residues of short-lived proteins act as degradation determinants (N-degrons). Substrates carrying N-degrons are recognized by N-recognins that mediate ubiquitylation-dependent selective proteolysis through the proteasome. Our previous studies identified the mammalian N-recognin family consisting of UBR1/E3α, UBR2, UBR4/p600, and UBR5, which recognize destabilizing N-terminal residues through the UBR box. In the current study, we addressed the physiological function of a poorly characterized N-recognin, 570-kDa UBR4, in mammalian development. UBR4-deficient mice die during embryogenesis and exhibit pleiotropic abnormalities, including impaired vascular development in the yolk sac (YS). Vascular development in UBR4-deficient YS normally advances through vasculogenesis but is arrested during angiogenic remodeling of primary capillary plexus associated with accumulation of autophagic vacuoles. In the YS, UBR4 marks endoderm-derived, autophagy-enriched cells that coordinate differentiation of mesoderm-derived vascular cells and supply autophagy-generated amino acids during early embryogenesis. UBR4 of the YS endoderm is associated with a tissue-specific autophagic pathway that mediates bulk lysosomal proteolysis of endocytosed maternal proteins into amino acids. In cultured cells, UBR4 subpopulation is degraded by autophagy through its starvation-induced association with cellular cargoes destined to autophagic double membrane structures. UBR4 loss results in multiple misregulations in autophagic induction and flux, including synthesis and lipidation/activation of the ubiquitin-like protein LC3 and formation of autophagic double membrane structures. Our results suggest that UBR4 plays an important role in mammalian development, such as angiogenesis in the YS, in part through regulation of bulk degradation by lysosomal hydrolases.
Keywords: cardiovascular system, ubiquitin ligase
The N-end rule pathway is a proteolytic system that controls the half-lives of proteins based on destabilizing activities of N-terminal residues (1–3). In mammals, known N-recognins bind to destabilizing N-terminal residues to induce selective proteolysis through the ubiquitin (Ub)-proteasome system (UPS). To date, N-recognins have not been implicated in bulk proteolysis by lysosomal hydrolases. Substrates of the N-end rule pathway include cytosolic short-lived proteins carrying type-1 (Arg, Lys, and His; positively charged) and type-2 (Phe, Tyr, Trp, Leu, and Ile; bulky hydrophobic) N-degrons (4–7). N-degrons can be exposed through N-terminal Met excision or proteolytic cleavage of otherwise stable proteins. The principal degron Arg can also be generated through posttranslational arginylation of Asp and Glu (and Cys in mammals as well) by ATE1-encoded ArgtRNA-protein transferases (R-transferases) (8–11). In addition, the arginylation-permissive pro–N-degrons Asp and Glu can be generated through deamidation of the secondary pro–N-degrons Asn and Glu (12). As the classical (type 1/2) N-end rule lists 13 of 20 amino acids of the genetic code, a vast number of cellular proteins or polypeptide fragments have potential to acquire N-degrons through posttranslational modifications of N-terminal residues. In addition to type-1 and type-2 N-degrons, a recent study reported that acetylated N-terminal residues, occurring at 80% of human proteins, can function as N-degrons (13). The functions of the N-end rule pathway in degradation of short-lived proteins carrying N-degrons have been characterized in various processes (2, 3). One outstanding question in the N-end rule pathway concerns differential functions of N-recognins that share binding specificity to N-terminal residues. Our previous studies identified the mammalian N-recognin family consisting of 200-kDa UBR1/E3α, 200-kDa UBR2, 570-kDa UBR4/p600, and 300-kDa UBR5 (4, 5, 7) (Fig. S1). These N-recognins share the UBR box, a ∼70-residue zinc finger domain that acts as a substrate recognition domain (14). Whereas the canonical N-recognins UBR1 and UBR2 have been characterized in various mammalian processes, functions and mechanisms of two noncanonical N-recognins, UBR4 and UBR5, in the N-end rule pathway remain poorly understood. Different from other N-recognins, UBR4 does not have a known ubiquitylation domain (7), suggesting that the action mechanism of UBR4 in N-end rule degradation could be different from those of canonical N-recognins. Recent studies using cultured cells suggest that UBR4 plays a role in membrane morphogenesis and extracellular matrix-mediated cell survival and signaling (15), human papillomavirus (HPV) E7-mediated cellular immortalization (16–18), and migration and development of neurons (19).
The autophagy-lysosome system (hereafter autophagy) mediates bulk degradation of cytoplasmic constituents such as proteins and organelles. Autophagy is induced by starvation or other cellular stresses to promote lysosomal digestion of nonessential cellular materials, providing fuels and amino acids and, thus, maintaining essential cellular processes. Substrates of autophagy are sequestered and delivered to autophagosomes whose fusion with lysosomes generates autolysosomes wherein the content is degraded by lysosomal hydrolases. The formation of autophagosomes involves Autophagy-related protein 12 (ATG12) recruitment to phagophores, which is conjugated to its substrate, ATG5. The ATG5–ATG12 conjugate promotes ligation of the ubiquitin-like protein LC3-I with phospholipid phosphotidylethanolamine (PE) to generate LC3-II whose PE moiety is anchored to phagophore membranes.
Studies in the late 70s by New and others demonstrated that the yolk sac (YS) is a primary supplier of amino acids for the embryo at early stages [until approximately embryonic day 9.5 (E9.5) in mice] and that these amino acids are generated through lysosomal digestion of endocytosed proteins (20, 21). However, the importance of the YS as a source of amino acids has received little attention since then. As illustrated in Fig. S2, the YS of mouse embryos at E8.5 can be divided into two developmentally distinct layers. The inner layer, derived from mesodermal cells, is a circulation organ in which blood vessels are developed through intercellular signaling of endothelial cells with vascular smooth muscle cells (SMCs) and surrounding mesenchymal cells. The outer layer, derived from endodermal cells, contains a highly specialized proteolytic system in which extracellular maternal proteins (e.g., albumins) in the YS cavity are internalized via endocytosis and delivered to multivesicular bodies (MVBs)/late endosomes (22) (Fig. S2B). Cargo-loaded MVBs are fused with autophagosomes which, in turn, are digested by hydrolases of professionally enlarged lysosomes (alternatively called vacuoles) of the YS endoderm. The resulting lysosome-derived amino acids with a maternal original are essential for protein translation during vascular development and other developmental processes in the YS and embryo. Because the placenta that transports maternal nutrients and gases to the embryos is not functional at E9.5, the embryo at this stage strictly depends on lysosome-generated amino acids from the YS endoderm. Therefore, in definition, the YS endoderm contains a tissue-specific autophagic system that mediates constitutive degradation of extracellular proteins as a major type of cargoes.
In this study, we show that UBR4 plays an important role in developmental processes during embryogenesis, in part through its function in autophagy. Our previous and current studies together reveal a dual function of UBR4 in selective proteolysis by the proteasome and bulk degradation by the lysosome.
Results
Mice Lacking UBR4 Die During Embryogenesis.
To determine the role of UBR4 in mammalian development, we constructed mice lacking UBR4. As the UBR box is a general substrate recognition domain for destabilizing N-terminal residues, a UBR-box–containing region spanning exons 36 through 42 was replaced with internal ribosome entry site (IRES)-translated tau-lacZ and a floxed tACE-Cre-Neo cassette in embryonic stem (ES) cells (Fig. S3). The selection marker Neo flanked by loxP sites was removed by the Cre recombinase expressed from the angiotensin-converting enzyme (tACE) promoter in spermatogonia of germ-line–transmitted F1 pups. Southern blot and reverse transcription-PCR (RT-PCR) analyses confirmed that UBR4 was properly inactivated (Fig. S4). Genotyping F2 C57BL/6J:129/Ola hybrid offspring from heterozygous parents yielded no viable UBR4−/− mice (Table S1), indicating that UBR4−/− mice die as embryos or neonates. By E9.0–9.5, UBR4−/− embryos were retrieved at Mendelian ratios but growth arrested at the 14- to 18-somite stage, some of which were found dead (Table S1). No live mutants were retrieved at E11.5 and afterward. Thus, mouse UBR4 is indispensable for survival during embryogenesis.
UBR4-Deficient Embryos Are Impaired in YS Vascular Development.
One prominent and invariable phenotype of UBR4−/− embryos was defective vascular development in the YS. UBR4-deficient YSs were apparently normal at E8.5 but became pale by E9.5 (Fig. 1A, arrow). The vascular defect was severer in the YS compared with the embryo proper, and the severity of YS vascular defect correlated to death and developmental arrest of the embryo. UBR4−/− YS showed wild-type rates in cell proliferation as determined by bromodeoxyuridine (BrdU) incorporation assay of pregnant females (Fig. 2L) and apoptosis as determined by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (Fig. 2M and Fig. S5A). As further discussed in SI Results, these results suggest that YS vascular defects are the primary cause of death in UBR4-deficient embryos.
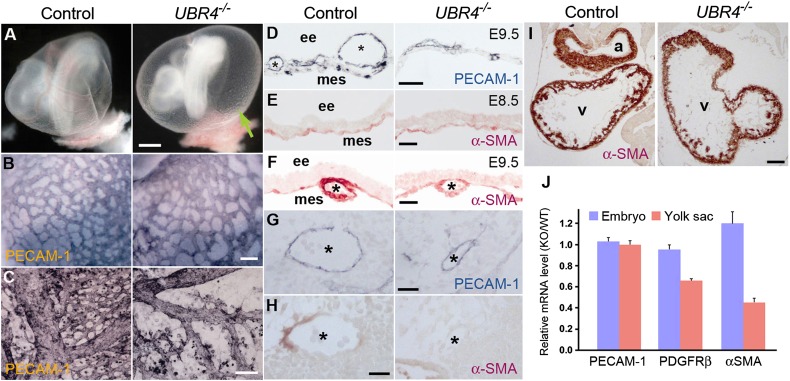
Fig. 1.
UBR4-deficient embryos die during embryogenesis with angiogenic arrest of primary capillary plexus in the YS. (A) Appearance of control and UBR4−/− embryos at E9.5 with the YS intact. Arrowheads, blood vessels. (B and C) Whole mount PECAM-1 staining of control and UBR4−/− YS at E8.5 (B) and E9.5 (C). (Scale bars, 100 μm.) (D) Cross-sections of +/+ and UBR4−/− YS at E9.5 stained for PECAM-1. (Scale bars, 50 μm.) *, vessels; ee, extraembryonic endodermal layer; mes, mesodermal layer. (E and F) Cross-sections of α–SMA-stained +/+ and UBR4−/− YS at E8.5 (E) and E9.5 (F). (Scale bars, 25 μm.) (G and H) Cross-sections of control and UBR4−/− embryos stained for PECAM-1 (G) and α-SMA (H). *, aorta. (Scale bars, 25 μm.) (I) Cross-sections of control and UBR4−/− embryonic hearts stained for α-SMA. a, atrium; v, ventricle. (Scale bar, 100 μm.) (J) Real-time PCR analysis of control and UBR4−/− YS and embryos at E9.5.
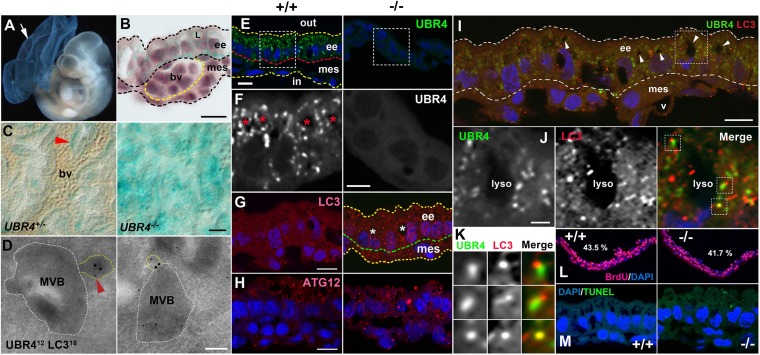
Fig. 2.
Expression of UBR4 in endoderm-derived, autophagy-enriched YS cells and its association with a constitutive autophagic pathway of the YS endoderm. (A) X-gal staining of E9.5 UBR4−/− embryo and YS. (B) Cross-section of X-gal–stained UBR4+/− YS. L, lysosome; ee, extraembryonic endoderm; mes, mesodermal layer; bv, blood vessel. (Scale bar, 10 μm.) (C) Whole-mount X-gal staining of UBR4+/− and UBR4−/− YS. Arrowhead, a relatively intense β-gal signal surrounding a blood vessel. (Scale bar, 25 μm.) (D) EM of HEK293–UBR4V5 cells labeled for UBR4 (12 nm; arrowhead) and LC3 (18 nm). (Scale bar, 200 nm.) (E) Immunostaining of UBR4 on cross-sections of +/+ and UBR4−/− YS at E9.5. (Scale bar, 10 μm.) (F) Enlarged views of areas indicated by rectangles in E. (Scale bar, 5 μm.) (G and H) Cross-sections of +/+ and UBR4−/− YS stained for LC3 (G) or ATG12 (H). (Scale bar, 10 μm.) *, lysosome. (I) Cross-sections of the YS costained for UBR4 and LC3. Arrowhead, lysosome; v, blood vessel. (Scale bar, 10 μm.) (J) Enlarged views of area indicated by rectangles in I. (Scale bar, 2 μm.) (K) Enlarged views of areas indicated by rectangles in J. (L) BrdU incorporation assay of control and UBR4−/− embryos at E9.5. S-phase indices were determined to be 43.5% and 41.7% for +/+ and UBR4−/− YS, respectively. (M) TUNEL assay of +/+ and UBR4−/− YS at E9.5.
Vascular Development in UBR4-Deficient YS Is Arrested During Angiogenic Remodeling of Primary Capillary Plexus.
To characterize vascular development in +/+ and UBR4−/− embryos, we examined expression of platelet endothelial cell adhesion molecule 1 (PECAM-1), a marker of differentiated endothelial cells. The mRNA expression of PECAM-1 and α-fetoprotein (AFP), a major plasma protein produced by the YS, was normal at E9.5 UBR4−/− YS (Fig. S5D). When visualized by whole-mount immunostaining, E8.5 UBR4−/− YS contained a wild-type level of PECAM-1+ cells around primary vessels, forming a network of PECAM-1+ primary vessels (Fig. 1B). Thus, UBR4 is not critical for vasculogenesis during which angioblasts proliferate and differentiate to endothelial cells. However, the same staining showed that the vascular network of UBR4−/− YS rapidly loses its integrity over a period of 24 h from E8.5 through E9.5 (Fig. 1 C and D and Fig. S5B). We therefore examined expression of α-smooth muscle actin (α-SMA), a marker of vascular and cardiac SMCs, in UBR4+/− and UBR4−/− YS at E8.5–E9.5 to monitor reorganization of primary capillary plexus. During angiogenic remodeling, endothelial cells recruit and induce the differentiation of mesenchymal cells to vascular SMCs, establishing cell-to-cell adhesion with each other and endothelial cells. Semiquantitative real-time PCR analysis showed that the mRNA level of α-SMA in UBR4−/− YS was markedly down-regulated by E9.5 (Fig. 1J). Immunostaining of UBR4−/− YS showed that α-SMA+ cells normally located in capillary vessels at E8.5 (Fig. 1E) but were markedly down-regulated in large vessels at E9.5 and, to a lesser degree, in small vessels (Fig. 1F). Down-regulation of α-SMA+ cells was also observed in arteries (Fig. 1 H vs. G), but not the hearts (Fig. 1I), of UBR4−/− embryos, which correlated to a moderately affected vascular network on the surface of the mutants (Fig. S5C). Thus, YS vascular development is arrested during angiogenic remodeling of primary capillary plexus.
In the YS, UBR4 Marks Endoderm-Derived, Autophagy-Enriched Cells That Coordinate Differentiation of Mesoderm-Derived Vascular Cells.
The expression of UBR4 at E8.5–E9.5 was prominent in the YS relative to the embryo (Fig. 2A, arrow), as determined by whole-mount staining of β-galactosidase (β-gal) derived from tau-lacZ of UBR4− allele. Whereas β-gal staining in UBR4+/− YS at E9.5 revealed a vascular network (Fig. 2C), this pattern was disrupted in UBR4−/− YS (Fig. 2C), similarly to a disorganized PECAM-1+ vascular network (Fig. 1 C and D). This prompted us to examine UBR4 expression on the cross-section of the YS comprising two developmentally distinct layers, the outer layer derived from the endoderm and the inner layer from the mesoderm (Fig. S2B). The β-gal staining was not readily detected in endothelial cells, vascular SMCs, or nearby undifferentiated mesenchymal cells (precursors of vascular SMCs) in the mesoderm (Fig. 2B). Instead, the staining was largely restricted to autophagy-enriched endodermal cells (Fig. 2B) that coordinate differentiation of mesoderm-derived vascular cells and supply lysosome-produced amino acids during early embryogenesis.
UBR4 Is Associated with a Tissue-Specific Constitutive Autophagic Pathway in the YS Endoderm.
To determine the function of UBR4 in the YS endoderm, we immunostained endogenous UBR4 on YS cross-sections at E9.5. In contrast to typical cytosolic proteins of the UPS that would appear to be diffusive, the staining appeared as cytosolic punctate signals (Fig. 2 E and F). Although morphologically heterogeneous, some had a round shape with diameters of 0.1–0.3 or ∼1.0 μm and codistributed with professionally enlarged lysosomes in a cytosolic compartment between the microvilli and nucleus (Fig. 2F). A significant portion of UBR4 puncta were at least partially associated with puncta positive for LC3, a hallmark of double membrane structures (Fig. 2 I–K). Despite colocalization, the shapes and sizes of UBR4 puncta were distinct from those of LC3 puncta (Fig. 2K). These results suggest that UBR4 is not a constitutive component of autophagic machinery but associated with autophagic cargoes. Next, we asked whether UBR4 loss affects autophagic activity. Immunostaining of LC3 on YS cross-sections from +/+ and UBR4−/− embryos at E9.5 indicated that UBR4−/− YS contained a markedly increased level of LC3 puncta in number and signal intensity (Fig. 2G), without significant changes in cell proliferation and death (Fig. 2 L and M). ATG12 puncta, a hallmark of phagophores, were also significantly induced in UBR4−/− YS, although less striking compared with LC3 (Fig. 2H). Given that MVBs carrying maternal proteins from the YS cavity are major autophagic cargoes in the YS endoderm, we asked whether UBR4 is associated with the MVB using the transmission electron microscopy (EM). The EM of UBR4 and LC3 in HEK293–UBR4V5 cells identified one or a few UBR4 dots (not as a cluster of aggregates) within the MVB or in the proximity of endosome-like cellular materials and LC3 at the “entrance” of the MVB (Fig. 2D). These results suggest that UBR4 plays a role in a tissue-specific constitutive autophagic pathway of the YS endoderm, through its association with autophagic cargoes. Given that the YS endoderm coordinates vascular development and supplies lysosome-produced amino acids, misregulation of autophagy in the YS endoderm is likely to contribute to the vascular failure in the YS mesoderm (SI Results).
UBR4 Plays a Role in Delivery of Autophagic Cargoes to Phagophores.
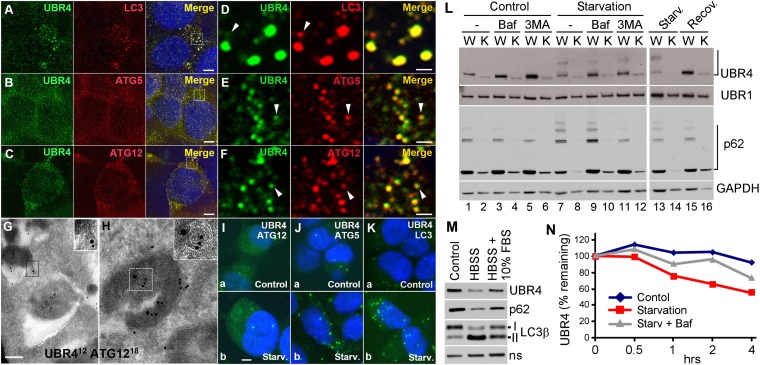
To test whether UBR4 has a general function in autophagy, we performed anti-UBR4 immunostaining in HEK293 cells expressing 570-kDa UBR4 with C-terminal V5 tag (UBR4V5). The staining revealed cytosolic punctate signals with diameters of 0.1–0.3 or ∼1.0 μm, typical for phagophores and autophagosomes, respectively (Fig. 3 A–F). Most UBR4 puncta colocalized at least partially with LC3 puncta (Fig. 3 A and D), a hallmark of phagophores and autophagosomes. UBR4 puncta with sizes of 0.1–0.3 μm also showed similar colocalization with ATG12 (Fig. 3 C and F) and ATG5 (Fig. 3 B and E) that mark phagophores. However, UBR4 puncta had shapes and sizes distinct from those of ATG12, ATG5, and LC3 (Fig. 3 D–F, arrowheads). In addition, a confocal distance measurement indicated that the center of UBR4 puncta was separated from that of LC3 puncta by ∼0.15 μm. To exclude the possibility that UBR4 in HEK293–UBR4V5 cells is misfolded and degraded by autophagy as aggregates, we fractionated the cells growing in normal or starvation media. The majority of UBR4 was retrieved from cytosolic and ER membrane fraction in hypotonic buffer and could be solubilized with a nonionic detergent, 1% Nonidet P-40 (Fig. S6). To directly determine the association of UBR4 with autophagic vacuoles, we performed the EM of UBR4V5 in HEK293–UBR4V5 cells. The analysis identified one or a few individual UBR4V5 dots (not as a cluster of aggregates) in association with cargo-like materials with a diameter of ∼0.1 μm and in the proximity of ATG12 on the surface or lumen of autophagic vacuoles (Fig. 3 G and H). As further discussed in SI Results, these results collectively suggest that UBR4 is specifically associated with (unidentified) cellular materials to form UBR4–cargo complexes which, in turn, are delivered to LC3+ autophagic vacuoles. However, we do not exclude the possibility that a fraction of overexpressed UBR4 molecules are misfolded and, thus, deposited to autophagic vacuoles.
Fig. 3.
UBR4 plays a role in delivery of autophagic cargoes to phagophores, and UBR4 subpopulation is degraded by autophagy through starvation-enhanced association with autophagic core machinery. (A–C) Costaining of UBR4 in comparison with LC3 (A), ATG5 (B), and ATG12 (C) in HEK293–UBR4V5 cells. (D–F) Enlarged views of areas indicated by rectangles in A–C. (G and H) EM of HEK293–UBR4V5 cells labeled for UBR4 (12 nm) and ATG12 (18 nm). (Scale bar, 200 nm.) (I–K) PLA assay of UBR4 in comparison with ATG12 (I), ATG5 (J), and LC3 (K) in HEK293–UBR4V5 cells. (Scale bar, 1 μm.) (L) Immunoblotting of +/+ (WT) and UBR4−/− (KO) MEFs growing in normal or starvation medium for 90 min in the presence or absence of 0.2 μM bafilomycin A1 or 10 mM 3-methyladenine. (M) Immunoblotting of MEFs cultured in normal media or HBSS for 90 min, with or without being supplemented by 10% (vol/vol) FBS. (N) Cyclohexamide-chase assay of UBR4 in MEFs. Cells were cultured in normal or starvation medium for 6 h in the presence of cycloheximide. Shown is a quantitation of a representative blot image from three independent experiments.
Finally, we tested whether recombinant expression of UBR4V5 affects formation of autophagic double membrane structures. Compared with parental HEK293 cells, HEK293–UBR4V5 cells generated increased numbers of ATG12, ATG5, and LC3 puncta (Fig. S7 A–C), suggesting that the level of UBR4 may need to be tightly regulated to maintain the homeostasis in UBR4-dependent autophagy and other processes.
UBR4 Subpopulation Is Degraded by Autophagy Through Starvation-Enhanced Association with Autophagic Core Machinery.
We established mouse embryonic fibroblasts (MEFs) from +/+ and UBR4−/− embryos at E8.5. The steady-state level of UBR4 was reduced when MEFs were starved in Hanks' balanced salt solution (HBSS) and recovered when supplemented with 10% FBS (Fig. 3M). The down-regulated level of UBR4 in nutrient-deprived MEFs (Fig. 3L, lane 13 vs. 1) was readily restored by serum resupplementation (Fig. 3L, lane 15 vs. 13). The level of UBR4 was increased by the treatment of bafilomycin A1, which inhibits the fusion of autophagosomes with lysosomes (Fig. 3L, lane 3 vs. 1) or 3-methyladenine that inhibits class III PI3K (Fig. 3L, lane 5 vs. 1). To examine whether the metabolic stability of UBR4 is sensitive to nutrient states, we monitored the decay of UBR4 in +/+ and UBR4−/− MEFs treated with cyclohexamide, a translation inhibitor. UBR4 was a relatively stable protein in untreated cells over 6 h (Fig. 3N). However, when autophagy was induced using starvation media, UBR4 became significantly short-lived with a half-life of ∼3.5 h (Fig. 3N). Starvation-induced decay was moderately, but significantly, inhibited by bafilomycin A1 (Fig. 3N and Fig. S7D). The relatively modest stabilization by autophagic inhibition may be because cyclohexamide partially inhibits autophagic flux. To examine whether nutrient deprivation promotes UBR4 association with autophagic core machinery, we used proximity ligation assay (PLA), which visualizes two proteins located in the proximity (<400 Å) based on the ability of antibody-conjugated oligonucleotides to hybridize with each other (SI Materials and Methods). The PLA of UBR4V5 and LC3 in HEK293–UBR4V5 cells revealed punctate signals with diameters of ∼1.0 μm (2.76 ± 0.28 per cell, n = 148), indicating multiple UBR4–LC3 PLA interactions within autophagosomes (Fig. 3 K, a). UBR4V5 showed similar PLA interactions with its upstream components, ATG12 (1.47 ± 0.33, n = 51) and ATG5 (3.36 ± 0.31, n = 77) (Fig. 3 I, a and J, a). Nutrient deprivation for 1 h caused a significantly increased correlation between UBR4 and LC3 (7.96 ± 0.39, n = 101) and, to a lesser degree, ATG12 (2.27 ± 0.30, n = 45) and ATG5 (5.04 ± 0.34, n = 111) as well (Fig. 3 I–K, Lower). These results together suggest that nutrient deprivation promotes the association of UBR4 with autophagic cargoes, leading to its degradation by autophagy.
UBR4 Loss Results in Misregulation of Autophagic Induction and Flux.
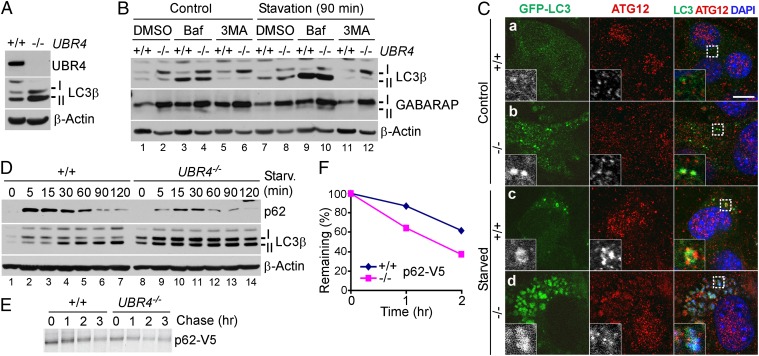
Given the enhanced formation of LC3 and ATG12 cytosolic puncta in UBR4−/− YS (Fig. 2G), we examined autophagic induction and flux in +/+ and UBR4−/− MEFs. Immunostaining showed that UBR4−/− MEFs contained a higher level of LC3 compared with +/+ cells (Fig. S8). Immunoblotting analysis confirmed that UBR4−/− MEFs contained significantly increased basal levels for LC3-I and LC3-II (Fig. 4 A and D, lane 8 vs. 1) as well as γ-Aminobutyric acid type A receptor-1 (GABARAP-1) and GABARAP-II (Fig. 4B, lane 2 vs. 1). Time-course immunoblotting of +/+ and UBR4−/− MEFs undergoing nutrient deprivation also indicated that UBR4−/− MEFs contained higher levels of both LC3-I and LC3-II at all time points until 120 min (Fig. 4D). Transient expression of green fluorescence protein (GFP)-tagged LC3 (LC3-GFP) confirmed that formation of cytosolic punctate structures was activated in UBR4−/− MEFs growing in normal as well as starvation media (Fig. 4C). These results suggest that UBR4 loss induces autophagy.
Fig. 4.
UBR4 loss results in multiple misregulations of autophagic induction and flux. (A) Immunoblotting of +/+ and UBR4−/− MEFs. (B) Immunoblotting of +/+ and UBR4−/− MEFs growing in normal or starvation medium for 90 min in the presence or absence of 0.2 μM bafilomycin A1 or 10 mM 3-methyladenine. (C) Immunostaining of transiently expressed LC3-GFP and endogenous ATG12 in +/+ and UBR4−/− MEFs cultured in normal or starvation medium for 90 min. (D) Time-course immunoblotting of +/+ and UBR4−/− MEFs following nutrient deprivation. Cells were cultured in normal or starvation medium for 90 min, followed by time-course immunoblotting of remaining proteins. (E) Pulse chase analysis of p62-V5 transiently expressed in HEK293 cells. (F) Quantitation of E.
To further characterize the role of UBR4 in autophagy, we transiently transfected HEK293 cells with siRNA against human UBR4. Transiently silencing of human UBR4 markedly induced the formation of LC3 puncta in HEK293 cells (Fig. S9). Moreover, transient overexpression of recombinant human UBR4V5 in UBR4-knockdown HEK293 cells reverted the induced LC3 puncta back to the basal level (Fig. S9). These results indicate that UBR4 plays an active role in autophagic flux. By contrast, transient reexpression of human UBR4V5 in UBR4−/− MEFs reduced only marginally the synthesis of LC3-I and the formation of LC3 foci (Fig. S8). This marginal effect is consistent with a possibility that autophagic induction in UBR4−/− MEFs is contributed by long-term cellular stress and/or impairment of organelles, which are not readily rescued by transient supplement of UBR4 for less than 24 h.
Interestingly, the transient expression of UBR4V5 resulted in formation of UBR4V5+LC3− puncta in both in MEFs (Fig. S8) and HEK293 cells (Fig. S9), which are likely to represent UBR4V5+ cellular cargoes that have not been delivered to autophagic vacuoles in the absence of autophagic induction.
Because LC3 puncta can accumulate by reduced flux, we examined the metabolic stability of the selective autophagic substrate p62 in +/+ and UBR4−/− MEFs. In both normal and starvation media, the level of p62 was significantly reduced in UBR4−/− cells compared with +/+ cells (Fig. 3L, lanes 2 vs. 1 and 8 vs. 7). Time-course immunoblotting following nutrient deprivation confirmed down-regulation of p62 at all time points until 120 min (Fig. 4D). The 1.5-h treatment of bafilomycin A1 or 3-methyladenine in starved UBR4−/− cells increased the level of p62 (Fig. 3L, lane 10 vs. 8). We therefore measured the turnover rate of p62 using the pulse chase analysis in which p62-V5 was expressed and labeled with 35S-Met/Cys for 20 min, followed by inhibition of protein synthesis by cycloheximide and the chase of 35S-labeled p62-V5 (Fig. 4 E and F). The half-life of the autophagic substrate was determined to be ∼140 min in control cells. Compared with controls, UBR4−/− cells showed a lower zero-time level, indicating accelerated acute decay during labeling for 20 min. UBR4−/− cells showed a comparable level of postpulse degradation with a half-life of ∼90 min. These results, together with the finding that the level of LC3 in UBR4−/− cells is sensitive to bafilomycin A1 (Fig. 4B, lanes 4 vs. 2 and 10 vs. 8), suggesting that UBR4−/− cells contain a largely normal range of autophagic flux.
Discussion
Since the discovery in 1986 (1), the N-end rule pathway has been characterized as a proteolytic system that controls the half-lives of short-lived proteins through selective proteolysis by the 26S proteasome. In this study we demonstrate that mice lacking UBR4, a newly identified N-recognin, die during embryogenesis associated with severe YS vascular defects. UBR4−/− YSs normally generate a honeycomb-like network of primary vascular plexus. However, angiogenic remodeling of the primary vessels are arrested over a period of 24 h from E8.5 through E9.5 as vascular SMCs fail to reorganize into mature vessels. As further discussed in SI Results, several lines of evidence suggest that YS vascular defects are the primary cause of the embryonic death.
Despite severe defects in vascular development of mesodermal cells, the expression of UBR4 is not readily detected in the YS mesoderm, a site of vascular development, but is prominent in a neighboring layer derived from the endoderm. The YS endoderm is a digestive organ wherein maternal proteins from the YS cavity are uptaken and digested into amino acids through the MVB–autophagosome–lysosome pathway (22). The results presented in this paper together suggest that UBR4 subpopulation in the YS endoderm is specifically associated with cellular cargoes to form UBR4–cargo complexes destined to autophagosomes and professionally enlarged lysosomes. Given that embryonic growth and vascular development in the YS mesoderm strictly requires lysosome-derived amino acids from the YS endoderm, it is likely that misregulation of autophagic pathways in the UBR4−/− YS endoderm significantly contributes to vascular failure in the mesoderm. One question that remains to be answered is whether UBR4 puncta in the YS endoderm carry maternal proteins as autophagic substrates. Another question is the mechanism by which an N-recognin, known to bind to substrates of selective proteasomal degradation, is also associated with substrates of bulk lysosomal degradation. UBR4 does not have a known ubiquitylation domain but does contain the UBR box, which is conserved in the UBR N-recognin family and binds to type-1 destabilizing N-terminal residues, including the primary degron Arg. It is therefore tantalizing to speculate that UBR4 is associated with cellular cargoes through binding of the UBR box to the N-terminal Arg exposed from the substrates.
Our results show that UBR4−/− YS and MEFs contain an increased level of LC3+ puncta compared with controls. This suggests that multiple misregulations of autophagy and other UBR4-dependent processes in UBR4−/− cells result in cellular stress (e.g., nutrient insufficiency, and/or accumulation of damaged organelles), which, in turn, stimulates the formation of autophagic vacuoles. Intriguingly, whereas UBR4-loss induces autophagy, UBR4 overexpression also results in autophagic induction, as determined by enhanced formation of UBR4+LC3+ puncta in HEK293–UBR4V5 cells (Fig. S7). This suggests that the level of UBR4 may need to be tightly regulated by regulated proteolysis to maintain the homeostasis of UBR4-dependent autophagy and other processes. A mutually nonexclusive, alternative possibility is that the overexpression of UBR4 for a long period may generate excessive UBR4+ cargoes destined to autophagy, which, in turn, induces formation of phagophores. Further investigation is needed to understand the mechanism by which UBR4 plays a dual role in selective degradation of short-lived proteins via the proteasome and bulk degradation of cellular materials via the lysosome.
Materials and Methods
For descriptions of materials and methods, including antibodies and PCR primers, see SI Results, SI Materials and Methods, and Tables S1 and S2. Immunostaining, quantitative RT-PCR, transmission electron microscopy, proximity ligation assay, immunoblotting, and mouse knockout were performed using standard techniques, which are described in SI Results and SI Materials and Methods. We cloned UBR4 from a bacterial artificial chromosome (BAC) library (CITB; Invitrogen) derived from the mouse ES cell line CJ7 using a mouse UBR4 cDNA fragment (nucleotides 4231–5430) as a probe. The targeting vector pUBR4KO-tauLacZ was constructed using gene recombineering (recombination-mediated genetic engineering), linearized with FseI, and electroporated into E14 ES cells (a gift from Peter Mombaerts, Rockefeller University, New York). The clone 2F12 ES cells were injected into C57/BL6J blastocysts to generate chimeric mice which, in turn, were used to obtain germ-line transmission of the UBR4− mutant allele. Animal studies were conducted according to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH publication no. 85–23, revised in 1996) and the protocols (0812811-A1) approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh. Euthanization involves inhalant anesthetic, isoflurane, followed by i.p. injection of xylazine (10 mg/kg) and ketamine (100 mg/kg) mixture.
Supplementary Material
Acknowledgments
We are grateful to David Rubinsztein at the University of Cambridge for critical comments on the manuscript; Donna Stolz at the University of Pittsburgh for help with transmission electron microscopy; and the Y.T.K. and B.Y.K. research team members at the University of Pittsburgh, Seoul National University, and Korea Research Institute of Bioscience and Biotechnology for helpful discussions. This work was supported in part by NIH Grant HL083365 (to Y.T.K.), in part by a Canadian Institute of Health Research Operating Grant (to M.D.N.), and in part by World Class University Grant R31-2008-000-10103-0 (to Y.T.K. and E.C.Y.) and World Class Institute Grant WCI 2009-002 (to B.Y.K.) through the National Research Foundation funded by the Ministry of Education, Science, and Technology, Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217358110/-/DCSupplemental.
References
- 1.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 2.Sriram SM, Kim BY, Kwon YT. The N-end rule pathway: Emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol. 2011;12(11):735–747. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- 3.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon YT, et al. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci USA. 1998;95(14):7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon YT, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23(22):8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol Cell Biol. 2001;21(23):8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tasaki T, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25(16):7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu RG, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437(7061):981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 9.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297(5578):96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 10.Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19(1):182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102(42):15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon YT, et al. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol Cell Biol. 2000;20(11):4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327(5968):973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasaki T, et al. The substrate recognition domains of the N-end rule pathway. J Biol Chem. 2009;284(3):1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakatani Y, et al. p600, a unique protein required for membrane morphogenesis and cell survival. Proc Natl Acad Sci USA. 2005;102(42):15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMasi J, Huh KW, Nakatani Y, Münger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc Natl Acad Sci USA. 2005;102(32):11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh KW, et al. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci USA. 2005;102(32):11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White EA, et al. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci USA. 2012;109(5):E260–E267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shim SY, et al. Protein 600 is a microtubule/endoplasmic reticulum-associated protein in CNS neurons. J Neurosci. 2008;28(14):3604–3614. doi: 10.1523/JNEUROSCI.5278-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman SJ, Lloyd JB. Evidence that protein ingested by the rat visceral yolk sac yields amino acids for synthesis of embryonic protein. J Embryol Exp Morphol. 1983;73:307–315. [PubMed] [Google Scholar]
- 21.New DA. Whole-embryo culture and the study of mammalian embryos during organogenesis. Biol Rev Camb Philos Soc. 1978;53(1):81–122. doi: 10.1111/j.1469-185x.1978.tb00993.x. [DOI] [PubMed] [Google Scholar]
- 22.Fader CM, Colombo MI. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 2009;16(1):70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.